BTNL2 associated with the immune response to hepatitis B vaccination in a Chinese Han population
Abstract
No response to hepatitis B vaccination is a complex phenomenon, which is induced by the combinations of environmental and genetic factors. The aim of the study was to investigate the association between the polymorphisms of the butyrophilin-like 2 (BTNL2) gene and the immune response to hepatitis B vaccination in a Chinese Han population. A total of 7 single nucleotide polymorphisms in the BTNL2 gene were analyzed in 566 non-responders and 1,040 high-responders to hepatitis B vaccination. The alleles T, T, C, A, G of rs3763316, rs3763311, rs9268494, rs3806156, and rs2076530 were associated with no response to hepatitis B vaccination (P = 0.015, odds ratio (OR) = 1.20; P = 0.029, OR = 1.18; P = 2.00E−07, OR = 1.58; P = 0.002, OR = 1.27; P = 2.90E−06, OR = 1.41, respectively). Whereas, the alleles T, C of rs9268501 and rs3763313 played significantly protective roles in the immune response to hepatitis B vaccination (P = 0.007, OR = 0.81; P = 0.004, OR = 0.74). Besides, the risks of no response to hepatitis B vaccination were increased significantly among individuals harbored the haplotypes of G-T-A-T-C-A-G (P = 0.038, OR = 1.48), G-T-A-T-C (P < 0.0001, OR = 2.34), A-A (P < 0.0001, OR = 4.08), and C-G (P < 0.0001, OR = 4.75). However, the haplotype of G-C-A-T-C (P = 1.00E−04, OR = 0.54) exhibited a protective role in the immune response to hepatitis B vaccination in the study. These findings suggest that polymorphisms in the BTNL2 gene might play a vital role in determining the outcome of the immune response to hepatitis B vaccination. J. Med. Virol. 86:1105–1112, 2014. © 2014 Wiley Periodicals, Inc.
INTRODUCTION
Infection with hepatitis B virus (HBV) is a worldwide public health problem, which causes more than half a million deaths each year. It is estimated that there are 350 million carriers of HBV all over the world, including approximately 93 million Chinese patients [Kane, 1995; Lu and Zhuang, 2009]. All the chronic HBV carriers suffer a 15–25% risks of dying from the consequences of the infection, such as cirrhosis and hepatocellular carcinoma (HCC) [Kao and Chen, 2002]. Hepatitis B vaccination is the dominant and effective method to prevent infection and transmission of HBV. However, there are still about 1–10% of recipients failing to elicit protective levels of antibody against the recombinant hepatitis B vaccines (anti-HBs) [Shokri and Amani, 1997; Shokri and Jafarzadeh, 2001; Zuckerman et al., 2001].
Environment factors and physical parameters, such as older age, male gender, higher body mass index (BMI), a history of smoking and alcohol consumption, are associated with low or no response to hepatitis B vaccination [Weber et al., 1985; Hollinger, 1989; Wood et al., 1993; Yu et al., 1998; Boland et al., 2003]. However, a twin study in Chinese infants reported that the heritability of poor response to HBV vaccination may be 91% [Yan et al., 2013]. Polymorphisms in genes of the major histocompatibility complex (MHC) [Godkin et al., 2005; Png et al., 2011], the T cell receptor (TCR)/CD3 complex [Pan et al., 2012], cytokines such as the interleukin-4 [Wang et al., 2004], costimulatory molecules such as Integrin alpha-L [Hennig et al., 2008], are associated with vaccine-induced immune response.
