Heterozygote of PLCE1 rs2274223 increases susceptibility to human papillomavirus infection in patients with esophageal carcinoma among the Kazakh populations
Abstract
The involvement of human papillomavirus (HPV) in the carcinogenesis of esophageal squamous carcinoma remains undetermined. However, three genome-wide association studies of esophageal cancer have identified a shared susceptibility locus at 10q23 (rs2274223: A5780G) in phospholipase C epsilon 1 (PLCE1). The current study aims to present a comprehensive and novel spectrum about the HPV genotype distribution of esophageal carcinoma in Kazakhs and assess its association with PLCE1 polymorphisms. The HPV genotypes in 183 patients with esophageal cancer and 89 controls selected from the Kazakh population were evaluated using the HPV gene chip. The PLCE1 rs2274223 variant was genotyped in esophageal carcinoma patients by MALDI-ToF Mass Spectrometry. The presence of seven HPV genotypes in esophageal carcinoma tissues—including HPV 16, 18, 35, 52, 6, 11, 43—was significantly higher at 31.7% than those in controls at 9.0% (P < 0.001). Such presence was strongly associated with increased risk of esophageal carcinoma (OR 4.70; 95% CI 2.13–10.36). Among all HPV genotypes detected, HPV16 was the most common genotype identified (29.0%, OR 4.13; 95% CI 1.87–9.13), which is significantly associated with well-differentiated esophageal carcinoma (P = 0.037). HPV-positive patients were generally younger than HPV-negative patients (70.1% vs. 29.3%, P = 0.013). PLCE1 rs2274223 genotypes AG and AG/GG were significantly associated with HPV-positive patients with esophageal carcinoma (OR 2.05, 95% CI 1.03–4.08 and OR 1.98, 95% CI 1.02–3.84, respectively). These findings suggest that heterozygote of PLCE1 rs2274223 increases susceptibility to HPV infection in patients with esophageal carcinoma among the Kazakh populations. J. Med. Virol. 86:608–617, 2014. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Esophageal cancer, one of the most aggressive malignancies originating in the gastrointestinal tract, results in more than 400,000 deaths annually. Esophageal cancer is the sixth leading causes of cancer death worldwide, with wide geographic, ethnic, and/or sociocultural variation [Parkin et al., 2005; Lambert and Hainaut, 2007]. Esophageal cancer comprises two major histological types: esophageal adenocarcinoma and esophageal squamous cell carcinoma. In China, esophageal squamous cell carcinoma is the predominant histological type of esophageal cancer [Kamangar et al., 2006a]. Comparing with other parts and ethnic groups of China, the Kazakh population, a nomadic tribe and mainly residing in the northwest part of Xinjiang Province, shows high incidence and mortality of esophageal squamous cell carcinoma, with an age-adjusted mortality of 90.7/100,000 [Zheng et al., 2010]. This rate is higher than that of any other ethnic groups. Despite epidemiological studies identifying excessive tobacco use, alcohol consumption [Holmes and Vaughan, 2007], micronutrient deficiency, and dietary carcinogen exposure [Wei et al., 2004] as cause of esophageal carcinoma prevalence, the accurate etiology of esophageal carcinoma remains undetermined.
Human papillomavirus (HPV), a nonenveloped double-stranded DNA virus with more than 90 genotypes, is a crucial tumor-related virus. HPV DNA has been detected in more than 90% of cervical cancer specimens [Abate et al., 2013]. Notably, several studies indicated that HPV infection was found in extragenital cancers, such as 30% of the human head and neck cancer [Gillison et al., 2000; Hansson et al., 2005]; however, the etiological implication of HPV in these malignancies remains controversial [Castillo et al., 2006]. The association between HPV and esophageal carcinoma was first reported by Syrjanen et al. [1982]. Subsequent studies conducted in different geographic areas and ethnic groups, such as Han ethnic groups in eastern China (Linxian and Anyang in Henan Province, Cixian in Hebei, Yangcheng in Shanxi, and the northern Jiangsu province) and Kazakh minority residing in the northwest part of China, widely vary in HPV, have shown large variations in the prevalence of HPV among esophageal carcinoma patients, ranging from 0% to 100% [Antonsson et al., 2010; Guo et al., 2012; Hu et al., 2012b; Liu et al., 2013a], resulting in contrasting views regarding the influence of HPV in the development of esophageal carcinoma. The discrepancies may be generally attributed to the small size of the samples, differences in immunological and molecular methods, as well as inter-laboratory variability in sample collection and handling [Hubbard, 2003; Kamangar et al., 2006b]. In previous studies, HPV infection rate in esophageal carcinoma in Kazakh patients varied from 18.6% to 40% [Hu et al., 2012b; Liu et al., 2013b]. However, there studies mainly focused on the high-risk HPV genotypes, such as HPV16 and 18. Studies on the involvement other HPV genotypes infection in the carcinogenesis of esophageal carcinoma in Kazakh patients are rarely reported.
