Absence of human herpesvirus 6B detection in association with illness in children undergoing cancer chemotherapy
Abstract
The lymphotropic herpesviruses, cytomegalovirus (CMV), Epstein-Barr virus (EBV), and human herpesvirus 6B (HHV-6B) can reactivate and cause disease in organ transplant recipients; the contributions of HHV-6A and HHV-7 to disease are less certain. Less is known about their pathogenic roles in children undergoing treatment for malignancies. Children with newly diagnosed cancer were followed for 24 months. Clinical information and blood samples were collected during routine visits and during acute visits for fever or possible viral infections. Lymphotropic herpesvirus DNA in blood was measured by polymerase chain reaction (PCR). Although HHV-6B DNA was detected at least once in about half of the patients; the other viruses were seldom detected. There was no association between HHV-6B detection and individual acute clinical events, however, HHV-6B detection was more common in children who experienced more frequent acute clinical events. In children being treated for various malignancies, HHV-6B detection was common, but was not associated with individual events of acute illness. Thus, if HHV-6B is not assessed longitudinally, clinical events may be misattributed to the virus. The elevated frequency of detection of HHV-6B in sicker children is consistent with prior reports of its detection during apparently unrelated acute clinical events. J. Med. Virol. 88:1427–1437, 2016. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Human herpesviruses 6A, 6B, 7 (HHV-6A, HHV-6B, HHV-7), human cytomegalovirus (CMV), and Epstein-Barr virus (EBV) are lymphotropic herpesviruses most often acquired during early childhood [Yoshikawa et al., 1989; Ward et al., 1993]. Primary infection with these viruses can cause febrile illness and a broad range of accompanying non-specific symptoms, but are often asymptomatic. Infection results in establishment of lifelong latency. These viruses reactivate intermittently throughout life, most often asymptomatically. Primary and reactivated infections can cause mild to severe disease in immune compromised individuals, such as hematopoietic and solid organ transplant recipients [Hall et al., 1994; Zerr et al., 2005; Hill and Zerr, 2014; Ward, 2014]. EBV has well-established etiologic roles in pediatric cancers such as Burkitt's lymphoma and X-linked lymphoproliferative disease, and there is inconclusive mechanistic, pathologic, and epidemiologic support for HHV-6 and CMV playing roles in the development of some malignancies [Dziurzynski et al., 2012; Faten et al., 2012; Wick and Platten, 2014; Pellett, 2015]. Relevant to our purposes, little is known about the behavior of these viruses during treatment of cancers that do not appear to be caused by any of the viruses. Although HHV-6 DNA has been detected during febrile episodes of children with cancer [Lyall and Cubie, 1995; Yoshida et al., 1996; Michalek et al., 1999; Michalek and Horvath, 2002; Yee-Guardino et al., 2008], asymptomatic activity has not been studied in such children and the specificity of the association remains uncertain.
The purpose of this study was to evaluate the association between lymphotropic herpesvirus infections and acute illness in pediatric oncology patients. Our study design involved longitudinal sampling at regular intervals over a period of 2 years, as well as during acute clinical events of possible viral origin. This enabled comparison of detection frequencies during routine clinic visits and acute visits for symptoms to assess whether virus detection was specifically associated with disease.
MATERIALS AND METHODS
Patient Enrollment
Patients ≤21 years old who were newly diagnosed with a malignancy on the Cleveland Clinic Children's Hospital Hematology/Oncology service, and who would require chemotherapy were eligible for the study. The Cleveland Clinic Institutional Review Board (IRB) approved the study. Informed consent was obtained from a parent or guardian, and assent was obtained per IRB guidance.
