Epidemiological and molecular analyses of a non-seasonal outbreak of acute icteric hepatitis E in Bangladesh
Abstract
Acute hepatitis due to hepatitis E virus (HEV) is endemic in Bangladesh, but its epidemiological characteristics and virological features remain obscure. An outbreak of acute icteric hepatitis E occurred in Rajshahi, Bangladesh during 2010 when 200 patients with visible jaundice visited physicians within a period of 1 month (January–February). Clinical and epidemiological data were collected from these patients using questionnaires. Nucleic acids were isolated from 15 patients who were selected at random to ascertain their HEV genotypes. Near-complete nucleotide sequences of the HEV genome were detected in two patients and partial ORF2 regions in the other 13 patients. All patients tested positive for IgM antibodies to HEV but negative for other hepatitis viruses. Most patients were icteric and complained of vomiting, fever, itching, and abdominal pain. All 15 HEV sequences formed a single cluster within genotype 1a. Two of the 7,186-nt HEV sequences were 99.8% identical. This is the first study to report the clinical, epidemiological, and molecular characterization of an outbreak of acute hepatitis E in Bangladesh. J. Med. Virol. 85:1369–1376, 2013. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Viral hepatitis caused by the hepatitis E virus (HEV) is mainly a waterborne infection in most of the developing countries in Asia and Africa [Balayan et al., 1983; Ray et al., 1991; Ticehurst et al., 1992; Shrestha et al., 2003]. Bangladesh, a Southeast Asian country with a population of 160 million, does not have a safe water supply, hygienic disposal of sewage, or the correct reporting of communicable diseases. Approximately 25% of the rural residents of Bangladesh express IgG antibodies to HEV in their sera [Labrique et al., 2009]. It has been reported that HEV is the main etiological agent of acute hepatitis [Al-Mahtab et al., 2009] and the single major cause of fulminant hepatitis [Sheikh et al., 2002]. However, there is a paucity of information about the epidemiological characteristics, clinical features, and genomic analysis of acute hepatitis due to HEV in Bangladesh.
An outbreak of HEV-induced acute icteric hepatitis was reported during January and February 2010 in Rajshahi, a Northern district of Bangladesh. Two hundred patients with acute jaundice visited physicians during this period. This is not a usual phenomenon in Bangladesh because endemic or epidemic outbreaks of acute hepatitis E are usually detected during the rainy season in August and September. No outbreaks of HEV were detected in any other regions of Bangladesh at that time. The epidemiology of this outbreak of sporadic acute hepatitis E was investigated by developing and administering a questionnaire to analyze clinical data. The sera from 15 patients with acute hepatitis E were also selected at random to determine their HEV genotypes. Finally, the almost complete nucleotide (nt) sequences of the HEV genomes from two patients with acute hepatitis E that had the highest levels of HEV RNA were determined and a phylogenetic tree was constructed.
MATERIALS AND METHODS
A total of 200 patients with acute icteric hepatitis E visited physicians in Rajshahi over a period of 1 month (January 2010–February 2010). All patients were positive for IgM antibodies to HEV (IgM anti-HEV) but negative for IgM type antibodies to hepatitis A virus (HAV), hepatitis B virus (HBV), and hepatitis C virus (HCV). Informed consent was obtained from all patients after explaining the nature and purpose of the study. The study protocol was also approved by the review board at the Viral Hepatitis Foundation of Bangladesh, Dhaka, Bangladesh. A questionnaire was prepared for patients to collect information about their age, gender, occupation, source of drinking water, and place of habitation (rural or urban). Additional information was obtained about each patient's history of anorexia, vomiting, diarrhea, pruritis, abdominal pain, and weight loss.
Assessment of Serological and Biochemical Markers
IgM antibodies to HEV, HAV, HBV, and HCV were measured using commercial kits (Atlas Medical, Cambridge, UK). The levels of alanine aminotransferase (ALT) and bilirubin in the sera were measured using commercial kits. The hepatitis B surface antigen (HBsAg) and antibodies to HCV (anti-HCV) were measured using an enzyme-linked immunosorbent assay. Ultrasonography was performed in all patients to assess whether they were suffering from chronic liver diseases or complications, such as liver cirrhosis or hepatocellular carcinoma.
