Molecular analysis of hepatitis C virus infection in Bulgarian injecting drug users†
Massimo Ciccozzi and Gianni Zehender are contributed equally to this work.
Abstract
Intravenous drug users constitute a group at risk for hepatitis C virus (HCV) infection. Today, no data are available on the molecular epidemiology of HCV in Bulgaria despite the fact that in recent years the incidence of acute hepatitis C infection among Bulgarian intravenous drug users increased sixfold and about 2/3 of them developed a chronic infection. The aim of this study was to determine the circulation of hepatitis C genotypes among drug users and to study the evolution and transmission history of the virus by molecular clock and Bayesian methods, respectively. Sequencing of NS5B gene showed that the genotype 3a was the most prevalent type among intravenous drug users. In the Bayesian tree, the 3a subtypes grouped in one main clade with one small cluster well statistically supported. The root of the tree was dated back to the year 1836, and the main clade from Bulgaria was dated 1960. The effective number of infections remained constant until about years 1950s, growing exponentially from the 1960s to the 1990s, reaching a plateau in the years 2000. The not significant intermixing with isolates from other countries may suggest a segregated circulation of the epidemic between 1940s and 1980s. The plateau reached by the epidemic in the early 2000s may indicate the partial success of the new preventive policies adopted in Bulgaria. J. Med. Virol. 83:1565–1570, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Hepatitis C virus (HCV) is the major cause of chronic hepatitis, cirrhosis of the liver and hepatocellular carcinoma in developed countries. About 170 million people are infected chronically with HCV worldwide. In recent years, the HCV epidemics worsened due to the virus dissemination by blood transfusions, blood-derived products, and unsafe medical practice. In industrialized countries, needle sharing is the major risk factor of viral dissemination among injecting drug users [Trepo and Pradat, 1999]. HCV is classified by phylogenetic methods into six major genotypes and many subtypes which differ in geographical distribution, and transmission route [Kuiken et al., 2009]. Genotype 1 and subtypes 1a and 1b are the most prevalent worldwide, though subtypes 3a and 1a are highly prevalent among injecting drug users [Kuiken et al., 2009]. Few reports are available on the molecular epidemiology of HCV infection among injecting drug users in Eastern Europe [Kalinina et al., 2001; Krekulova et al., 2001, 2005] and none on injecting drug users in Bulgaria. Therefore, a molecular analysis of HCV from drug users infected chronically was carried out to determine the epidemiology of the viral subtypes. Indeed, it has been reported that some geographical differences may exist as reported in Czech Republic, where genotype 1b is the most prevalent subtype among drug users [Krekulova et al., 2001]. In this study, the transmission history of HCV in Bulgaria was investigated by Bayesian statistical inference framework to have the simultaneous reconstruction of the temporal and spatial history of the epidemic based on isolates randomly sampled at known times in different places [Lemey et al., 2009]. A comprehensive phylodynamic [Grenfell et al., 2004] study of HCV diversity in Bulgaria may also provide information about the demographic, social, and biological factors that have given rise to the current epidemiological patterns in this geographical area.
MATERIAL AND METHODS
Study Group
Serum samples were obtained from 32 drug users infected chronically with hepatitis C and admitted at the National Center for Addictions, Sofia, Bulgaria. After collection, the serum was stored at −80°C until analysis.
HCV Genotyping
HCV RNA was extracted from 140 µl serum samples with the QIAamp Viral RNA Mini kit (Qiagen, Milan, Italy), then 10 µl of RNA were reverse transcribed by random examer and cDNA amplified by a heminested PCR with primers targeting the NS5B as reported in Laperche et al. [2005].
The amplified products were purified with the QIAquick PCR purification kit (Qiagen) and sequenced using the fluorescent dye terminator technology (BigDye Terminator v3.1 Cycle Sequencing kit; Applied Biosystem, Foster City, CA) according to the manufacturer's instructions and run on an ABI 3100 genetic analyzer (Applied Biosystems). Sequences were aligned using CLUSTAL X software [Thompson et al., 1994], then manually edited with the Bioedit software [Hall, 1999]. HCV genotype was assessed by phylogenetic analysis of the NS5B sequences. The ModelTest [Posada and Buckley, 2004] was used to select the simplest evolutionary model that fitted adequately the sequence data.