Study Design
Patients were followed prospectively from the time of diagnosis for 24 months. Clinical information and blood samples were collected during routine visits at baseline (enrollment), every 2 weeks for the first 6 months, monthly for the next 6 months, and then every 3 months for the next year. The same data were also collected during acute unscheduled visits for fever or possible infection. If a child relapsed and chemotherapy was re-started, the patient was re-enrolled (after consent/assent) and data and labs were collected as if newly enrolled according to the more frequent schedule of visits. The total follow-up, however, remained 24 months from initial enrollment. For every visit data collected included: interim history, physical exam, medications, blood product administration, and laboratory data (including cultures) ordered as part of routine care. All of these data were entered into the electronic medical record (EMR) by a research nurse using a custom study form. Based on review of the EMR, a senior pediatric infectious diseases physician (JG) assigned a severity score (defined below) to each acute visit and determined the probable or definite etiology.
Data Collection and Management
The custom electronic medical records (EMR) workflow for the study was exercised by the study team prior to initiation of patient enrollment. An Oracle-based web-accessible data management system was developed to allow easy access to tools for data entry and reporting. Study data were periodically queried to perform accuracy and consistency checks on key variables and between forms. At the conclusion of the study, the senior physician (JG) reviewed the EMR to identify and repair errors. A data translation process was created to transform the EMR data into a database that was imported into Research Electronic Data Capture (RedCap) for statistical analysis [Harris et al., 2009].
Definitions
Routine visit: a visit scheduled for routine care on the schedule described. Acute visit: a visit during which a child was evaluated with fever (>38°C) or an unexpected visit to evaluate the child for a possible infection. Routine visits that were accompanied by an acute illness within 7 days of the visit were analyzed as routine visits. HHV-6A and HHV-6B viremia were analyzed under two different definitions for a positive test: any positive HHV-6 DNA and ≥500 copies/ml. HHV-7 results were reported as either positive or negative. CMV and EBV were also assessed under two different thresholds for positivity: any positive viral DNA and a higher standard of ≥350 copies/ml for CMV and ≥500 and copies/ml for EBV. Neutropenia was defined as <500 polymorphonuclear cells/microliter and lymphopenia as <500 lymphocytes/microliter. To assess the association between viral DNA loads and clinical outcomes, HHV-6A and HHV-6B DNAemia were analyzed under two different definitions for a positive test.
Clinical severity of each acute encounter was determined using a score based on NCI Common Terminology Criteria for rating severity of illness related to infection (http://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/ctcaev3.pdf). The scoring system was from 0 to 5, as follows: 0 if the child was well; 1 for fever without localizing signs or other abnormal vital signs; 2 for a mildly ill child with localized findings such as otitis media or fever and malaise with normal blood pressure and no need for fluid resuscitation; 3 for an acute illness requiring fluid resuscitation with rapid improvement and no need for intensive care acutely; 4 included clinical signs of sepsis: tachycardia, tachypnea and failure to respond to first fluid bolus; there could also be abnormal blood gas, decreased oxygen saturation, widened or low pulse pressure, and the child was admitted to PICU; 5 for acute fatal illness. Scores were assigned by a research nurse and reviewed by the physician who provided the etiologic assessment for each acute visit. CDC definitions were used as follows: Urinary tract infections: patient has at least one of the following signs or symptoms: fever (<38 degrees, or for infants: fever or hypothermia, apnea, bradycardia), urgency, frequency, dysuria or tenderness and a positive urine culture ≥105; bacteremia: pathogen cultured from one or more blood cultures or if a non-pathogen: more than one blood culture with the same organism with fever, or chills or hypotension; wound infection: purulent drainage from the site with signs of infection such as pain, tenderness, localized swelling; pneumonia: fever or signs of sepsis such as chills, hypotension with shortness of breath and findings on chest exam and or X-ray to suggest lung disease.
PCR Methods
DNA for PCR was extracted from whole blood on the MagNaPure automated extractor (Roche Diagnostics, Inc. Indianapolis, IN) and eluted in 50 µl of buffer. Five microliter of eluted DNA was analyzed in each reaction. Real time PCR was used for the quantitative detection of HHV-6A, HHV-6B, CMV, EBV (RealArt™ LC Assays, Qiagen GmbH, Hamburg, Germany). Qualitative detection of HHV-7 was accomplished by real time PCR using previously described primers and hybridization probes [Safronetz et al., 2003]. PCR was performed on a LightCycler (Roche Diagnostics). To differentiate between HHV-6 variants, the system uses the specific melting temperatures of the probes. Sample series from each patient were tested together in the same run.