RNA Extraction, cDNA Synthesis, Amplification, and Sequencing
Isolation of RNA, genotyping of HEV, and near-complete nt sequencing of HEV genomes were performed according to previously described methods, with some modifications [Takahashi et al., 2009]. Nucleic acids were extracted from sera using a QIAamp MinElute Virus Spin Kit (Qiagen GmbH, Hilden, Germany). HEV RNA genomes were reverse transcribed and cDNA was amplified using PCR with primers specific for 23 overlapping regions in the HEV genome (Table I). The reverse transcription and first-round PCR were conducted using a PrimeScript II High Fidelity One Step RT-PCR Kit (Takara Bio, Inc., Shiga, Japan), while the second-round PCR was conducted using PrimeSTAR GXL DNA Polymerase (Takara Bio, Inc.). A 3′-Full RACE Core set (Takara Bio, Inc.) was used to amplify the core 3′ sequences. The final products were sequenced using a 3,100 DNA sequencer with a BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Genetic analyses of HEV sequences were conducted using the neighbor-joining method with Genetyx-Mac Version 13 (Genetyx Corporation, Tokyo, Japan).
| Primers | Stage-polarity | Nucleotide sequence (5′–3′) |
|---|---|---|
| Primer set 1 | 1st sense | GCAGACCACRTATGTGKTCG |
| 2nd sense | GCAGACCACRTATGTGKTCG | |
| 1st antisense | TGGAGGGCAGCRTAAAGCCG | |
| 2nd antisense | GTCATRCCATGGCGGAACAT | |
| Primer set 2 | 1st sense | CCTAATGTGGTCCACCGCTG |
| 2nd sense | GACCTCGGTRGACCGGGGGTA | |
| 1st antisense | GTTCAGCGYTGGTATACTGC | |
| 2nd antisense | TGGCTCCGGGGCKGCCGTGAG | |
| Primer set 3 | 1st sense | GCAGGCGYGTTGTGGTGACG |
| 2nd sense | CATCCCCTTRGATATAGCCTG | |
| 1st antisense | GAGGGTGAYACYAGTGCTGG | |
| 2nd antisense | GCGGCAGTRATGACAGCTGT | |
| Primer set 4 | 1st sense | ACCTCATGCTCCACTAAGTC |
| 2nd sense | TCCTGRCCMAGCCACTTCAT | |
| 1st antisense | GTCCCTGYCCATATYTGGGA | |
| 2nd antisense | CARTGGCAGGGGGCCGACTC | |
| Primer set 5 | 1st sense | CTTYGAGAAGTCCGGCCGTGA |
| 2nd sense | GTCTCRCARTCGATCCGCCC | |
| 1st antisense | GCYCAGTGYAGGCGCTGGCT | |
| 2nd antisense | TTTACTGTGGCGGTCAGCCG | |
| Primer set 6 | 1st sense | GCCTATGAGGGGTCYGATGT |
| 2nd sense | TTAGCCGACTCCCARACATG | |
| 1st antisense | AGTGACATMTCTGGGTCCTA | |
| 2nd antisense | AGYCCCTCAGGRTGGAACCA | |
| Primer set 7 | 1st sense | AGTCAGAGCACTATGGCCGC |
| 2nd sense | CGACTCRAACAGCGAGCCGGC | |
| 1st antisense | CGCTATGTYGCTGCCGGGCT | |
| 2nd antisense | CCRYCTGGTGGGTTATGGCC | |
| Primer set 8 | 1st sense | GAGAGCACACTYTACACCCG |
| 2nd sense | TATATGCCGGTCCCGAGGAG | |
| 1st antisense | ACTTGGTCGGARGTTGATGC | |
| 2nd antisense | CTTTGGGTTATGTTCCAACCTATA | |