Hepatitis C Virus Dataset
Two hepatitis C data sets were built. The first one contained reference sequences downloaded from the Los Alamos HCV sequence database (http://hcv.lanl.gov/content/index) on the basis of the following inclusion criteria: (1) sequences already published in peer-reviewed journals, except the new sequences described below; (2) no uncertainty about the subtype assignment; (3) known city/state of origin as established in the original publication. Nine reference sequences related to the HCV genotypes and subtypes 1a, 1b, 3a, 3b, 4a, and 6a (AB444473, 157781216, AB444480, AB444481, AB444482, AF169005, D90208, EU255952, FJ469164) were selected and added to the newly generated NS5B HCV gene sequences from Bulgaria. The second data set contained sequences from the most representative genotype of this study (genotype 3a) including sequences isolated from Western, Central, and East European countries. The sampling locations of the isolates were Bulgaria (n = 13); Cyprus (n = 8); Estonia (n = 33); Austria(n = 22); France (n = 58); Ireland (n = 1); Lithuania (n = 22); Spain (n = 4).
Evolutionary Rate Estimates and Time-Scaled Phylogeny Reconstruction
A statistical method, based on coalescent theory, was used to estimate the epidemic history of HCV subtype 3a from the viral gene sequences obtained as well as to describe the relationship between the demographic history of a population and the genealogy of individuals sampled randomly from it [Holmes et al., 1995; Pybus et al., 2000; Pybus and Rambaut, 2002].
As sampling dates were not available for all sequences included in the analysis, the evolutionary rates were estimated using a data set containing the same 342 nt NS5B fragment following the external calibration approach used previously by other authors [Pybus et al., 2009]. The phylogeny obtained from the original data set was calibrated by adjusting the mean substitution rate to the mean external evolutionary rate estimates. The external sequences sample included 148 dated HCV NS5B sequences of genotype 3a from Western, Central, and Eastern European countries, which were retrieved from the Los Alamos HCV Sequence Database, as well as the new Bulgarian isolates. The sampling dates ranged from 2000 to 2009.
Evolutionary rates were estimated using a Bayesian–Markov–Chain–Monte–Carlo (MCMC) method implemented in BEAST package 1.5.3 [Drummond et al., 2005; Drummond and Rambaut, 2007] and both a strict and relaxed clock with an uncorrelated log normal rate distribution. As coalescent priors, three parametric demographic models of population growth (constant size, exponential, and logistic growth) and a Bayesian skyline plot (BSP, a non-parametric piecewise-constant model) were compared. Analysis was performed as follows. To reconstruct the time-scaled phylogeny of the main data set, the same Bayesian MCMC method [Drummond et al., 2005; Drummond and Rambaut, 2007], molecular clock and demographic models, assuming the GTR + G + I model of nucleotide substitution, were used. Statistical support for specific clades was obtained by calculating the posterior probability of each monophyletic clade.
The MCMC chains were run for at least 50 million generations, and sampled every 5,000 steps. Convergence was assessed on the basis of the effective sampling size (ESS) after a 10% burn-in [Drummond and Rambaut, 2007] using Tracer version 1.5 (http://tree.bio.ed.ac.uk/software/tracer/). Only ESS values of >250 were accepted. Uncertainty in the estimates was indicated by 95% highest posterior density (95% HPD) intervals, and the best fitting models were selected using a Bayes factor (BF, using marginal likelihoods) implemented in BEAST [Drummond and Rambaut, 2007]. According to Suchard et al. [2001], the strength of the evidence against H0 was evaluated as follows: 2lnBF <2 = no evidence; 2–6 = weak evidence; 6–10 = strong evidence; and >10 = very strong evidence. A negative 2lnBF indicates evidence in favor of H0. Only values of ≥6 were considered significant.
Population dynamics were also analyzed in the Bulgarian clade on an individual basis by comparing the three coalescent models (constant, exponential, and BSP) and implementing a relaxed molecular clock model under the conditions described above. This analysis was not applied to genotype 1a because of the small sample size (six sequences).
RESULTS
Of the 32 injecting drug users examined, NS5B gene sequences were obtained from 20 of them. The maximum-likelihood tree showed that 13 patients were infected by 3a subtype, one by 1b subtype and six by 1a subtype, Figure 1. The distribution of the different subtypes by age, sex, mode of transmission, residence is reported in Table I.