HHV-6 Serology
HHV-6 serology was performed using a commercial kit (Human Herpesvirus-6 IgG Enzyme Immunoassay, Biotrin (Diasorin), Dublin, Ireland). Approximately 10–12 plasma specimens were selected per patient. In the absence of clinical events, sampling intervals for specimens from routine visits were approximately 3–4 weeks for the first 6 months of enrollment, 2–3 months for months 6–12, and 3–6 months for the second year. Specimens were tested from acute time points if plasma was available. Routine specimens obtained two or more weeks before and after acute events were also included. To ensure visualization of antibody fluctuations, patients were screened for potential assay saturation. To do this, the last sample of each patient series was tested using the kit-recommended dilution of 1:100. If the OD value was ≤2.5, that patient's sample series was tested at a dilution of 1:100; if the screening OD was ≥2.5, the patient's series was tested at a dilution of 1:500. All samples collected from the same patient were tested on the same run. Longitudinal HHV-6 serology was evaluated longitudinally to look for evidence of host response to infection during periods of DNAemia. The serologic assay used does not discriminate between HHV-6A and HHV-6B antibodies, and it has a low level of cross-reactivity with HHV-7.
Statistical Methods
The study cohort was described using means, standard deviations, and ranges for continuous characteristics and counts and percentages for categorical characteristics. Patients who were reenrolled in the study due to relapse were analyzed as a single patient. The primary analysis was longitudinal with each subject serving as their own control: for each virus, a logistic regression model with generalized estimating equations (GEE) was used to assess the association between PCR results, classified as positive or negative, and visit type, categorized as routine or acute, adjusting for the correlation between visits of the same subject. GEE correlation structures were chosen for each model using the quasi-likelihood information criterion (QIC). The associations between other demographic and clinical variables and HHV-6 PCR positivity were also studied using univariable and multivariable logistic regression models with GEE. A secondary, patient-level analysis was performed to assess whether positive PCR tests were more likely in patients with at least one acute visit; for each virus, negative binomial regression (Poisson regression corrected for overdispersion) models for the number of positive tests predicted by whether or not a subject had at least one acute visit, adjusting for the number of PCR tests performed, were constructed. Longitudinal analyses were performed for presence of viruses with at least 10 positive PCR results in at least 10 subjects, and patient—viral load level analyses were performed for viruses with positive PCR results in at least 10 subjects. Analyses were performed with SAS 9.4 software (SAS Institute, Cary, NC). All analyses were two-tailed and performed at a significance level of 0.05. Graphs were produced using R version 3.1.1(R Project for Statistical Computing, Vienna, Austria).
RESULTS
Study Design and Participants
Seventy-seven patients were prospectively enrolled with a new diagnosis of a malignancy from September 2005 until February 2009 at Cleveland Clinic Children's Hospital. Two children withdrew from the study prior to obtaining evaluable data, and one child was removed from the study because of concerns about the validity of the informed consent. One child was found to have chromosomally integrated HHV-6B and was removed from the analysis. The remaining 73 subjects were used in the analysis. Fourteen patients died during the study: eight within 1 year of enrollment and six between 1 and 2 years after enrollment. Data were collected continuously except for children who had bone marrow transplants outside of our hospital; they were followed until the transplant and then after they returned to our care. Data could not be collected while they were away from our institution. Eight children relapsed and were re-enrolled.
