Molecular characterization of measles virus strains causing subactute sclerosing panencephalitis in France in 1977 and 2007
Abstract
Measles virus strains from two subacute sclerosing panencephalitis (SSPE) cases diagnosed in 1977 (Laine strain) and in 2007 (Hoedts strain) were studied. Phylogenetic analysis based on C-terminal part of the nucleoprotein and the entire H gene showed that Hoedts strain, circulating in France presumably in the 1980s, belonged to genotype C2. However, Laine strain, suspected to have circulated between 1940s and 1960s, could not be assigned to any known measles virus genotypes. Sequences analysis of the Laine strain suggested that it originated from a measles virus that may have circulating at the same period as the Edmonston strain. The analysis of the whole genome of both SSPE strains revealed biased hypermutations in M, F, and H gene. Some of these mutations like the L165P found in the M protein sequence of the Laine strain, the amino acid position 94, where a mutation M94V was found in the F protein sequence of the Hoedts strain are known to play an important role in the glycoprotein interaction and to impair the ability of measles virus strain to produce cell-free infectious viral particles.This is the first study on molecular characterization of the entire coding region of measles virus isolated from SSPE cases in France. J. Med. Virol. 83:1614–1623, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Subacute sclerosing panencephalitis (SSPE) is a fatal disease of the central nervous system that generally develops 7–10 years after infection by the measles virus. However, even with the elimination of measles, cases of SSPE may still occur 20–30 years later because of the skew of the latency distribution. Despite the availability of efficient vaccines and widespread vaccination, measles remains a major cause of child mortality worldwide. An estimated 164,000 people died from measles in 2008 [WHO, 2009]. SSPE is caused by a persistent measles virus infection of the brain. According to the WHO, the incidence of SSPE is approximately 4–11 cases per 100,000 cases of measles [WHO, 2006]. Clinical manifestations of SSPE include behavioral abnormalities, cognitive decline, myoclonic jerks, seizure, and abnormalities in vision [Garg, 2008; Mahadevan et al., 2008]. Death generally occurs 1–3 years after onset of symptoms. The reason why measles virus persists in some individuals is unknown, but is likely to be host related.
Measles virus belongs to the paramyxovirus family and is a member of the Morbillivirus genus. It is an enveloped virus whose genome contains six genes that encodes for six structural proteins: nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), haemagglutinin (H), and large protein (L). The P gene also encodes several other proteins C, V, and W. Sequence analysis of SSPE viruses indicate that they differ from wild-type viruses due to the introduction of several mutations that mainly affect the matrix, haemagglutinin, nucleocapsid, and fusion genes [Ayata et al., 2007; Jiang et al., 2009]. These genetic mutations in SSPE virus result in poor expression of envelope proteins. Consequently, the SSPE virus is able to maintain a persistent infection in neuronal cells of the brain but is unable to produce transmissible infectious viral particles [Oldstone et al., 2005].
In France, a nationwide case-based mandatory reporting of measles cases was established in 2005. The vaccination coverage was approximately 87% at 24-month of age in 2005 [Waku-Kouomou et al., 2010]. This is lower than the 95% requested to stop the circulation on measles virus in the population. As result, a number of measles outbreaks were reported in recent years [Waku-Kouomou et al., 2006, 2007, 2010; Zandotti et al., 2004]. Although the surveillance of measles is now well established in France, information regarding SSPE cases is very rare. From 1980 to 1996, around 10–30 cases of SSPE were reported each year by the Renaroug network [Ministère-de-la-santé-DGS, 2008]. With the introduction of the vaccination campaign in 1983, the number of measles cases was reduced drastically and SSPE cases also dropped from 25 in 1980 to 3 cases in 1996. Recently, molecular biology techniques were used to help in the diagnosis of an SSPE case [Souraud et al., 2009]. However, up until the present, there has been no molecular data regarding measles strains causing SSPE in France.