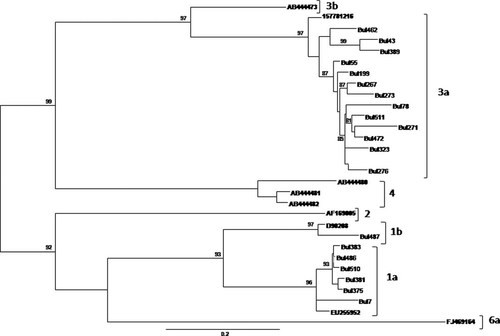
HCV genotype was assessed by phylogenetic analysis of NS5B sequences. The phylogenetic analysis was performed on the 20 sequences of HCV-infected patients included in the study, adding to the alignment 9 reference strains for HCV genotypes 1a, 1b, 2, 3a, 3b, 4, 4a, and 6a sub-genotypes (AB444473, 157781216, AB444480, AB444481, AB444482, AF169005, D90208, EU255952, FJ469164).
| Patient | HCV genotype | Sex | Age | Transmission | City |
|---|---|---|---|---|---|
| 462 | 3a | Male | 28 | Intravenous drug use | Botevgrad |
| 43 | 3a | Male | 28 | Intravenous drug use | Sofia |
| 389 | 3a | Female | 19 | Intravenous drug use | Sofia |
| 55 | 3a | Female | 23 | Intravenous drug use | Burgas |
| 199 | 3a | Female | 30 | Intravenous drug use | Sofia |
| 267 | 3a | Male | 26 | Intravenous drug use | Sofia |
| 273 | 3a | Female | 25 | Intravenous drug use | Sofia |
| 78 | 3a | Male | 25 | Intravenous drug use | Radnevo |
| 511 | 3a | Male | 42 | Intravenous drug use | Sofia |
| 271 | 3a | Male | 22 | Intravenous drug use | Pasargik |
| 472 | 3a | Female | 24 | Intravenous drug use | Sofia |
| 323 | 3a | Male | 23 | Intravenous drug use | Pleven |
| 276 | 3a | Male | 29 | Intravenous drug use | Sofia |
| 487 | 1b | Male | 57 | Intravenous drug use | Sofia |
| 383 | 1a | Male | 26 | Intravenous drug use | Sofia |
| 486 | 1a | Female | 39 | Intravenous drug use | Sofia |
| 510 | 1a | Male | 25 | Intravenous drug use | Sofia |
| 381 | 1a | Male | 34 | Intravenous drug use | Sofia |
| 375 | 1a | Male | 29 | Intravenous drug use | Plovdiv |
| 7 | 1a | Male | 46 | Intravenous drug use | Svoge |
In Figure 2 are shown the genotype 3a sequences analyzed to infer the demographic history in Bulgaria. The reconstruction of a Bayesian phylogenetic tree of HCV 3a subtype, in a calendar timescale, showed that the majority of the Bulgarian strains (11 sequences) segregated together and, with the exception of only three sequences from France, formed a highly significant clade (posterior probability > than 90%). Two sequences clustered instead with sequences from Cyprus. The molecular clock analysis estimated that the HCV 3a subtype was present in Bulgaria since the early 1960s for the main clade, with a lower limit estimate for the first introduction around the 1940s. Two sequences only suggested a more recent introduction of the virus around the end of the 60s. Based on the approximate marginal likelihoods of the six different demographic models used, the BF favored models enforcing a relaxed molecular clock over strict clock models. Models assuming exponential population growth performed always better than models assuming constant population size. In contrast, the BF was not significant when exponential models were compared with BSPs (data not shown) suggesting that both the parametric and the non-parametric models fit the data equally well. The tMRCA of all internal nodes was estimated using a relaxed molecular clock model. The root of the tree was dated back to the year 1836 (95%HPD 1743–1933), the main clade from Bulgaria was dated to 1960, (95%HPD 1948–1980), whereas the small cluster was dated to 1968 (95%HPD 1955–1987), Figure 2. The comparison between parametric and non-parametric models by BF showed that the BSP was preferred. The BSP showed that the effective number of infections remained constant until about the 1950s, growing exponentially from the 1960s to the 1990s, and reaching a plateau in the years 2000, Figure 3.
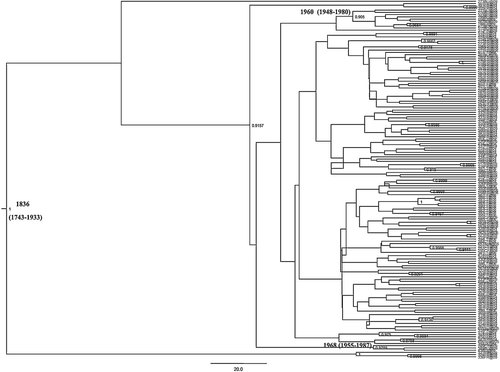
Bayesian time-scaled tree of Bulgarian and European HCV NS5B genotype 3a sequences. The tMRCA, with the credibility interval based on 95% highest posterior density interval (HPD), of the root and of the two statistically supported (posterior probability > 1) internal nodes was reported in years.