Characteristics of the study population are summarized in Table I. The mean age at enrollment was 10 years (range: 7 months–21 years; S.D. 6.4 years). Diagnoses included a variety of malignancies, a third of which were leukemias. There were 1,291 clinical encounters (1,111 routine and 180 acute). Among the 73 subjects there were a mean of 17.7 study encounters (SD 7.6, median 18, range 1–41), and a mean of 2.5 acute encounters (SD 2.6, median 2.0, range 0–11). Twenty-three (32%) of the subjects had no acute events, 10 (14%) had one, and 40 (54%) had at least two. One child was hospitalized at the time of enrollment and remained acutely ill until his death; all of his encounters were coded as acute. As part of routine clinical care, non-study infections were detected during 26% of acute clinical events.
| Total patients enrolled | 73 |
|---|---|
| Gender | 29 (53%) |
| Male | 34 (47%) |
| Female | |
| Age at enrollment | |
| Mean (SD) | 10.1 years (6.4) |
| Range | 0.6–21 years |
| Died during the study | 14 (19%) |
| Diagnoses | n |
| Leukemia | 25 |
| Brain tumor | 6 |
| Non-Hodgkin's lymphoma | 6 |
| Rhabdomyosarcoma | 6 |
| Osteosarcoma | 5 |
| Ewing's sarcoma | 4 |
| Hodgkin's lymphoma | 4 |
| Langerhans cell histiocytosis | 4 |
| Neuroblastoma | 3 |
| Retinoblastoma | 3 |
| Other | 7 |
| Visits with data | n |
| Total | 1,291 |
| Acute | 180 (14%) |
| Routine | 1,111 (86%) |
| Major interventions | |
| Chemotherapy | 924/1,285 (72%) |
| Blood products | 320/1,243 (26%) |
| Antivirals | 97/1,284 (8%) |
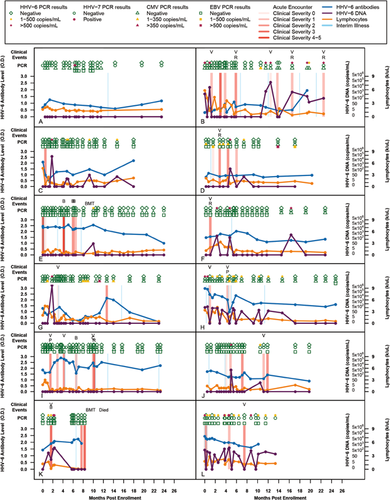
Figure 1 includes representative examples of encounter timing and frequencies, as well as the nature of the clinical and laboratory data collected, relative to acute clinical events of possible viral origin. Panel A shows the course of a patient who had a relatively uneventful follow-up period. Lymphocyte levels were relatively constant and HHV-6 antibody levels changed in small increments. Only HHV-7 DNA was detected, and at only one timepoint.
Serologic Responses to HHV-6 After Initiation of Cancer Treatment
Serological responses to HHV-6 have not previously been studied longitudinally in a pediatric oncology cohort, and the relationship between DNAemia and antibody responses have not been studied outside the context of primary infection. As can be seen in the longitudinal plots (Fig. 1), there was no discernible relationship between HHV-6B PCR positivity and HHV-6 antibody levels (the test used does not discriminate HHV-6A from HHV-6B antibodies). As an example, the patient illustrated in Figure 1H was PCR positive at 12 of 22 visits, but their HHV-6 antibody levels declined gradually over most of the study period.
HHV-6B PCR Positivity Is Not Associated With Acute Events
The overall frequencies of virus detection in the 73 subjects are summarized in Table II. No child had HHV-6A. HHV-6B was detected at least once in 39 (53%) of the subjects, and in 114 (9.3%) of 1,223 specimens. Factors associated with HHV-6B PCR positivity in both single-variable and multivariable logistic regression models included younger age, chemotherapy, higher lymphocyte counts, and steroid administration; there was no association with gender or administration of blood products (Table III). All children had HHV-6 antibody at enrollment.