Activation of CD4+ Th cells plays a pivotal role in the immune protection triggered by hepatitis B vaccine [Milich and Leroux-Roels, 2003; Goncalves et al., 2004]. The interaction between the TCR/CD3 complex and the MHC-II–peptide complex, which is promoted by CD4, triggers the first signal for the activation of CD4+ Th cells [Portoles and Rojo, 2009]. However, a full-force activation of CD4+ Th cells requires a second indispensable signal initiated by the interactions of costimulatory molecules with their receptors. For example, CD80/CD86 and their receptor CD28/CTLA4 are the well-known costimulatory signals. They trigger the positive/negative regulation of CD4+ Th cells activation [Bretscher, 1999; Alegre et al., 2001]. In addition to the classic costimulatory molecules, others such as B7RP-1 and its receptor ICOS, B7h, PDL1/PDL2 and their receptor PD-1, butyrophilin-like 2 (BTNL2) are also of importance to the activation of CD4+ Th cells [Swallow et al., 1999; Yoshinaga et al., 1999; Nguyen et al., 2006; Sage et al., 2013]. A genome-wide association study of HBV vaccine-induced response in an Indonesian population suggested that the BTNL2 gene might be associated with the immune response to hepatitis B vaccination. Researchers found that tag single nucleotide polymorphisms (SNPs) rs9277535 and rs3135363, encompassing the HLA-DP, HLA-DR, and the BTNL2 gene, were the first to emerge from the conditional analysis and exhibited strongest association with the immune response to hepatitis B vaccination [Png et al., 2011].
In the present study, subjects were recruited to evaluate whether the polymorphisms in the BTNL2 gene were associated with different outcomes of the immune response to hepatitis B vaccination in the Chinese Han population. The findings suggested that polymorphisms in the BTNL2 gene might affect the efficacy of hepatitis B vaccination. Furthermore, the results might be helpful in identifying specific genes which could be used as targets in the development of novel and more effective vaccines for prevention of HBV infection.
MATERIALS AND METHODS
Subjects
The study subjects were recruited from a large hepatitis B vaccination campaign in Shandong province in 2009. After signing written informed consent, participants were also required to complete a questionnaire including questions about demographic information, smoking history, vaccination history, chronic diseases, and immunosuppressive disease/medications. All individuals were tested for five markers of hepatitis B using an Abbott i2000 detection kit (Abbott Laboratory, Chicago, IL). Individuals who were negative for the five markers of hepatitis B were tested further for HBV DNA and for anti-HCV and anti-HIV. The exclusion criteria of participants and the method of administration were same as former study [Pan et al., 2012]. Participants who showed high levels of anti-HBs (≥1,000 mIU/ml) were designated as high-responders, whereas participants who showed very low levels of anti-HBs (<10 mIU/ml) were designated as non-responders [Davila et al., 2010]. The definition of non-responders and high-responders was based on the level of anti-HBs 1 month after the third dose of vaccine. A total of 1,606 high-quality individuals were recruited in this study, including 1,040 high response subjects and 566 no response subjects.
The study was performed in accordance with the guidelines of the Helsinki Declaration and was approved by the Ethics Committee of the Institute of Basic Medical Sciences, Chinese Academy of Medical Science.
SNP Selection and Genotyping
The selected SNPs in the BTNL2 gene were based on their potential function, Chinese Han race in Beijing (CHB) and a minor allele frequency (MAF) >0.20. A full list of the 13 selected SNPs was given in Table S1. The SNP ID numbers and detailed sequence information were available at http://www.ncbi.nlm.nih.gov/SNP/. Within the 13 selected SNPs, only 7 SNPs were genotyped, because rs3763311 was in linkage disequilibrium (LD) with rs9268492 and rs9268493, rs9268494 was in LD with rs9268499 and in complete LD with rs9268497, rs9268501 was in LD with rs3763317 and in complete LD with rs6926737 (HapMap2 and Fig. S1). The degree of LD was analyzed for the selected SNPs in the study with HapMap2 genotyping results and 45 CHB genotyping results in Reference Genome Orientation (NCBI). It was difficult to produce a probe for rs2076523, rs3806156 was genotyped instead of rs2076523 because of the complete LD between the two SNPs.