In addition to infectious agents, either direct carcinogens or promoters, genetic susceptibility contributes to the high incidence of esophageal carcinoma, as suggested by a strong tendency toward familial aggregation of esophageal carcinoma in high-risk areas [Guohong et al., 2010]. Three genome-wide association studies (GWAS) reported recently on a shared susceptible locus at 10q23 (rs2274223: A5780G, exon 26) in the phospholipase C epsilon 1 (PLCE1) gene that is associated with risk of esophageal cancer [Abnet et al., 2010; Wang et al., 2010; Wu et al., 2011]. This finding was confirmed in a related study among the Chinese Han population [Gu et al., 2012; Hu et al., 2012a; Cui et al., 2013]. More importantly, the overexpression of PLCE1 was involved in metastasis and aggressiveness of esophageal carcinoma in ethnic Kazakh patients from our previous research [Chen et al., 2013]. PLCE1, a unique member of the phospoholipase C family of proteins, contains Ras-binding domains for small G-proteins of the Ras family. PLCE1 acts as an effector of GTPases and influences cell growth regulation, differentiation, apoptosis and angiogenesis [Bourguignon et al., 2006]. PLCE1 significantly affects carcinogenesis and progression of a series of human cancers, such as cancers of the intestine, skin, bladder, colorectal, and head and neck [Bai et al., 2004; Cheng et al., 2011; Ling et al., 2011]. Especially, PLCE1 significantly affects skin chemical carcinogenesis by augmenting TPA-induced inflammation [Bai et al., 2004; Hu et al., 2010] and promotes intestinal tumorigenesis of ApcMin/+ mice by inflammation augmentation and angiogenesis [Li et al., 2009]. Inflammatory esophageal disease increases the risk of developing cancer [Mandard et al., 2000]; thus, the occurrence and progression of esophageal carcinoma can be regulated by their local environment, where inflammation possibly induced by PLCE1 is a critical constituent [Gu et al., 2012].
HPV induced inflammation may also significantly affect the immune response to infection, such as prevention and clearing out the initial infections. Dysregulation of immune response increases the persistence of HPV infection, thereby promoting lesion progression and HPV related neoplasia. Given the involvement of PLCE1 in inflammation, rs2274223 causes a change in amino acid from histidine to arginine in calcium-dependent lipid-binding (C2) domain of the PLCE1 protein. The C2 domain is the core catalytic domain that contributes to intracellular signaling. The effect of the novel PLCE1 variant rs2274223 in the involvement of HPV in esophageal tissues needs further investigation.
Therefore, in the present study, the comprehensive and novel spectrum of the HPV genotype distribution, as well as the clinicopathologic features of patients with HPV genotypes, was evaluated in Kazakh patient with esophageal carcinoma from the Yili, an area with high incidence of esophageal carcinoma. The HPV DNA gene chip used in the evaluation effectively detects as many as 23 different HPV genotypes and exhibits higher sensitivity to paraffin-embed tissue than do other traditional HPV detection methods. Additionally, the putatively functional PLCE1 rs2274223 polymorphisms were genotyped in the esophageal carcinoma patients by the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-ToF–MS). Subsequently, a case-to-case comparison based on the genotyping results was conducted to address the possible function of the rs2274223 variant in the involvement of HPV in Kazakh patients with esophageal cancer.