| HHV-6 | HHV-7 | CMV | EBV | Any virusa | HHV-7,CMV, or EBVa | More than one virusa | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Participants evaluated | 73 | 73 | 63 | 57 | 73 | 73 | 73 | |||
| Threshold for positivity (copies/ml) | >0 | >500 | >0 | >0 | >350 | >0 | >500 | >0 | >0 | >0 |
| Patients with >1 positive PCR | 39 (53%)b | 26 (36%)b | 4 (5%)b | 12 (19%)b | 3 (5%)b | 9 (16%)b | 5 (9%)b | 50 (68%)b | 21 (29%)c | 10 (14%)c |
| Number of positive PCRs per patient:b, c | ||||||||||
| Mean (SD) | 2.9 (3.2) | 1.7 (1.4) | 1.5 (0.6) | 2.4 (1.7) | 1.3 (0.6) | 2.6 (2.0) | 2.2 (2.2) | 3.3 (3.3) | 2.6 (2.2) | 2.4 (0.7) |
| Median | 1.0 | 1.0 | 1.5 | 2.0 | 1.0 | 2.0 | 1.0 | 2.0 | 2.0 | 2.0 |
| Range | 1–15 | 1–7 | 1–2 | 1–6 | 1–2 | 1–7 | 1–6 | 1–15 | 1–7 | 2–4 |
| Number of specimens | 1,223 | 1,214 | 859 | 814 | 1,277 | 1,256 | ||||
| Specimens with positive PCR (%) | 114 (9.3) | 43 (3.5) | 6 (0.5) | 29 (3.4) | 4 (0.5) | 23 (2.8) | 11 (1.4) | 167 (13.1) | 54 (4.3) | |
- a Using the >0 copies/ml threshold for all viruses.
- b Means, medians, and ranges for numbers of positive tests are for patients with at least one positive PCR.
- c Means, medians, and ranges for numbers of positive tests are for patients with at least one positive PCR for two different viruses, not necessarily at the same visit.
| Factora | HHV-6 PCR resultsa | Univariable models | Multivariable modelb | |||
|---|---|---|---|---|---|---|
| Negative (N = 1,109) | Positive (N = 114) | Odds ratioc (95% CI): | P-value | Adjusted odds ratioc (95% CI): | Adjusted P-value | |
| Female gender, No. (%) | 490 (88) | 66 (12) | 1.7 (0.74, 4.1) | 0.23 | 1.6 (0.76, 3.2) | 0.25 |
| Age at study enrollment (years), mean ± SD | 10.4 ± 6.0 | 5.3 ± 4.6 | 0.85 (0.80, 0.90)d | 0.002 | 0.86 (0.81, 0.91)d | 0.002 |
| Chemotherapy received, No. (%) | 775 (89) | 98 (11) | 2.8 (1.4, 5.4) | 0.004 | 2.0 (1.04, 3.8) | 0.030 |
| Steroids received, No. (%) | 383 (87) | 57 (13) | 1.9 (1.3, 2.8) | 0.013 | 1.6 (1.08, 2.5) | 0.024 |
| Blood products received, No. (%) | 276 (91) | 28 (9) | 1.00 (0.64, 1.6) | 0.99 | 1.00 (0.62, 1.6) | 0.99 |
| Abs. lymphocyte count, mean ± SD | 1.3 ± 1.3 | 1.8 ± 1.5 | 1.1 (1.04, 1.2)e | 0.008 | ||
| Abnormal Abs. lymphocyte count, No. (%) | 737 (92) | 61 (8) | 0.57 (0.36, 0.91) | 0.055 | 0.56 (0.35, 0.89) | 0.033 |
| Antiviral medications received, No. (%) | 84 (95) | 4 (5) | 0.45 (0.12, 1.6) | 0.20 | 1.4 (0.36, 5.5) | 0.65 |
| Acute visit type, No. (%) | 154 (89) | 20 (11) | 1.3 (0.78, 2.2) | 0.33 | 1.4 (0.77, 2.5) | 0.33 |
- a PCR tests results and risk factors not available for all visits. Counts and percentages reflect missing data.
- b Multivariable logistic regression model containing all risk factors, using GEE to adjust for encounters onsame patient.
- c Odds ratios for outcome PCR+ estimated by logistic regression models using GEE to adjust forencounters on same patient.
- d Odds ratio for 1 year increase in age at enrollment.
- e Odds ratio for 0.5 unit increase in lymphocyte value.