Genomic DNA was extracted from peripheral blood using the phenol–chloroform method. The candidate SNPs were genotyped using the TaqMan-MGB (Genecore Biotech, Shanghai, China) or TaqMan-BHQ (Sangon Biotech, Shanghai, China) probe-based real-time polymerase chain reaction (PCR). The primer and probe sequences are shown in Table S2. Amplification and detection were conducted using a Bio-Rad iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA).
Statistical Analysis
The demographic characteristics of the participants were compared according to the status of the immune response to hepatitis B vaccination using χ2-test for categorical variables and t-test for continuous variables. A χ2 goodness-of-fit test was used to examine whether the genotype distributions of each SNP conformed to Hardy–Weinberg equilibrium (HWE) in the study subjects. The EPI software (version 5.0) was used to calculate the statistical power. The allele frequencies for each SNP were compared between the high- and non-responders using χ2-test. The calculation was conducted with EPI software (version 5.0). LD of the SNPs was tested using HaploView (version 4.2). The analyses of association of SNP genotype distributions and haplotypes with HBV vaccine-induced response were performed using SNPstats software, available at http://bioinfo.iconcologia.net/SNPstats. Multiple odds ratios (ORs) were adjusted further for the potential non-genetic risk factors of gender, age, BMI, smoking, alcohol consumption, and vaccines. Frequency threshold for rare haplotypes is 0.05. A value of P < 0.05 was considered statistically significant.
RESULTS
Demographic Characteristics of the Study Subjects
For a case–control study, a total of 1,606 participants were recruited in the Chinese Han population. Among the 1,606 participants, 566 individuals showed very low levels of anti-HBs (<10 mIU/ml) and were assigned to the non-responder group, whereas the other 1,040 individuals namely high-responders (anti-HBs ≥1,000 mIU/ml), were designed as a control group that had the similar age and gender ratios with the non-responder group.
The demographic details of the study subjects are summarized in Table I. There were no significant differences in the demographic characteristics and the diverse vaccine responses between high- and non-responder groups in the case–control study (P > 0.05).
| HR, n = 1,040 (100%)b | NR, n = 566 (100%)c | P | |
|---|---|---|---|
| Age (years)a | 41.3 ± 5.61 | 41.5 ± 7.35 | 0.492 |
| Gender | |||
| Male | 493 (47.4) | 296 (52.3) | 0.061 |
| Female | 547 (52.6) | 394 (47.7) | |
| BMI (kg/m2) | 24.5 ± 3.10 | 24.7 ± 3.07 | 0.076 |
| Smoking | |||
| Yes | 167 (16.1) | 100 (17.7) | 0.407 |
| No | 873 (83.9) | 466 (82.3) | |
| Drinking | |||
| Yes | 156 (15.0) | 79 (14.0) | 0.572 |
| No | 884 (85.0) | 487 (86.0) | |
| Vaccinesd | |||
| 1 | 409 (39.3) | 247 (30.1) | 0.695 |
| 2 | 369 (35.5) | 208 (25.3) | |
| 3 | 262 (25.2) | 159 (19.4) | |
- a Values are mean ± SD.
- b HR: high-responder (anti-HBs ≥1,000 mIU/ml).
- c NR: non-responder (anti-HBs <10 mIU/ml).
- d Vaccines: (1) recombinant yeast-derived hepatitis B vaccine, 3 × 20 μg, (2) recombinant CHO-derived hepatitis B vaccine, 3 × 20 μg, (3) recombinant yeast-derived hepatitis B vaccine, 3 × 10 μg.