| Primer set 9 | 1st sense | TAATGTTGACCAYCGCCCTGG |
| 2nd sense | GGCACACCTGCRGTAAACTG | |
| 1st antisense | TGCTGCCTCTTTTGTGATGC | |
| 2nd antisense | CCCGGCCGACRTCTGTGGC | |
| Primer set 10 | 1st sense | TTTGACGCCTGGGAGCGGAA |
| 2nd sense | TGATGGCGGGRACGAGCCC | |
| 1st antisense | CARATGGTTYGAGGCCAATA | |
| 2nd antisense | ACGGTGGCGGCGCGCTGCAT | |
| Primer set 11 | 1st sense | GGACGTTGTYGTGGTYCCGAC |
| 2nd sense | CCTGGTGCGTCAATGATGAC | |
| 1st antisense | TGCGTAAYGCCTGGCGCCG | |
| 2nd antisense | ACTTCTCAGTGTGGCGCGTC | |
| Primer set 12 | 1st sense | GATYCAGACCACTAGYCGGGT |
| 2nd sense | CRACACTATCACAGGYGGTG | |
| 1st antisense | GAAAYTAGTGTTCACCCAGGC | |
| 2nd antisense | AGGTAGAGRAGGCCCTGTTC | |
| Primer set 13 | 1st sense | AATGTYGACACCYTGGCTGCC |
| 2nd sense | TCGAGCTCAAGGACGGCGGAG | |
| 1st antisense | CCCNCCGTCTTGCCAGATTAG | |
| 2nd antisense | TGGCCCTTCTCRACCATGGC | |
| Primer set 14 | 1st sense | CGAGNCAGCGCAAGGCCGT |
| 2nd sense | TCCWCCATAATAGCACACTC | |
| 1st antisense | GTYCGCGACTCTCTCGCCCG | |
| 2nd antisense | CAGARAARTCATTCTCAAACACCAT | |
| Primer set 15 | 1st sense | CATGGYAAAGTGGGYCAGGG |
| 2nd sense | ACGRTACTCACTGCAAAGCAC | |
| 1st antisense | TTCCGYGCTATTGAGAAGGC | |
| 2nd antisense | CGAATCATCACCTTTAAARGC | |
| Primer set 16 | 1st sense | GATYYTGCAGGCCCCGAAGGA |
| 2nd sense | ATRCCAATCAGGTTATGAAC | |
| 1st antisense | CACTCCGGTGAGCCYGGCAC | |
| 2nd antisense | GGGGARACCCCATARACACG | |
| Primer set 17 | 1st sense | GGTGTTGTGGTGGCCCCCGG |
| 2nd sense | GGCGAAGGGGTTGGTTGGATG | |
| 1st antisense | ACYGAGAAGAATTGGGGCCC | |
| 2nd antisense | TAGGGGATTGCGAAGGGCTGAG | |
| Primer set 18 | 1st sense | GGGTGGAATGAATAACATGTCT |
| 2nd sense | CAACCCGGTACTGGGCATAA | |
| 1st antisense | ATGCGCCCTCGGCCTATTTTG | |
| 2nd antisense | ATGRGTATTGGTRCCGTCCTG | |
| Primer set 19 | 1st sense | TTGGCGTGACCAGGCCCAGCG |
| 2nd sense | GCCATGTATRCARAGCATAACAAG | |
| 1st antisense | GTAGACCTRCCACAGCTGGG | |
| 2nd antisense | GCCTCCTCCTCMGCCACCCC | |
| Primer set 20 | 1st sense | ACCACCACCCCGACGTCCGT |
| 2nd sense | TGTCAGCAAGGTTRAACAGGG | |
| 1st antisense | ATAACYTCGACKGATGTTCG | |
| 2nd antisense | GCTATCCCGCGGCCGATCTC | |
| Primer set 21 | 1st sense | CGCAACCTYACCCCYGGTAA |
| 2nd sense | GGACTGGTCATACTCGGCAGC | |
| 1st antisense | GTCTCCCGTTAYTCCAGCAC | |
| 2nd antisense | AGAGARAGCCAAAGCACATC | |
| Primer set 22 | 1st sense | ACAGAATTGATTTCGTCGGC |
| 2nd sense | TAATTATAAGGGTACCCGG | |
| 1st antisense | CTGTAGAGAATGCTCAGCAGG | |
| 2nd antisense | CGRAGCGGCAGGACAAAGAA | |
| Primer set 23 | 1st sense | TCYGACTCTGTGACCTTGGT |
| 2nd sense | CAAGCAAATAAACTATAACTCCCG | |