MATERIALS AND METHODS
Patients and Specimens
A total of 183 esophageal carcinoma specimens in formalin-fixed paraffin-embedded archival tissues were collected from the Xinjiang Yili Prefecture Friendship Hospital and the People's Hospital of Xinjiang Uygur Autonomous Region located in Xinjiang Province, northwestern China with between May 2011 and January 2012. No patients had received chemo- and radiotherapy before endoscopies and surgery. All samples were surgically resected and fixed in 10% buffered-formalin, routinely processed, and paraffin-embedded. The following parameters were evaluated according to the World Health Organization histological classification criteria: anatomic site (upper, middle, or lower esophagus); histological grade (well-differentiated, moderately differentiated, or poorly differentiated); depth of invasion; and lymph node metastases, Tumor-node-metastasis stages were evaluated according to Cancer Stage Manual 7th Edition 2009 issued in 2009 by the American Joint Committee on Cancer. Biopsy samples of normal esophageal squamous epithelium were available from 89 control patients who participated in the project of Early Diagnosis and Treatment of Esophageal Cancer in Xinjiang Province. Each control patient had undergone upper gastrointestinal endoscopy, and the esophageal tissue samples were confirmed as histologically normal. Informed consent of each patient was obtained, and study protocol was approved by the Institutional Review Board at Shihezi University School of Medicine.
The various clinic-pathological characteristics of Kazakh patients with esophageal carcinoma and controls were investigated as followed: The mean age for the 183 esophageal carcinoma patients was 59.38 ± 7.71 years (mean ± SD), and 36% of the patients were female. The mean age for the 89 esophageal tissue controls was 58.98 ± 6.93 years (mean ± SD), and 34% of the controls were female. No difference in age (P = 0.29) and gender (P = 0.79) was found. Histopathologically, the cases included 28 (15.3%) well-differentiated patients (group G1), 113 (61.7%) moderately differentiated patients (G2), and 42 (23.0%) poorly differentiated patients (G3), respectively. Other salient clinicopathologic features were also evaluated, including the tumor location, invasion depth, lymphatic invasion, and tumor-node-metastasis stage.
DNA Extraction and PCR Amplification
The formalin-fixed paraffin-embedded samples were cut into 10 µm slices, and four sections per sample were collected in microcentrifuge tubes according to the previously described method [Greer et al., 1994]. Genomic DNA was isolated using standard proteinase K digestion and a tissue DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. As an internal control, all purified genomic DNA samples were successfully tested by polymerase chain reaction (PCR) with a human β-actin primer set (forward: 5′-CAGACACCATGGTGCACCTGAC-3′ and reverse: 5′-CCAATAGGCAGAGAGAGTCAGTG-3′), indicating that the quality and quantity of DNA were suitable for detecting the presence of HPV. PCR analysis was then performed using a modified HPV MY09 and MY11 biotinylated consensus primer set (MY09: 5′-biotin-CGTCCMARRGGAWACTGATC-3′, MY11: 5′-biotin-GCMCAGGGWCATAAYAATGG-3′ [M = A/C, W = A/T, Y = C/T, R = A/G]) and β-actin biotinylated primer set (forward: 5′-CTTAGTTGCGTTACACCCTT-3′ and reverse: 5′-GTCACCTTCACCGTTCC-3′) for amplification of HPV and β-actin, respectively. The MY09/11 primer set was used to amplify a fragment of about 150 bp in the L1 open reading frame of the HPV. The quality of specimen DNA was validated by amplification of a 155 bp fragment of β-actin as internal control.
HPV Typing Using HPV Gene Chip
HPV genotyping of the PCR products of HPV and β-actin was conducted using the commercial HPV genotyping gene chip (Yaneng, Shenzhen, China). The HPV gene chip contains 23 type-specific probes, including the high risk types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, 82, 83) and the low risk type (HPV6, 11, 42, 43, and 81), as well as an internal control. The chip membrane was hybridized with 5–6 ml hybridization buffer A[2× SSC contained 0.15 M NaCl and 0.015 M sodium citrate, 0.1% sodium dodecyl sulfate (SDS), pH 7.4] and the denatured amplicons (HPV and β-actin PCR product) at 51°C for 1.5 hr. The membrane was washed in washing buffer B (0.5× SSC, 0.1% SDS, pH 7.4) for 5 min at 51°C and washing buffer (peroxides isozyme) at room temperature for 30 min and then washed twice in hybridization buffer A for 5 min by shaking. The membrane was then washed in washing buffer C (0.1 mol/L sodium citrate, pH 5). NBT/BCIP (5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium) was subsequently added and incubated for 30 min at 25°C. The reaction was stopped by aspiration of the substrate solution and addition of distilled water. The HPV types displayed on the chip were determined by a standard visual assessment protocol (Fig. 1a).