Association Between SNPs in the BTNL2 Gene and the Immune Response to Hepatitis B Vaccination
A total of seven SNPs in the BTNL2 gene were analyzed in the study. The average success rate for genotyping was 99.6% (ranging from 99.2% to 99.9%). The genotype distributions of all the SNPs studied were in HWE. The allele frequencies of the SNPs were compared between the high- and non-responder groups and the results are summarized in Table II. All SNPs genotyped in the study showed significant association with the immune response to hepatitis B vaccination. As shown in Table II, the frequencies of the minor alleles T, T, C, A, G of rs3763316, rs3763311, rs9268494, rs3806156, and rs2076530 were significantly higher in the non-responders than in the high-responders (P = 2.00E−07, OR = 1.58; P = 0.002, OR = 1.27; P = 2.90E−06, OR = 1.41; P = 0.015, OR = 1.20; P = 0.029, OR = 1.18; respectively). The frequencies of the minor alleles T, C of rs9268501 and rs3763313 were significantly higher in the high-responders than in the non-responders (P = 0.007, OR = 0.81; P = 0.004, OR = 0.74). Given that the occurrence of the minor alleles described herein ranged from 16.5% to 44.1%, the present study demonstrated a statistical power to detect allelic association of 99.96% to 99.99% with an OR of 2.0 at a significance level of 0.05.
| SNP | Alleles | Pc | OR (95% CI) | ||
|---|---|---|---|---|---|
| HRa | NRb | ||||
| rs9268501 | n = 2,076 (%) | n = 1,122 (%) | |||
| G | 1,254 (60.4) | 732 (65.2) | 0.007 | 0.81 (0.70–0.95) | |
| T | 822 (39.6) | 390 (34.8) | |||
| rs3763316 | n = 2,074 (%) | n = 1128 (%) | |||
| C | 1,681 (81.1) | 824 (73.0) | 2.00E−07 | 1.58 (1.32–1.88) | |
| T | 393 (18.9) | 304 (27.0) | |||
| rs3763313 | n = 2,068 (%) | n = 1,126 (%) | |||
| A | 1,727 (83.5) | 983 (87.3) | 0.004 | 0.74 (0.59–0.91) | |
| C | 341 (16.5) | 143 (12.7) | |||
| rs3763311 | n = 2,060 (%) | n = 1,126 (%) | |||
| C | 1,281 (62.2) | 637 (56.6) | 0.002 | 1.27 (1.09–1.48) | |
| T | 779 (37.8) | 489 (43.4) | |||
| rs9268494 | n = 2,076 (%) | n = 1,132 (%) | |||
| A | 1,173 (56.5) | 542 (47.9) | 2.90E−06 | 1.41 (1.22–1.64) | |
| C | 903 (43.5) | 590 (52.1) | |||
| rs3806156 | n = 2,070 (%) | n = 1,122 (%) | |||
| C | 1,249 (60.3) | 627 (55.9) | 0.015 | 1.20 (1.05–1.39) | |
| A | 821 (39.7) | 495 (44.1) | |||
| rs2076530 | n = 2,078 (%) | n = 1,128 (%) | |||
| A | 1,316 (63.3) | 670 (59.4) | 0.029 | 1.18 (1.01–1.37) | |
| G | 762 (36.7) | 458 (40.6) | |||
Note
- The number of subjects for each SNP in the table was inconsistent because some genotyped failure were excluded in process of statistic.
- a HR: high-response group (anti-HBs ≥1,000 mIU/ml).
- b NR: no-response group (anti-HBs <10 mIU/ml).
- c P values for χ2-test.