| 1st antisense | ACCGGCGCGCAGGCCGTTG | |
| 2nd antisense | GTTTTACCCACCTTCATCTTAAGG |
RESULTS
All 200 patients in this cohort were negative for IgM antibodies to HAV, HBV, and HCV but all expressed IgM anti-HEV, which indicated that all were suffering from acute hepatitis E. Table II shows the age and gender distribution, and the occupations and residences of the patients. The patients had used drinking water from multiple sources (supply water, pond water, tube well water, and water from rivers). Approximately 90% of the patients visited local, unqualified doctors (without medical graduation) when they first noticed jaundice or symptoms of acute hepatitis. After there were no improvements, they visited qualified physicians (medical graduates) in Rajshahi.
| Parameters | Data |
|---|---|
| Age (years) | 29.3 ± 10.1 (range: 3–60 years) |
| Sex | |
| Male | 177 |
| Female | 23 |
| Occupation | |
| Student | 94 |
| Service holder | 33 |
| Farmer | 24 |
| Businessman | 25 |
| House wives | 24 |
| Residence | |
| Urban | 121 |
| Rural | 79 |
Patient symptoms were recorded on a questionnaire by the attending physicians and nurses during the patient's first visit. All patients had visible jaundice. Anorexia, nausea, and vomiting were reported by most of the patients. The clinical features of the patients are summarized in Table III [A].
| Parameters | Numbers of patients |
|---|---|
| [A] Clinical profiles of the patients | |
| Anorexia | 200 |
| Nausea | 200 |
| Vomiting | 199 |
| Abdominal pain | 198 |
| Pruritis | 32 |
| Loose motion | 88 |
| Weight loss | 30 |
| Parameters | Mean and standard deviation |
|---|---|
| [B] Laboratory profiles of the patients | |
| Serum bilirubin (mg/dl) | 5.9 ± 3.0 (range: 1.6–20.2 mg/dl) |
| Serum alanine aminotransferase (IU/L) | 733 ± 533 (range: 41–2,971 IU/L) |
| IgM type antibody to | |
| Hepatitis A virus | 0 |
| Hepatitis B virus | 0 |
| Hepatitis C virus | 0 |
| Hepatitis E virus | 200 |
| Hepatitis B surface antigen | 20 |
| Anti-hepatitis C virus | 14 |
Biochemical analyses showed that the levels of bilirubin were above the upper normal limit (normal limit = 0.2–0.8 mg/dl) in all patients (Table III [B]). The levels of ALT varied from 41 to 2,971 IU/L (723 ± 532 IU/L, N = 200). Abdominal ultrasonography detected no features of liver cirrhosis or hepatocellular carcinoma in the patients. One patient died of acute liver failure 3 days after presentation. This patient was a 21-year-old woman who had given birth to her first child 2 months earlier. Twenty patients expressed HBsAg and 14 were positive for IgG anti-HCV.