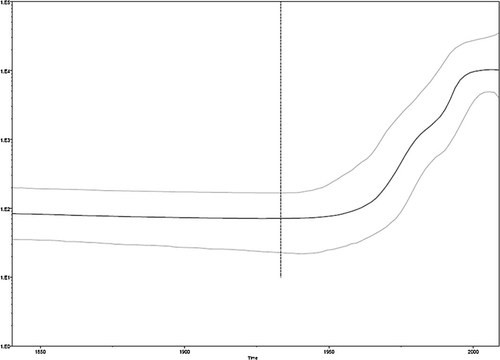
Bayesian skyline plot (BSP) of Bulgarian and European HCV NS5B genotype 3a sequences. The effective number of the infections is reported on the Y axis. Time is reported in the X axis. The grey area corresponds to the credibility interval based on 95% highest posterior density interval (HPD).
DISCUSSION
Hepatitis C chronic infection is a global health problem which affects about 3% of the world population [http://www.who.int/csr/disease/hepatitis/whocdscsrlyo2003/en/index1.html]. HCV genotypes 1, 2, and 3 constitute more than 90% of world chronic infections. Among them, subtypes 1a and 3a are common in injecting drug users [Kuiken et al., 2009], although specific geographical situations have been reported [Krekulova et al., 2005]. A detailed investigation of the molecular epidemiology of HCV among Bulgarian injecting drug users is described.
The molecular monitoring showed that the circulation of HCV subtypes in this risk group reflects that reported by a number of groups worldwide [Kuiken et al., 2009]. Analysis of the NS5B gene of the HCV 3a subtype, the most prevalent subtype identified in this study, allowed us to reconstruct the epidemiological history of HCV 3a subtype in Bulgaria. The Bulgarian sequences were analyzed together with those obtained from neighboring Mediterranean or Eastern countries. The phylogenetic, phylodinamic, and coalescent-based analyses provided a clear picture of its history. Indeed, Bayesian methods have been used widely because of their ability to integrate information about mean evolutionary rates, dated phylogeny and coalescent population dynamics.
The evolutionary rate of the NS5B gene detected in Bulgarian injecting drug users was estimated using an external data set aligned and cropped with the Bulgarian sequences, as described previously [Pybus et al., 2009]. It was found that it fell between previously calculated values [Power et al., 1995; Salemi and Vandamme, 2002; Tanaka et al., 2002; Magiorkinis et al., 2005, 2009; Ciccozzi et al., 2011].
It was estimated that HCV 3a subtype was present in Bulgaria since the early 1960s even though two sequences suggested the possibility of a more recent introduction of the virus around the end of the 60s.
The coalescent-based population dynamics analysis of the HCV 3a subtype done using a non-parametric coalescent model (BSP), made possible to estimate past changes in effective population size [Drummond et al., 2005]. The analysis showed a 2-log increase in the effective number of HCV 3a subtype infections between the end of 1940s (when the exponential growth began), and the end of 1990s, when the plot reached a plateau that still persists today. However, this result does not reflect probably the specific course of the Bulgarian outbreak, but also that of the HCV subtype 3a in Europe, as reported by Pybus et al. [2005]. Furthermore, this period coincides with a considerable increase of the incidence of HCV infection especially among injecting drug users (46%) in Bulgaria as estimated in 2000–2006 [Boykinova et al., 2009]. Thus, the estimate seems to reflect the growth of the injecting drug users risk group rather than the innate transmission potential of the virus. Therefore, it cannot be excluded that the study population is not truly representative of all HCV cases in Bulgaria. Based on this analysis, the growth of the epidemic was relatively explosive and associated with the expansion of intravenous drug use [Magiorkinis et al., 2009; Pybus et al., 2005].
In conclusion, the Bayesian framework, which allows a spatial-temporal reconstruction of a phylogeny and an estimate of the population dynamics using a coalescent-based approach, indicates that the HCV 3a subtype epidemic entered in Bulgaria first in the 1960s as a result of a main ancestral event involving most of the cases. The slight and not significant intermixing with isolates from other countries may suggest a segregated circulation of the epidemic in the country between 1940s and 1980s, due probably to unsafe parenteral medical procedures but with drug addiction playing a relatively important role. The plateau reached by the epidemic in the early 2000s seems to indicate the partial success of the new preventive policies adopted in Europe and Bulgaria.