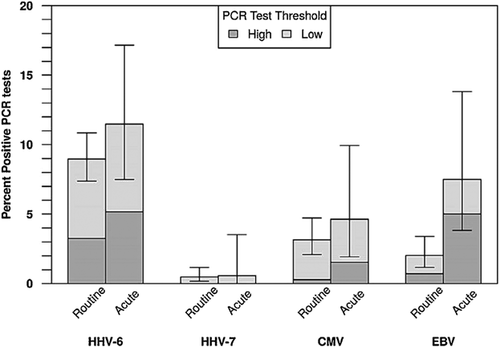
As can be seen in the longitudinal plots (Fig. 1), when considered in the context of the entire time series, scattered acute events seem to have plausible connections to HHV-6B activity (e.g., the first acute event in panel K). In contrast, the patient shown in panel L was positive for HHV-6B DNA at 15 of their 20 study visits but they were positive for the virus at just two of their four acute events. The absence of a clear association between acute clinical events and detection of HHV-6B is apparent upon inspection of the plot shown in Figure 1. Including all subjects, the low threshold HHV-6B PCR was positive at 11.5% of acute visits and 9% of routine visits (P = 0.33) (Table IV). In addition, no associations between detection of HHV-6B and acute events were identified after restricting the analysis to the 49 subjects who had at least one acute and one routine visit, or by considering only events where HHV-6B levels were at least 500 copies/ml (Table IV and data not shown). In conclusion, we found no temporal association between HHV-6B PCR positivity and acute illness. The 50 subjects with at least one acute visit had higher proportions of PCR tests positive for HHV-6B both at the low and high thresholds than the 23 subjects who had no acute visits: the incidence of positive HHV-6 tests was 4.8 times higher in patients with at least one acute visit than in those with no acute visits. (P = 0.001, Table V; Fig. 3).
| All subjectsa | Subjects with both acute and routine visitsb | |||||||
|---|---|---|---|---|---|---|---|---|
| Routine (N = 1,111) N (%) | Acute (N = 180) N (%) | Adjusted odds ratioc (95% CI): acute versus routine | Adjusted P-value | Routine (N = 831) N (%) | Acute (N = 173) N (%) | Adjusted odds ratioc (95% CI): acute versus routine | Adjusted P-value | |
| HHV-6 PCR > 0 copies/ml | 1.3 (0.78, 2.2) | 0.33 | 1.03 (0.61, 1.8) | 0.90 | ||||
| Negative | 955 (91.0) | 154 (88.5) | 701 (89.0) | 148 (88.6) | ||||
| Positive | 94 (9.0) | 20 (11.5) | 87 (11.0) | 19 (11.4) | ||||
| HHV-6 PCR > = 500 copies/ml | 1.3 (0.60, 2.8) | 0.56 | 1.08 (0.51, 2.3) | 0.85 | ||||
| Negative | 1,015 (96.8) | 165 (94.8) | 757 (96.1) | 159 (95.2) | ||||
| Positive | 34 (3.2) | 9 (5.2) | 31 (3.9) | 8 (4.8) | ||||
| HHV-7 PCR > 0 copies/ml | d | d | ||||||
| Negative | 1,035 (99.5) | 173 (99.4) | 767 (99.5) | 166 (99.4) | ||||
| Positive | 5 (0.48) | 1 (0.57) | 4 (0.52) | 1 (0.60) | ||||
| CMV PCR > 0 copies/ml | 1.2 (0.41, 3.6) | 0.77 | d | |||||
| Negative | 706 (96.8) | 124 (95.4) | 540 (97.3) | 120 (97.6) | ||||
| Positive | 23 (3.2) | 6 (4.6) | 15 (2.7) | 3 (2.4) | ||||
| CMV PCR > = 350 copies/ml | d | d | ||||||
| Negative | 727 (99.7) | 128 (98.5) | 554 (99.8) | 123 (100.0) | ||||
| Positive | 2 (0.27) | 2 (1.5) | 1 (0.18) | 0 (0.0) | ||||
| EBV PCR > 0 copies/ml | d | d | ||||||
| Negative | 680 (98.0) | 111 (92.5) | 506 (98.4) | 111 (98.2) | ||||
| Positive | 14 (2.0) | 9 (7.5) | 8 (1.6) | 2 (1.8) | ||||
| EBV PCR > = 500 copies/ml | d | d | ||||||
| Negative | 689 (99.3) | 114 (95.0) | 511 (99.4) | 113 (100.0) | ||||
| Positive | 5 (0.72) | 6 (5.0) | 3 (0.58) | 0 (0.0) | ||||
- a PCR tests results were not available for all viruses at every visit. Total number of PCR results: HHV-6: 1,223; HHV-7: 1,124; CMV: 859; EBV, 814.