After adjusting for gender, age, BMI, smoking, alcohol consumption and vaccines, the TC, TT, and TC + TT genotype carriers of the rs3763316 had increased risks of no response to hepatitis B vaccination compared to the CC genotype carriers in the codominant and dominant models, the adjusted ORs were 1.45 (95% CI 1.16–1.81), 3.21 (95% CI 1.98–5.20), and 1.60 (95% CI 1.29–1.98), respectively. The TT and CT + TT genotype carriers of the rs3763311 had increased risks of no response to hepatitis B vaccination compared to the CC genotype carriers in the codominant and dominant models, the adjusted ORs were 1.56 (95% CI 1.16–2.10) and 1.32 (95% CI 1.07–1.64). The AC, CC, and AC + CC genotype carriers of the rs9268494 had increased risks of no response to hepatitis B vaccination compared to the AA genotype carriers in the codominant and dominant models, the adjusted ORs were 1.33 (95% CI 1.03–1.71), 2.08 (95% CI 1.54–2.81), and 1.52 (95% CI 1.20–1.93), respectively. Besides, the AA genotype carriers of the rs3806156 and the GG genotype carriers of the rs2076530 had increased risks of no response to hepatitis B vaccination compared to the CC genotype carriers and the AA genotype carriers in the codominant model, the adjusted ORs were 1.46 (95% CI 1.07–2.00) and 1.42 (95% CI 1.03–1.95). However, the GT, TT, and GT + TT genotypes of the rs9268501 played significantly protective roles in the immune response to hepatitis B vaccination compared to the GG genotype in the codominant and dominant models, the adjusted ORs were 0.77 (95% CI 0.61–0.96), 0.66 (95% CI 0.47–0.93), and 0.74 (95% CI 0.60–0.92), respectively. The AC, CC, and AC + CC genotypes of the rs3763313 played significantly protective roles in the immune response to hepatitis B vaccination compared to the AA genotype in the codominant and dominant models, the adjusted ORs were 0.74 (95% CI 0.58–0.95), 0.48 (95% CI 0.21–0.83), and 0.72 (95% CI 0.57–0.91), respectively (Table III).
| SNP | Model | Genotype | HRa | NRb | ORc (95% CI) |
|---|---|---|---|---|---|
| rs9268501 | Codominant | GG | 361 (34.8%) | 235 (41.9%) | 1.00 |
| GT | 528 (50.9%) | 262 (46.7%) | 0.77 (0.61–0.96) | ||
| TT | 149 (14.3%) | 64 (11.4%) | 0.66 (0.47–0.93) | ||
| Dominant | GG | 361 (34.8%) | 235 (41.9%) | 1.00 | |
| GT-TT | 677 (65.2%) | 326 (58.1%) | 0.74 (0.60–0.92) | ||
| rs3763316 | Codominant | CC | 675 (65.1%) | 305 (54.1%) | 1.00 |
| TC | 331 (31.9%) | 214 (37.9%) | 1.45 (1.16–1.81) | ||
| TT | 31 (3%) | 45 (8%) | 3.21 (1.98–5.20) | ||
| Dominant | CC | 675 (65.1%) | 305 (54.1%) | 1.00 | |
| TC-TT | 362 (34.9%) | 259 (45.9%) | 1.60 (1.29–1.98) | ||
| rs3763313 | Codominant | AA | 719 (69.5%) | 428 (76%) | 1.00 |