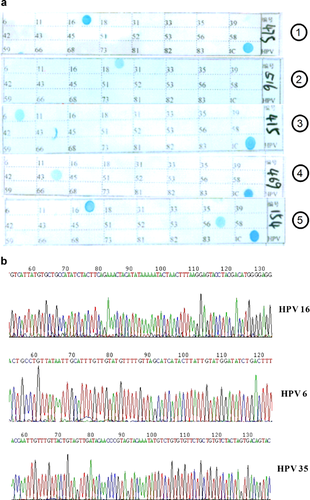
To confirm the accuracy of the HPV genotyping by the HPV gene chip, 55 samples were tested by the HPV gene chip as singer HPV genotype infection were performed by direct sequencing of PCR products. The DNA sequence was then identified by NCBI Blast (Fig. 1b). A 100% match was identified between the results of DNA sequence displayed in the Genbank and HPV genotype detected by the HPV gene chip.
Genotyping of PLCE1 rs2274223 Variant by MALDI-ToF-MS
PLCE1 rs2274223 genotyping was performed on the esophageal carcinoma patients who had undergone HPV genotyping. SNP genotyping was done using the Sequenom MassARRAY platform (Sequenom, San Diego, CA) by MALDI-ToF–MS at Key Laboratories for Xinjiang Endemic and Ethnic Diseases (Shihezi University School of Medicine), as described previously [Schaeffeler et al., 2008]. Primers were designed using the proprietary software Assay Design 3.1 provided by Sequenom, Inc. and synthesized by Invitrogen. The primer sequences were as follows: (forward: 5′-ACGTTGGATGTCCACAACTGCAAAACGAAG-3′, reverse: 5′-ACGTTGGATGTCCATCGAAACACCCTGAAC-3′, extend: 5′-tgccTACAAGATCTTCGAAGTGAACG-3′). Genotype calling was analyzed using Mass-ARRAY Typer software version 4.0 (Sequenom). For repeated genotyping, 10% of the samples were randomly selected, and the results were 99.95% concordant. A total of 169 Kazakh patients with esophageal cancer cases were included in the final analysis after data cleaning.
Statistical Analysis
All statistical analyses were performed using SPSS version13.0, U.S. The presence of HPV DNA in different groups was analyzed by chi-square test. Correlation between HPV-16 infection and the clinical characteristics of esophageal carcinoma patients, as well as that between HPV 6 infection and the clinical characteristics of the patients, was analyzed by Fisher's exact test or Spearman correlation analysis as appropriate. The association of HPV positivity in esophageal cancer patients with genotypes of PLCE1 rs2274223 polymorphisms was estimated by computing the odds ratios (ORs) and 95% confidence intervals (95% CIs). Both univariate and multivariable logistic regression models were performed for the analyses. The multivariable logistic regression models were fully adjusted with age, sex, smoking status, and alcohol intake. All statistical analyses were two-sided, and P < 0.05 was considered statistically significant.
RESULTS
Prevalence of HPV Infection in Esophageal Carcinoma Patients
HPV DNA gene-chip testing evaluated the prevalence of HPV in 183 patients with esophageal carcinoma and 89 controls, and the results are shown in Table I and Figure 2. The prevalence of HPV DNA was higher in esophageal carcinoma cases than that in the control group (31.7% vs. 9.0%, P < 0.001). Seven HPV genotypes were identified in both esophageal cancer tissue and normal esophageal tissue, including HPV16, 18, 6, 11, 43, 35, and 52. HPV DNA was identified in 58 of 183 tumor tissue samples. Six esophageal tumors showed double-infection with other high-risk/low-risk HPV types, including four coinfections with HPV-16 and 6, 1 coinfection with HPV-16 and 35, and 1 coinfection with HPV-6 and 52. HPV-16 (29.0% in cases vs. 9.0% in controls) was the predominant high risk HPV type identified in all samples. HPV-6 (5.0%) was the predominant low risk HPV type identified in tumor cases. No low risk HPV DNA was identified in controls group.