- b PCR tests results were not available for all viruses at every visit. Total number of PCR results: HHV-6: 955; HHV-7: 938; CMV: 678; EBV, 627.
- c Odds ratios for outcome PCR+ estimated by logistic regression models using GEE to adjust for encounters on same patient.
- d Odds ratios were estimated for viruses for which there were at least 10 positive PCR results and at least 10 subjects with a positive PCR.
| Subjects’ PCR results | Subjects with no acute visits (N = 23) | Subjects with at least one acute visit (N = 50) | Incidence rate ratioa(95% CI) for number of positive PCR tests | Adjusted P-valuea |
|---|---|---|---|---|
| At least 1 HHV-6 + PCR, No. (%) | 5 (22) | 34 (68) | ||
| Percent positive HHV-6 PCR tests, median [Q1, Q3] | 0% [0%,0%] | 5% [0%,16%] | 4.8 (1.8, 13.0) | 0.001 |
| At least 1 HHV-6 + PCR > = 500, No. (%) | 3(13) | 23 (46) | ||
| Percent positive HHV-6 PCR tests (> = 500), median [Q1, Q3] | 0% [0%,0%] | 0% [0%,8%] | 4.0 (1.08, 15.2) | 0.022 |
| At least 1 HHV-7 + PCR, No. (%) | 1 (4) | 3 (6) | ||
| Percent positive HHV-7 PCR tests, median [Q1, Q3] | 0% [0%,0%] | 0% [0%,0%] | b | b |
| At least 1 CMV + PCRc, No. (%) | 3 (14) | 9 (21) | ||
| Percent positive CMV PCR testsc, median [Q1, Q3] | 0% [0%,0%] | 0% [0%,0%] | 0.75 (0.16, 3.5) | 0.72 |
| At least 1 CMV + PCR > 350c, No. (%) | 1 (5) | 2 (5) | ||
| Percent positive CMV PCR tests (>350)c, median [Q1, Q3] | 0% [0%,0%] | 0% [0%,0%] | b | b |
| At least 1 EBV + PCRd, No. (%) | 4 (21) | 5 (13) | ||
| Percent positive EBV PCR testsd, median [Q1, Q3] | 0% [0%,0%] | 0% [0%,0%] | b | b |
| At least 1 EBV + PCR > = 500d, No. (%) | 2 (11) | 3 (8) | ||
| Percent positive EBV PCR tests (> = 500)d, median [Q1, Q3] | 0% [0%,0%] | 0% [0%,0%] | b | b |
- a Incidence rate ratios and adjusted P-values for outcome number of PCR+ tests estimated by negative binomial (Poisson corrected for overdispersion) regression models with offset for number of PCR tests performed.
- b Incidence rate ratios were estimated for viruses for which there were at least 10 subjects with a positive PCR.
- c 10 subjects had no CMV PCR tests performed. Percentages calculated out of 63 subjects.
- d 16 subjects had no EBV PCR tests performed. Percentages calculated out of 57 subjects.
CMV, EBV, and HHV-7 Results
CMV, EBV, and HHV-7 DNA were detected much less frequently than HHV-6B DNA (Table II). There was no association between an acute visit and the rate of positive CMV PCR tests. There were not enough positive test results to assess this for HHV-7 or EBV.