Patients With Acute Hepatitis E Were Infected With HEV Genotype 1a
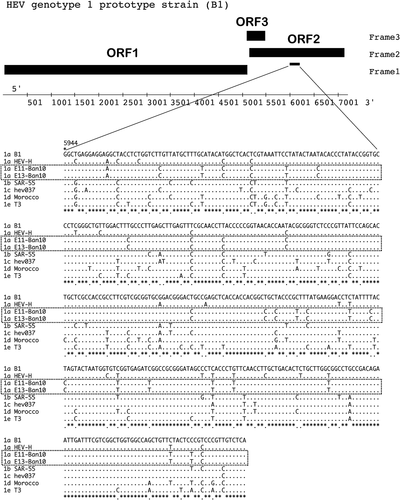
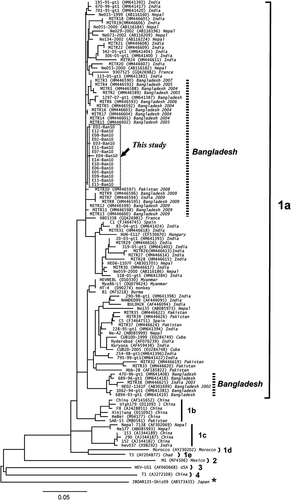
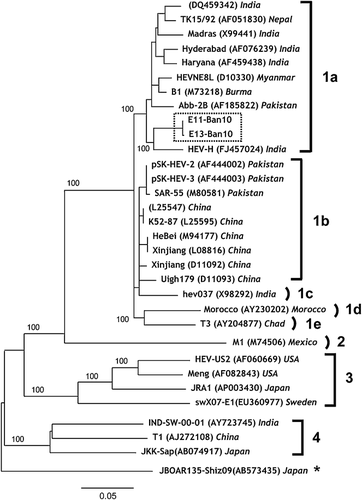
In 15 patients, the HEV genotype was determined by analyzing the nt sequences of the HEV genome. An image representing HEV genome is shown in Figure 1. The sequence analysis based on the PCR-amplified products indicated that all 15 HEV isolates belonged to HEV genotype 1a. A phylogenetic tree of HEV was constructed based on the 266-nt sequences of ORF 2 from the 15 HEV isolates (E01-Ban 10 to E15-Ban10, DDBL/EMBL/GenBank accession numbers AB720034–AB720048) collected in this study (Fig. 2). These 15 patients formed a single cluster in the phylogenetic tree (indicated by the dotted square in Fig. 2). Romanò et al. [2011] reported that the HEV genotypes of Italians who developed acute hepatitis E after travelling to Bangladesh. The HEV sequences of 15 HEV isolates from Rajshahi, Bangladesh shared close homology with the HEV isolates of Italian travelers infected with HEV in Bangladesh (MITR 1–17) (Fig. 2). The HEV sequences of the 15 isolates from Rajshahi were also similar to HEV isolates from India, Burma, China, and Nepal (Fig. 2).
Comparison of the Almost Complete nt Sequences of Two HEV Isolates From Bangladesh and Other Reported HEV Isolates
The almost complete genome sequences of the two HEV isolates with high levels of HEV RNA (E11-Ban 10 and E13-Ban10) were evaluated (Fig. 3). These patients who were the sources of these isolates presented to the physician on February 10, 2010 (E11-Ban10) and February 11, 2010 (E13-Ban10) and they lived in close proximity. The nucleotide sequences of both HEV isolates contained 7,186 nt, which comprised the 5′UTR (nt 1–5), ORF1 (nt 6–5,087 for 1,693 amino acids [aa]), ORF2 (nt 5,125–7,107 for 660 aa), ORF3 (nt 5,084–5,455, for 122 aa), 3′UTR (nt 7,108–7,172), and poly-A tail (nt 7,173–7,186). The almost complete genome sequences had 14 mismatched nts in the two Bangladeshi isolates, E11-Ban10 and E13-Ban10. These two Bangladeshi isolates shared 92.9–94.5% sequence homology with HEV genotype 1a isolates from India (DQ459342, X99441, AF076239, and FJ457024). They also shared 91.9–92.3% sequence homology with HEV genotype 1b isolates from Pakistan (AF444002, AF444003, and M80581) and China (L25547, L25595, M94177, L08816, D11092, and D11093). The Bangladeshi HEV isolates shared 91.9% sequence homology with HEV genotype 1c isolates (X98292, India), 88.2–88.3% sequence homology with HEV genotype 1d (AY230202, Morocco), 88.7% sequence homology with HEV genotype 1e (AY204877, Chad), 75.8% sequence homology with genotype 2 (M74506, Mexico), 73.9% sequence homology with genotype 3 (AF060669, USA), and 74.3% sequence homology with HEV genotype 4 (AY723745, India). An outgroup HEV genotype that shared 73.7% sequence homology is shown at the end of Figure 3 (AB573435, Japan), which was isolated from a wild boar [Takahashi et al., 2010].