| AC | 289 (27.9%) | 127 (22.6%) | 0.74 (0.58–0.95) | ||
| CC | 26 (2.5%) | 8 (1.4%) | 0.48 (0.21–0.83) | ||
| Dominant | AA | 719 (69.5%) | 428 (76%) | 1.00 | |
| AC-CC | 315 (30.5%) | 135 (24%) | 0.72 (0.57–0.91) | ||
| rs3763311 | Codominant | CC | 413 (40.1%) | 191 (33.9%) | 1.00 |
| CT | 455 (44.2%) | 255 (45.3%) | 1.24 (0.98–1.56) | ||
| TT | 162 (15.7%) | 117 (20.8%) | 1.56 (1.16–2.10) | ||
| Dominant | CC | 413 (40.1%) | 191 (33.9%) | 1.00 | |
| CT-TT | 617 (59.9%) | 372 (66.1%) | 1.32 (1.07–1.64) | ||
| rs9268494 | Codominant | AA | 318 (30.6%) | 130 (23%) | 1.00 |
| AC | 537 (51.7%) | 282 (49.8%) | 1.33 (1.03–1.71) | ||
| CC | 183 (17.6%) | 154 (27.2%) | 2.08 (1.54–2.81) | ||
| Dominant | AA | 318 (30.6%) | 130 (23%) | 1.00 | |
| AC-CC | 720 (69.4%) | 436 (77%) | 1.52 (1.20–1.93) | ||
| rs3806156 | Codominant | CC | 366 (35.4%) | 171 (30.5%) | 1.00 |
| CA | 517 (50%) | 285 (50.8%) | 1.18 (0.94–1.50) | ||
| AA | 152 (14.7%) | 105 (18.7%) | 1.46 (1.07–2.00) | ||
| Dominant | CC | 366 (35.4%) | 171 (30.5%) | 1.00 | |
| CA-AA | 669 (64.6%) | 390 (69.5%) | 1.25 (1.00–1.56) | ||
| rs2076530 | Codominant | A/A | 409 (39.4%) | 197 (34.9%) | 1.00 |
| A/G | 498 (47.9%) | 276 (48.9%) | 1.15 (0.92–1.44) | ||
| G/G | 132 (12.7%) | 91 (16.1%) | 1.42 (1.03–1.95) | ||
| Dominant | A/A | 409 (39.4%) | 197 (34.9%) | 1.00 | |
| A/G-G/G | 630 (60.6%) | 367 (65.1%) | 1.20 (0.97–1.49) |
- a HR: high-response group (anti-HBs ≥1,000 mIU/ml).
- b NR: no-response group (anti-HBs <10 mIU/ml).
- c Adjusted for gender, age, BMI, smoking, alcohol consumption, and vaccines.
Association of Haplotypes With the Immune Response to Hepatitis B Vaccination
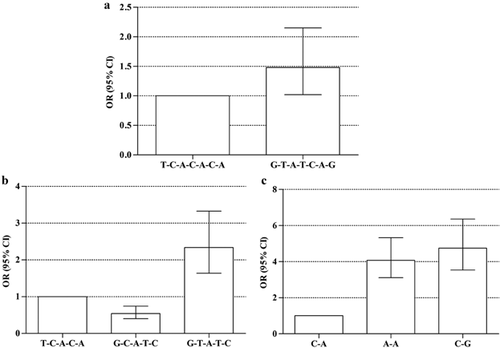
Given that there were different levels of LD between SNPs analyzed in the present study. Haplotype analysis was just performed with rs9268501, rs3763316, rs3763313, rs3763311, rs9268494, rs3806156, and rs2076530. Association analyses revealed that the risks of no response to hepatitis B vaccination were increased significantly among individuals harbored the haplotypes of G-T-A-T-C-A-G (P = 0.038, adjusted OR = 1.48 and 95% CI 1.02–2.15), G-T-A-T-C (P < 0.0001, adjusted OR = 2.34 and 95% CI 1.64–3.33), A-A (P < 0.0001, adjusted OR = 4.08 and 95% CI 3.12–5.33), and C-G (P < 0.0001, adjusted OR = 4.75 and 95% CI 3.54–6.36) compared with those carried the T-C-A-C-A-C-A, T-C-A-C-A and C-A haplotypes. However, the haplotype of G-C-A-T-C (P = 1.00E−04, adjusted OR = 0.54 and 95% CI 0.40–0.76) was a protective construction of the immune response to hepatitis B vaccination compared with the T-C-A-C-A haplotype in the study (Fig. 1, Table S3).