HHV-7 was found in only six specimens from four patients and was not related to the presence of HHV-6B. CMV and EBV were detected in 12 and 9 subjects respectively during the study and were also not associated with positive HHV-6B. One of these children was being treated for an EBV-related hemophagocytic syndrome; he was persistently positive for EBV and was positive once each for HHV-6B and CMV. As can be seen in the longitudinal plots (Fig. 1), with the exception of EBV in the patient with EBV-related hemophagocytic syndrome, only scattered specimens were positive for any of the study viruses, and there was no statistical support for an association between any of these viruses and acute events (Table IV and Fig. 2).
Ten children were PCR positive for more than one virus during the study period (Table II), but more severe illness was not related to detecting more than one virus (data not shown).
HHV-6 PCR Positivity was More Frequent in Sicker Children
The 50 subjects with at least one acute visit had higher proportions of PCR tests positive for HHV-6B both at the low and high thresholds than the 23 subjects who had no acute visits: the incidence of positive HHV-6 tests was 4.8 times higher in patients with at least one acute visit than in those with no acute visits. (P = 0.001, Table V; Fig. 3). However, there was no association between an acute visit and the rate of positive CMV PCR tests. There were not enough positive test results to assess this for HHV-7 or EBV.

DISCUSSION
Longitudinal cohort studies can play important roles in defining the biological properties and etiologic roles of persistent and highly prevalent viruses (10). Here, we employed such a study design to assess the incidence and significance of detection of lymphotropic herpesviruses (CMV, EBV, HHV-6A, HHV-6B, and HHV-7), with an emphasis on HHV-6B, in children on therapy for malignancy. Children were followed from the time of their cancer diagnosis, throughout and beyond their period of chemotherapy, and were evaluated for the study viruses at visits for both routine care and acute illness. We found no association of HHV-6B detection with acute illness by either categorical or longitudinal analysis. Our major conclusion is that if HHV-6B is not assessed longitudinally, clinical events will be misattributed to the virus.
HHV-6B DNA was detected in about half of the children at some time during the 2 year follow up period but was not associated with acute disease. We found it as often in children who were ill as during routine visits to the clinic. Detection of HHV-6B DNA correlated with younger age, chemotherapy, steroids, and higher lymphocyte counts, but not with acute illness. As with many viruses, reactivation is more likely to occur during immune suppression. However, as clearly demonstrated in our study, detection of HHV-6B is not synonymous with acute disease. Acute illness should not be attributed to this virus without understanding the longitudinal course of the illness and virus activity; although some episodes may be causal, others will not be related to the virus. As younger age was a risk factor for activation, a weakness of our study may have been the small number of infants in our population. We also had no children with acute primary infection; all children had antibody present at enrollment into the study, again a reflection of the older age range.
Although we found no temporal association between detection of HHV-6B DNA and acute illness, HHV-6 PCR positivity was more frequent in sicker children. This may reflect the frequent HHV-6 activity seen in patients with multiple organ failure [Desachy et al., 2001] and intensive care unit patients [Razonable et al., 2002]. Similar to our observations, HHV-6 activity was not associated with additional morbidity or mortality in those critically ill patients. Roa et al. [2015] recently found frequent detection of HHV-6B DNA in critically ill adults, but in contrast to the other reports, simultaneous activity of both CMV and HHV-6 correlated with worse clinical outcomes. Although we did not detect clear evidence of a relationship between the study viruses and disease in children with cancer who are undergoing chemotherapy, it remains possible that such associations might be detected in other and larger populations of such patients.
ACKNOWLEDGMENTS
The authors thank the children and their parents for their participation in the study, and for the study nurses and laboratory personnel whose contributions were essential to completion of the study. This work was supported by the Thrasher Research Fund and the National Institutes of Health Supported in part by UL1TR000439, National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health via the Cleveland Clinic's General Clinical Research Center and its successor Clinical Translational Science Center, which provided access to tools and methods for collecting, storing, managing, and assessing data for correlations to disease data.