DISCUSSION
Hepatitis caused by HEV of Bangladeshi origin was first reported among patients from The Netherlands who developed acute hepatitis E after traveling to Bangladesh [Zaaijer et al., 1993]. Greater attention was focused on HEV in Bangladesh when United Nations peace-keepers from Bangladesh exhibited acute hepatitis E during their assignment in Haiti, while patients from Japan, Spain, and Italy also developed acute hepatitis E after traveling to Bangladesh [Drabick et al., 1997; Sanayama et al., 2008; Fogeda et al., 2009; La Rosa et al., 2011; Romanò et al., 2011]. However, the epidemiology of HEV-induced acute hepatitis E and the genomic features of HEV in Bangladesh have not been investigated adequately.
The present study provides some important insights into the epidemiology of acute hepatitis E in Bangladesh. It is not clear why patients with acute hepatitis E, rather than acute hepatitis A, visited physicians at Rajshahi during January and February in 2010 but there may be two main reasons. First, only patients with severe acute hepatitis visited physicians so other patients with acute hepatitis and comparatively mild symptoms may not have visited physicians. The second cause may be related to the timing of the outbreak. In general, waterborne acute icteric hepatitis outbreaks caused by HEV and HAV occur during August or September in Asian developing countries at the end of the monsoon season, or during/after flooding [Hlady et al., 1990; Naik et al., 1992; Purcell and Emerson, 2000]. However, the present episode of acute hepatitis E was detected during the winter (January and February 2010) and there were no other acute hepatitis epidemics in Bangladesh at that time. Thus, the non-seasonal outbreak of acute hepatitis E that started in January 2010 and ended in February 2010 (winter season) in Rajshahi may have been due to the contamination of a common source by HEV.
The genomic analysis showed that HEV isolates from 15 patients with acute hepatitis E belonged to HEV genotype 1a, which formed a single cluster in the phylogenetic tree (Fig. 2). The almost complete nt sequences of the HEV genomes recovered from two patients had 14 mismatched nts (E11-Ban 10 and E13-Ban10). Thus, it is possible that HEV strains with high pathogenic potential are endemic in and around the Rajshahi area. They may have experienced natural mutation at a rate of 1.40 × 10−3 base substitutions per site per year, as previously described [Takahashi et al., 2004]. This may explain the 14 mismatched nts between the two HEV isolates. The two Bangladeshi HEV isolates also shared more than 90% sequence homology with a HEV genotype 1a isolate from India, HEV genotype 1b isolates from Pakistan, China, and India, and a HEV genotype 1c isolate from India. HEV genotypes 1a is highly prevalent in India [Kumar et al., 2011] and Pakistan [Iqbal et al., 2011], two close neighbors of Bangladesh.
In addition to the information provided in this study of HEV in Bangladesh, this study also detected a high prevalence of HBV and HCV in patients with acute hepatitis E in this cohort (Table III [B]). This indicates the challenging issue of coinfection with HEV, HBV, and HCV in Rajshahi, Bangladesh.
In conclusion, this study showed that a sudden outbreak of acute icteric hepatitis due to HEV occurred in the winter in Bangladesh, but not after flooding. The HEV genotype was assessed in this study. This appears to be the first study to evaluate the almost complete gene sequences of HEV from Bangladeshi patients. This study was conducted in Bangladesh so it may help policy-makers in Bangladesh to recognize the extent of the HEV-related public health problem.