| Types | Cases (n = 183), n (%)a | Control (n = 89), n (%) | χ2 | OR (95% CI) |
|---|---|---|---|---|
| High risk | 56 (30.6) | 8 (9.0) | 15.54*** | 4.46 (2.03–9.85) |
| HPV16 | 53 (29.0) | 8 (9.0) | 13.73*** | 4.13 (1.87–9.13) |
| HPV18 | 1 (0.5) | |||
| HPV35 | 1 (0.5) | |||
| HPV52 | 1 (0.5) | |||
| Low risk | 8 (4.4) | 0 (0.0) | 4.009* | 1.51 (1.38–1.64) |
| HPV6 | 6 (3.3) | |||
| HPV11 | 1 (0.5) | |||
| HPV43 | 1 (0.5) | |||
| Total HPV positive | 58 (31.7) | 8 (9.0) | 16.80*** | 4.70 (2.13–10.36) |
- a The total number of HPV positives was smaller than the total number of HPV genetypes positives due to the multiple infections. Four coinfections with HPV-16 and 6, 1 coinfection with HPV-16 and 35, and 1 coinfection with HPV-6 and 52 were confirmed in cases.
- * P < 0.05.
- *** P < 0.001.
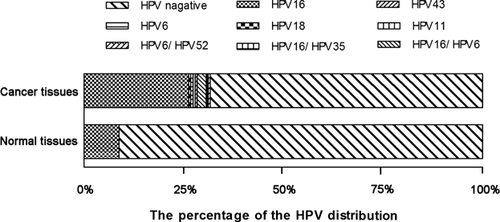
By using the unconditional logistic regression models, the association between HPV infection and Kazakh patients with esophageal cancer was evaluated. Detection of high risk HPV DNA (OR 4.46; 95% CI 2.03–9.85), including the HPV-16 (OR 4.13; 95% CI 1.87–9.13), exhibited a strong positive association with the risk of esophageal carcinoma in Kazakhs. However, a relationship between low-risk HPV 6 (OR 1.51; 95% CI 1.38–1.64) and esophageal carcinoma was also indicated. A higher OR value (OR 4.70; 95% CI 2.13–10.36) was observed in patients who tested positive for HPV DNA.
Association Between HPV Infection and Clinico-Pathological Characteristics of Patients With Esophageal Carcinoma
To identify the cofactors that increase the risk of HPV infection in Kazakh patients with esophageal carcinoma, the association of HPV infection with various clinicopathological and demographic characteristics was examined (Table II). HPV infection was mostly identified in younger patients (41 of 58 [70.7%] vs. 17 of 58 [29.3%], P < 0.05). More than 70% of HPV infection was found in patients with poorly differentiated or moderately differentiated esophageal carcinoma. The difference between well-differentiated, moderately differentiated, and poorly differentiated esophageal carcinoma patients was of marginal statistical significance (P = 0.069). No significant difference in clinicopathological characteristics was indicated between the HPV-positive esophageal cancer patients and the HPV-negative group.
| Characteristics | N° | HPV+ patientsa (n = 58), n (%) | HPV− patientsa (n = 125), n (%) | P-value |
|---|---|---|---|---|
| Age (years) | 0.013 | |||
| <60 | 105 (57.4) | 41 (70.7) | 64 (51.2) | |
| ≥60 | 78 (42.6) | 17 (29.3) | 61 (48.8) | |
| Gender | 0.308 | |||
| Male | 117 (63.9) | 34 (58.6) | 83 (66.4) | |
| Female | 66 (36.1) | 24 (41.4) | 42 (33.6) | |
| Location of the tumor | 0.579 | |||
| Upper + middle | 74 (68.5) | 16 (64.0) | 58 (69.9) | |
| Lower | 34 (31.5) | 9 (36.0) | 25 (30.1) | |
| Histologic grade (G) | 0.069 | |||
| G1 | 28 (15.3) | 13 (22.4) | 15 (12.0) | |
| G2/G3 | 155 (84.7) | 45 (77.6) | 110 (88.0) | |
| Invasion depth | 0.356b | |||
| T1 | 2 (1.1) | 1 (1.7) | 1 (0.9) | |
| T2 | 51 (28.8) | 21 (35.6) | 30 (25.4) | |
| T3 | 121 (68.4) | 36 (61.0) | 85 (72.0) | |
| T4 | 3 (1.6) | 1 (1.7) | 2 (1.7) | |
| Lymphatic invasion | 0.551 | |||
| Negative | 87 (55.4) | 30 (58.8) | 57 (53.8) | |
| Positive | 70 (44.6) | 21 (41.2) | 49 (46.2) | |
| Tumor-node-metastasis stage | 0.584b | |||
| I | 8 (6.5) | 4 (11.1) | 4 (4.6) | |
| II | 80 (65.0) | 22 (61.1) | 58 (66.7) | |
| III | 29 (23.6) | 8 (22.2) | 21 (24.1) | |
| IV | 6 (4.9) | 2 (5.6) | 4 (4.6) |
- a Numbers may not sum to total due to missing data.