The purpose of this study was to describe the detection and molecular characterization of measles viruses isolated in two SSPE cases diagnosed in 1977 (Laine strain) and in 2007 (Hoedts strain) and also to document molecular epidemiological data of measles virus strains in France. In the present study the sequences of the whole coding region of the two SSPE measles virus strains isolated from brain specimens were sequenced and analyzed.
Phylogenetic analysis showed that Hoedst strain belonged to genotype C2 while Laine strain could not be related to any known measles virus genotype. The analysis of the whole genome of both SSPE strains revealed biased hypermutations in M, F, and H gene. This study describes for the first time molecular characterization of the entire coding region of measles virus isolated from SSPE cases in France.
MATERIALS AND METHODS
Patient, Specimen, Cells, and Viruses
Brain biopsy specimens obtained from two patients were investigated. In both cases, diagnosis was confirmed clinically, by magnetic resonance imaging (MRI) of the brain or by the presence of measles antibody in cerebrospinal fluid (CSF). Clinical and virological studies information of these cases were published previously [Souraud et al., 2009; Wild et al., 1979].
Patient 1
A 38-year-old male patient who died 3 months after developing clinical symptoms. The CSF globulin level was elevated, constituting 45.5% of the total protein. In the brain biopsy, measles antibodies were found. A measles virus strain (Laine strain), was isolated by co-culture of the brain biopsy with vero cells [Wild et al., 1979]. This measles strain was stocked in liquid nitrogen since his isolation in 1979 and thaw only recently for sequence analysis.
Patient 2
In a 25-year-old male patient who died after 2 months course of SSPE, the MRI of the brain showed hyperintensity in the grey matter and the subcortical white matter [Souraud et al., 2009]. Measles antibody in the CSF was excessively high at 14,000 UI/L. Measles virus sequences were obtained from the brain biopsy by PCR (Hoedts strain).
It is assumed that the patients were infected during their childhood. However, virus sequences corresponding to these periods are not available, so the Laine strain was compared to the Edmonston wild-type strain while the Hoedts strain which was a measles virus circulating in the 1980s was compared with a wild-type strain in the corresponding genotype. Measles virus strains analyzed in this study are summarized in Table I.
| Measles virus strains | Lab name | Description | Genotype | Genebank accession number |
|---|---|---|---|---|
| Edmonston-wt-USA/54 | Edmonston | Wild-type MV strain isolated in USA in 1954 | A | AF266291 |
| Mvi/Lyon.FRA/77a | Laine | SSPE MV strain isolated in France in 1977 | Unknown | This study |
| Mvs/Toulon.FRA/08.07a | Hoedts | SSPE MV strain isolated in France in 2007 | C2 | This study |
| Mvi/Temara.MOR/24.03b | M185 | Wild-type MV strain isolated in Morroco in 2003 | C2 |
Alla et al. [2006 ], and this study |
- a Entire viral genome sequenced in this study.
- b Entire viral genome sequenced in this study except H gene [Alla et al., 2006].
RNA Extraction and Genome Amplification
Viral RNA from SSPE cases was extracted either directly from clinical specimens (brain biopsy for Hoedts strain) or from infected vero cells (Laine strain) using the RNA Now kits (Biogentex, Inc, Seabrook, South Carolina) in accordance with the manufacturer's protocol.
Measles virus RNA was reverse-transcribed at 42°C for 30 min followed by a denaturation step for 5 min at 85°C using iScript cDNA Synthesis Kit (Biorad, Marnes la Coquette, France). The resulting cDNA was used as a template for PCR amplification of N, P, M, F, and H genes-specific sequences. The H gene was amplified as described previously [Kouomou et al., 2002]. To amplify F and N genes, PCR were performed starting by a denaturation step at 94°C for 5 min, followed by 35 cycles of denaturing at 94°C for 30 sec, annealing at 56°C for 45 sec, and extension at 72°C for 2 min with a final extension at 72°C for 7 min. The PCR cycling program for P and M genes differed from that of F gene by only the annealing temperature, which was 59°C for P gene and 55°C for M gene.