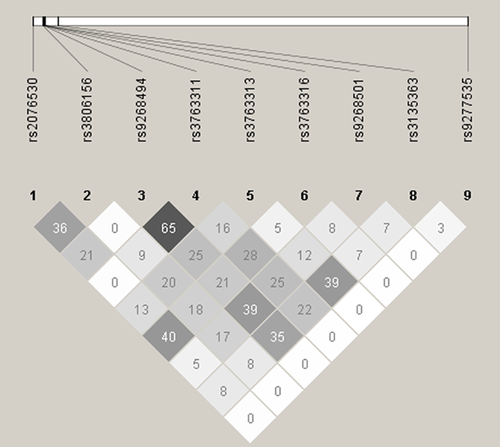
The Relationship Between the HLA and the BTNL2 Gene
The rs9277535 and rs3135363, encompassing the HLA-DP, HLA-DR, and the BTNL2 gene, were associated significantly with the immune response to hepatitis B vaccination in a genome-wide association study [Png et al., 2011]. Therefore, it was essential to give clear information about the relationship between rs9277535, rs3135363 and the BTNL2 gene and preclude the effect of rs9277535 and rs3135363 on the association of SNPs in the BTNL2 gene with the immune response to hepatitis B vaccination. The rs9277535 and rs3135363 were examined whether they were in LD with rs2076530, rs3806156, rs9268494, rs3763311, rs3763313, rs3763316, or rs9268501 of the BTNL2 gene. The method was supplied as Supplementary Material. The LD analysis showed that rs9277535 and rs3135363 were not in LD with any of the seven SNPs in the BTNL2 gene genotyped in the study subjects (Fig. 2).
DISCUSSION
The immune response to hepatitis B vaccination producing effective and protective anti-HBs antibodies was a complex process which was controlled by numerous factors. In addition to environmental and host-related physical factors, genetic variations have certain effects on the immune response to hepatitis B vaccination [Godkin et al., 2005; Chen et al., 2011; Pan et al., 2012]. Mutations or polymorphisms in the genes, which participated in the regulation of CD4+ Th cells activation, might affect the magnitude of the immune response to hepatitis B vaccination. For example, the delayed-type hypersensitivity (DTH) response and the cytolytic T lymphocyte (CTL) activity were enhanced significantly when B7-2 expression plasmids were coinoculated with a plasmid encoding HBV pre-S2 + S HBV DNA vaccine in the gastrocnemius muscles [Wang et al., 2002]. The s595 polymorphism, which was a non-synonymous coding change in exon 21 (R791T) and lied within the integrin-alpha-2 domain of the TGAL gene, was predicted to affect splicing regulation. The coding change in ITGAL (R719T) with possible function relevance was associated with increased peak anti-HBs level in response to HBV vaccination [Hennig et al., 2008].
BTNL2 gene was located in the MHC region of chromosome 6 in human [Stammers et al., 2000]. Its function was to inhibit T cells activation and was associated with inflammatory diseases [Valentonyte et al., 2005; Nguyen et al., 2006; Arnett et al., 2007]. In vitro, proliferation of purified CD4+ T cells stimulated by anti-CD3 mAb could be inhibited by BTNL2-Ig [Arnett et al., 2007]. BTNL2-Ig inhibited T cells proliferation with moderate decrease of IL-2 secretion. Furthermore, the inhibition of T cells proliferation by BTNL2-Ig was reversed hardly by exogenous IL-2 [Nguyen et al., 2006]. Peripheral blood lymphocytes from non-responders to hepatitis B vaccine failed to undergo a proliferative response to recombinant hepatitis B vaccine in vitro. The cell cultures of non-responders did not secrete IL-2 in response to hepatitis B vaccine stimulation and exogenous recombinant IL-2 did not restore the proliferative response of T cells in recombinant hepatitis B vaccine-pulsed cultures of non-responders as well [Chedid et al., 1997]. Recently, a study showed that BTNL2 modulated B7 costimulation to induce Foxp3 expression and regulatory T cells (Treg) development [Swanson et al., 2013]. These results might provide clues to how BTNL2 affected the outcome of the immune response to hepatitis B vaccination.