- b Fisher's exact test.
Given that the majority of HPV genotypes detected were HPV 16 and HPV 6, the influence of HPV-16 or HPV-6 on esophageal malignant progression was determined by examining the clinical characteristics of patients with esophageal cancer. As shown in Table III, HPV-16 infection was detected in 46.4% of well-differentiated tumor tissues exhibited a low positive correlation with histologic grade of esophageal carcinoma (P = 0.037). Similarly, double-infection with HPV genomes was more frequent in well differentiated tissues than in moderately differentiated tissues (G1:14.3% vs. G2:1.8%). The observed association was statistically significant according to Fisher' exact test (P = 0.009). No relationship was found between invasion depth and lymph-node metastasis.
| Characteristics | No. | Single infection | Double infectiond | ||||
|---|---|---|---|---|---|---|---|
| HPV-16a, n (%) | P-valueb | HPV-6a n (%) | P-valueb | No = 6 | P-valueb | ||
| Histologic grade (G) | 0.037 | 0.118c | 0.009c | ||||
| G1 | 28 | 13 (46.4) | 3 (10.7) | 4 (14.3) | |||
| G2 | 113 | 27 (23.9) | 3 (2.7) | 2 (1.8) | |||
| G3 | 42 | 9 (21.4) | 2 (4.7) | 0 | |||
| Invasion depth | 0.140 | 0.759 | 0.269 | ||||
| T1 | 2 | 1 (50.0) | 0 | 0 | |||
| T2 | 51 | 18 (35.3) | 3 (5.9) | 3 (5.9) | |||
| T3 | 121 | 30 (24.8) | 5 (4.1) | 2 (1.7) | |||
| T4 | 3 | 2 (66.7) | 0 | 0 | |||
| Lymphatic invasion | 0.113 | 0.243c | 0.408c | ||||
| Negative | 70 | 25 (35.7) | 5 (7.1) | 4 (5.7) | |||
| Positive | 87 | 21 (24.1) | 2 (2.3) | 2 (2.3) | |||
- a Indicates positive on HPV DNA chip testing.
- b P-values for the correlation between HPV-16 or/and HPV6 infection and clinical characteristics of patients with oesophageal squamous carcinoma were calculated by Spearman correlation or Fisher exact test as appropriate.
- c Fisher's exact test.
- d The co-infection contained that four coinfections with HPV-16 and 6, 1 coinfection with HPV-16 and 35, and 1 coinfection with HPV-6 and 52.
Relation of HPV Positivity of Kazakh Patients With Esophageal Carcinoma to PLCE1 Rs2274223 Genotype
MassARRAY genotyping technology was performed on 183 DNA samples from primary Kazakh patients with esophageal cancer to analyze the association between the rs2274223 polymorphism and HPV-associated Kazakh patients with esophageal cancer. The results of PLCE1 rs2274223 genotype distributions and allele frequencies in HPV-positive and HPV-negative patients are summarized in Table IV. The rs2274223 G allele was significantly more common in HPV-positive patients (33.7%) than that in HPV-negative patients (24.8%), suggesting that the G alleles may be associated with HPV-positivity in Kazakh patients with esophageal cancer. Unlike the wild-type AA homozygote, the combined AG/GG or AG variant genotypes were significantly associated with HPV-positive with esophageal cancer patients (OR 1.98, 95% CI 1.02–3.84; and OR 2.05, 95% CI 1.03–4.08, respectively), although the homozygous mutant type of PLCE1 rs2274223 (G/G) exhibited no association with HPV-positive esophageal cancer patients (OR 1.60, 95% CI 0.44–5.83).
| Genotype | HPV+ patients (n = 52) | HPV− patients (n = 117) | Adjusted ORb |
|---|---|---|---|
| n (%) | n (%) | 95% CI | |
| PLCE1 rs2274223 | |||
| AAa | 21 (40.4) | 67 (57.3) | 1.0 (reference) |
| AG | 27 (51.9) | 42 (35.9) | 2.05 (1.03–4.08) |
| GG | 4 (7.7) | 8 (6.8) | 1.60 (0.44–5.83) |
| Combined variant genotypes | |||
| AG/GG | 31 (59.6) | 50 (42.7) | 1.98 (1.02–3.84) |
- a Reference group.