In order to amplify the L gene, measles virus RNA was reverse-transcribed at 42°C for 90 min followed by a denaturation step for 5 min at 85°C using iScript Select cDNA Synthesis Kit (Biorad). The L gene was amplified as seven overlaping fragments. The PCR Program consisted of a denaturation step at 94°C for 5 min, followed by 35 cycles of 30 sec at 94°C, 45 sec at 57°C, 2 min at 72°C, with a final extension at 72°C for 7 min. Primers sequences used in this study are listed in Table II.
| Gene | Primers name | Primers sequences | PCR product size (pb) |
|---|---|---|---|
| N | N1 bis(+) | 5′ GATCCTATTATCAGGGACAAGAGC 3′ | 1,650 |
| N4 bis(−) | 5′ GATGTTGTTCTGGTCCTCGGCCTC 3′ | ||
| P5′s2(+) | 5′ GGGAAGATCTTCCAGCCAACCAACCATC 3′ | 1,014 | |
| p | Pseq 3(−) | 5′ GATGTCTTGGACATCGGAGAAC 3′ | |
| Pseq2(+) | 5′ TGTGAGCAATGCCGCACTGATAC 3′ | 730 | |
| P3′(−) | 5′ GAAGATCTTCCGGCAGGTAAGTTGAGC 3′ | ||
| M1(+) | 5′ CTTAGGAGCAAAGTGATTGCCTC 3′ | 582 | |
| M | M2(−) | 5′ GACCGATCTGAATTCCAGCATTC 3′ | |
| M3(+) | 5′ GTTAATCTGATACCGCTCGATACC 3′ | 633 | |
| M4bis(−) | 5′ CGCTTGGTCCGTGGAGTCTTTCG 3′ | ||
| F | MF1(+) | 5′ CCCAGAATCAAGACTCATCC 3′ | 880 |
| MF2(−) | 5′ CGTCGGATAGGCTATACTGAGGAC 3′ | ||
| MF3(+) | 5′ GGCATCTTAGAGAGCAGAGG 3′ | 932 | |
| MF4(−) | 5′ CGAAGAGGAGACTTGTGGGAAC 3′ | ||
| H | gh004(+) | 5′ GTGCAAGATCATCCACAATGTCACC 3′ | 1,251 |
| mh1251(−) | 5′ CGTATGAAGGAATCCTGTTATC 3′ | ||
| gh1029(+) | 5′ CCAACCGACATGCAATCCTGG 3′ | 914 | |
| mh1922(−) | 5′ GTATGCCTGATGTCTGGGTGAC 3′ | ||
| L | L1s(+) | 5′ GTGAAATAGACATCAGAATTAAG 3′ | 1,092 |
| L1as(−) | 5′ GTCAGATGTATGTCATCAGTTATG 3′ | ||
| L2s(+) | 5′ GCTTTACTGAAATACATGATGTTCTTGAC 3′ | 1,127 | |
| L2as(−) | 5′ GCCTCTGTGCAAACAAGCTGATGGTC 3′ | ||
| L3s(+) | 5′ GACCAAGACACTGATCATCCG 3′ | 1,121 | |
| L3as(−) | 5′ GAGGAGTCTAGTGATGCTCTGGACACATAC 3′ | ||
| L4s2(+) | 5′ GATTCTCGCCTCACTAATGCC 3′ | 718 | |
| L4as2(−) | 5′ GTTTCCTTGTCAATATCATCCAG 3′ | ||
| L4s3(+) | 5′ GTGTGGATCAGTCAACTACG 3′ | 911 | |
| L4as3(−) | 5′ GAATAATCTTGGCTCTATGAGC 3′ | ||
| L5s(+) | 5′ GCTAAGTCCACAGCACTATCTATG 3′ | 1,094 | |
| L5as(−) | 5′ CTCTTTATAAGTGATCAACATAGAACC 3′ | ||
| L6s(+) | 5′ CTGTTGAGATATCAACATTAATTAGGAG 3′ | 1,308 | |
| L6as(−) | 5′ GCAAATAATGCCTAACCACCTAGGGCAG 3′ |
- (+) and (−) indicated, respectively, the forward and the reverse primers.