The major novel findings in the present study indicated that the polymorphisms in the BTNL2 gene were associated significantly with the immune response to hepatitis B vaccination in Chinese Han population (Tables II and III and Fig. 1). As risk genetic variations of the immune response to hepatitis B vaccination, the minor alleles T, T, C of rs3763316, rs3763311, rs9268494 and the haplotype of G-T-A-T-C were suspected to enhance the BTNL2 gene transcription. In contrast, as protective genetic variations of the immune response to hepatitis B vaccination, the minor allele T, C of rs9268501, rs3763313 and the haplotype of G-C-A-T-C might reduce the BTNL2 gene transcription. To demonstrate if these minor alleles and haplotypes could affect the BTNL2 gene transcription, reporter plasmids encompassing different haplotypes were constructed and used for analyzing luciferase activity with Dual-Luciferase Reporter Assay System. However, no active promoter was found within −4,156 to +91 bp of the BTNL2 gene (data not showed). The G to A transition of rs2076530 led to the use of a cryptic splice site located 4 bp upstream of the affected wild-type donor site, which formed a premature stop in the spliced mRNA. The resulting protein lacked the C-terminal IgC domain and transmembrane helix, thereby disrupting the membrane localization of the protein. Evidence indicated that the G to A transition of rs2076530 was associated with the sarcoidosis [Valentonyte et al., 2005]. In contrast, transcript of the G allele could translate a complete protein with the normal immune regulation. However, it might be harmful to vaccine-induced immune response and the production of anti-HBs antibodies. Furthermore, because rs2076523 was in complete LD with rs3806156, functional and structural changes of the BTNL2 protein with the A to G transition of rs2076523 were discussed as follow: rs2076523 was located in the first IgC domain of the BTNL2 protein. The A to G transition of rs2076523 led to a missense mutation of the BTNL2 protein in position 196 (lysine to glutamic acid). K196E might not change the 3D structure of the BTNL2 binding domain (Supplementary Material and Fig. S2). Nevertheless, K196E might change the charged nature of the BTNL2 protein and the affinity of the BTNL2 protein and its unknown receptor, thereby affecting its regulation of T cells activation and the immune response to hepatitis B vaccination.
To determine whether or not comparison of several vaccination protocols in donors with various BTNL2 polymorphisms would increase significantly the value of aforementioned results. Comparing 3 hepatitis B vaccination protocols in donors with various BTNL2 polymorphisms, the results indicated that the recombinant yeast-derived hepatitis B vaccine (3 × 20 μg) compared with various BTNL2 polymorphisms in the study would increase significantly the value of aforementioned results. However, the recombinant CHO-derived hepatitis B vaccine (3 × 20 μg) and the recombinant yeast-derived hepatitis B vaccine (3 × 10 μg) compared with various BTNL2 polymorphisms in the study had all most no significant value (Table S4). Moreover, to understand if the association between the polymorphisms of the BTNL2 gene and the immune response to hepatitis B vaccination was resulted from the rs9277535 and rs3135363 that encompassed the HLA-DP, HLA-DR, and the BTNL2 gene and were associated with the immune response to hepatitis B vaccination in a genome-wide association study [Png et al., 2011]. Then, the relationship between rs9277535, rs3135363, and the BTNL2 gene was examined. The result showed that rs9277535 and rs3135363 were not in LD with any of the seven SNPs in the BTNL2 gene we genotyped in the study (Fig. 2). It was suggested that the association between the polymorphisms of the BTNL2 gene and the immune response to hepatitis B vaccination might not be contributed by the HLA-DP or HLA-DR, and that the BTNL2 gene might be a critical susceptibility gene of hepatitis B vaccine-induced immunity.
In summary, this study reveals that genetic variants in the BTNL2 gene are associated strongly with the immune response to hepatitis B vaccination in a Chinese Han population. Considering the negative immune regulation of the BTNL2 protein, the findings suggest that the BTNL2 protein may be critical for the immune response to hepatitis B vaccination and the low levels of anti-HBs (<10 mIU/ml). Furthermore, the findings may be helpful in identifying specific genes which could be used as targets in the development of novel and more effective vaccines for prevention of HBV infection.
ACKNOWLEDGMENTS
The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank all the individuals who participated in this study.