- b Adjusted for age, sex, smoking, and alcohol use in a logistic regression model.
DISCUSSION
The HPV DNA gene chip method was adopted to assess the HPV prevalence in the 183 Kazakh patients with esophageal cancer and 89 cancer-free controls obtained from several hospitals in the Ili Kazakh Autonomous Prefecture, an area with high esophageal cancer incidence located in northwestern China. The distribution of PLCE1 rs2274223 was also detected in the esophageal cancer specimens. The aim of the present study was to explore the association of HPV infection with PLCE1 rs2274223 polymorphism in esophageal squamous carcinoma in Kazakh patients, which could influence the carcinogenesis of esophageal carcinoma.
The significantly high prevalence of HPV (31.7%) in the Kazakh patients with esophageal carcinoma indicated that HPV infection significantly affect the carcinogenesis of the esophageal carcinoma in Kazakh population. In addition, the increase in risk of HPV-associated esophageal carcinoma was significantly greater in Kazakh with PLCE1 rs2274223 variant genotypes (AG) than those with the AA genotype. This finding suggests that heterozygotes of PLCE1 rs2274223 genotypes are risk genotypes for HPV-associated esophageal carcinoma. PLCE1 polymorphism may indicate genetic susceptibility to HPV-associated esophageal cancer.
The study is the first to use the HPV DNA gene chip to test HPV genotype prevalence in a large sample size obtained from the Kazakh population and its association with PLCE1 polymorphism. The involvement of HPV in etiology of esophageal carcinoma [Syrjanen et al., 1982] has drawn interest in the past three decades following the initial suggestion of the phenomenon in 1982 [Chang et al., 2000; Peixoto Guimaraes et al., 2001; Kamangar et al., 2006b; Guo et al., 2012; Hu et al., 2012b]. However, the incidence of HPV infection in esophageal cancer varies from the high-incidence to the low-incidence areas [Chang et al., 2000; Li et al., 2001; Peixoto Guimaraes et al., 2001; Kamangar et al., 2006a; Antonsson et al., 2010; Zheng et al., 2010; Goto et al., 2011; Hu et al., 2012b; Feng et al., 2013], unlike that of HPV infection in cervical carcinoma. This finding suggests that HPV infection exerts a complex effect on carcinogenesis of the esophageal carcinoma.
In a previous study, a PCR analysis detected only HPV 16 in esophageal specimens, and the incidence of esophageal carcinoma in Kazakh was found to be significantly higher (41%) than in the control group (14%) [Hu et al., 2012b]. Another study investigated the viral load of HPV in Kazakh patients with esophageal carcinoma and found that 18.6% of the 102 patients had HPV infections [Liu et al., 2012]. However, in the present study, HPV infection rates in Kazakh patients with esophageal carcinoma were higher than those obtained from the same region and people of the same ethnicity. On the basis of the INNO-LiPA HPV genotyping assay, the study of 67 Kazakh patients with primary esophageal carcinoma also revealed that the presence of HPV types in Xinjiang Kazakhs patients was 21% [Lu et al., 2008]. HPV was detected in only 9% of the normal esophageal tissue. This increased rate determined in the present study should be attributed to heterogeneity, especially that caused by genetic variation, in the large number of cases (183 Kazakh patients with esophageal squamous carcinoma), including the use of HPV gene chip, which exhibited a higher sensitivity compared with that of other traditional method of HPV detection in paraffin embed tissue and capability to identify as many as 23 different HPV genotypes. The frequency of HPV infection in the tumors observed in the current study is similar to the average rate from a systematic review and a formal meta-analysis of the literature on HPV detection in 10,234 esophageal carcinoma cases from 1954 to 2012 [Syrjanen, 2013]. The HPV genotype distribution in esophageal carcinoma that was found in the present study was consistent with the observations that the most predominant type of HPV associated with esophageal carcinoma is HPV-16, followed by the low risk type HPV6. Other HPV types, such as HPV18, 35, 52, 11, and 43, are rarely associated with esophageal carcinoma [Liu et al., 2012]. Double HPV infections were also identified in the current study population, which shows consistency with other reports on esophageal carcinoma in other areas of China [Li et al., 2001].
The data confirmed a significant association between the presence of HPV 16, particularly in double infection, and well-differentiated esophageal carcinoma, which is consistent with the similar study [Castillo et al., 2006]. Well-differentiated tumors tended to have either HPV16 or HPV16/6 infection, suggesting that HPV can initiate the development of esophageal carcinoma and explaining the higher survival rate indicated in HPV-positive patients with esophageal carcinoma. Other studies have shown that HPV infection more frequently occurs in well-differentiated head and neck squamous cell carcinoma [Fakhry and Gillison, 2006], which influence the treatment outcome [Datta et al., 2006]. Cisplatin sensitivity was also shown to increase in HPV 16 transfected ovarian cancer cells [Pestell et al., 2000]. Nevertheless, other study reported that a positive HPV16/18 rate and viral load were more often in poorly differentiated esophageal carcinoma cases, although no significant correlation was confirmed [Liu et al., 2012]. The discrepant results in the literature may be attributed to the difference in detection methods, sample size, and patient heterogeneity because of the variety of ethnicities in the sample although the samples cases were obtained from the same region. Thus, the conclusions concerning the influence of HPV on various differentiations of esophageal carcinoma warrant confirmation. In addition, patients with HPV-positive esophageal carcinoma were younger than their HPV-negative counterparts, which were in accordance with other findings [Antonsson et al., 2010].
Epidemiologic and molecular studies have shown that all virus agents and the genetic factors are involved in the initiation and progression of esophageal carcinoma [Hu et al., 2012b]. Another important finding in the present study is that the heterozygote of PLCE1 rs2274223 (AG) genotypes were associated with a twofold higher risk for the development of HPV-associated esophageal carcinoma, whereas the GG genotype was not associated with the risk of developing HPV-associated esophageal carcinoma. This finding suggests that the PLCE1 rs2274223 polymorphism interacts with HPV in Kazakh patients with esophageal carcinoma, thereby increasing susceptibility to HPV infection. Two carcinogenic mechanisms explain why PLCE1 polymorphism and HPV may be cofactors in the etiology of HPV-associated esophageal carcinoma. First, the p53 codon 72 arginine genotype was reported as a high-risk factor in the development of HPV-associated cervical carcinoma [Storey et al., 1998] because the arginine allele was more susceptible to HPV-E6 protein degradation, compared with the proline allele. Consequently, individuals who are arginine homozygotes for the codon 72 polymorphism were more likely to develop esophageal carcinoma [Kawaguchi et al., 2000]. The p53 protein, as a tumor suppressor, can initiate cell cycle arrest, thereby promoting DNA repair, triggering apoptosis, as well as inducing growth arrest and senescence. Similarly, PLCE1, a key mediator integrating multiple signaling pathways, is involved in regulating cell growth, differentiation, apoptosis and angiogenesis [Bourguignon et al., 2006]. The rs2274223 variants in the PLCE1 causes an amino acid change from histidine to arginine in the PLCE1 protein calcium-dependent lipid-binding (C2) domain. Therefore, arginine can influence HPV involvement in esophageal carcinoma [Abnet et al., 2010; Wang et al., 2010; Wu et al., 2011]. However, further investigation need to be conducted to determine whether an exact interaction between PLCE1 and HPV. Second, several studies have reported that PLCE1 influences skin chemical carcinogenesis by inflammation [Bai et al., 2004; Hu et al., 2010], and inflammatory esophageal disease increases the risk of cancer development [Mandard et al., 2000]. Thus, persistent HPV-induced inflammation can significantly affect the ability of the immune response to infections. The PLCE1 rs2274223 heterozygote may contribute to dysregulated immune response to infection, thereby promoting lesion progression and HPV related neoplasia. These findings need to be validated in further studies with larger samples although no studies have been conducted on the associations between the PLCE1rs2274223 polymorphism and HPV-associated esophageal carcinoma.
In conclusion, the present study demonstrated that the heterozygote of PLCE1 rs2274223 increased susceptibility to HPV infection in patients with esophageal squamous cell carcinoma among the Kazakh populations. This finding can be used for identifying potential approaches to prevention and controlling the incidence of esophageal carcinoma in Kazakh population. However, given that the mechanisms by which PLCE1 polymorphisms may influence the course of HPV infections remain under investigation, further studies need to be conducted to elucidate the influence of PLCE1 polymorphisms in HPV-associated esophageal carcinoma in the Kazakh population.