Nucleotides Sequences Determination and Analysis
PCR products were separated by electrophoresis using a 1.2% agarose gel and then purified using the nucleospin Extract II kit (Macherey-Nagel, Düren, Germany) following the manufacturer's instructions. Sequencing was performed using an ABI 3730 (Applied Biosystems, Langen, Germany). The nucleotides sequences of the N, P, M, F, H, and L gene were aligned and analyzed phylogeneticaly using the Molecular Evolutionary Genetics Analyses (MEGA) software version 4 [Tamura et al., 2007]. Phylogenetics trees were constructed by comparison of the C-terminal part of the N gene and the entire H gene of the sequences derived from the SSPE strains, with the references strains defined by the WHO [2005] using the neighbor-joining method. The reliability of each tree was estimated using 1,000 bootstraps replicates. The nucleotide sequences obtained in this study were deposited on Genbank under accession numbers HM562894-96 (F genes), HM562897-98 (H genes), HM562899-HM562901(L genes), HM562902-04(M genes), HM562905-07 (N genes), and HM562908-10 (P genes).
RESULTS
Phylogenetic Analysis
Two SSPE cases were diagnosed in France; one in 1977 (Laine strain) and the other in 2007 (Hoedts strain). The coding regions of their entire genome were sequenced, compared to other measles virus sequences available on GenBank and used for phylogenetic analysis.
The sequence comparison of the C-terminal part of the N gene with other measles virus sequences available in Genbank showed that the Laine strain is related most closely to the SSPE measles strain circulating in the United kingdom in 1956 (Mvs/Belfast1.UNK/1956-SSPE (AF504045) [Jin et al., 2002] and Edmonston strain with 97% identity. Phylogenetic analysis based on the C-terminal part of the N gene showed that the Laine strain could not be assigned to any known measles virus genotype (Fig. 1). This result was confirmed using the sequence of the entire H gene (Fig. 2). To assign a new genotype, the minimum nucleotide divergence should be 2.5% for C-terminal part of the N gene (450 nt) and 2% for the full length H gene open reading frame from the next most closely related strain [WHO, 2001a]. In this study, the nucleotide divergence between the Laine strain and all measles reference strains was calculated using the nucleotide sequence of the C-terminal part of the N gene and the entire H gene. The results showed that for the N gene, the nucleotide divergence varied from 2.7% with the genotype A to 5.8% with genotypes C2, D10, E, H2. Using H gene, the nucleotide divergence varied from 2,3% with the genotype A reference strain to 5.5% with genotype H1 reference strain. These results therefore suggested that the Laine strain is closer to the genotype A than to any other known measles virus genotype.

Phylogenetic tree based on the sequences of the hyper variable region of the N gene, showing the two SSPE strains (Laine and Hoedts) isolated in France in 1977 and 2007, respectively. Other SSPE strain (Patient B) and wild-type measles strains (M185) of genotype C2, were also included. Sequences analyzed in this study are in bold. Significant bootstrap values (>80) are indicated.
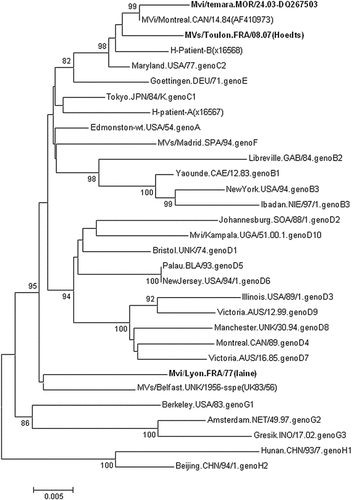
Phylogenetic tree based on the sequences of entire H gene, showing the two SSPE strains (Laine and Hoedts) isolated in France in1977 and 2007, respectively. Other SSPE strain (Patient B) and wild-type measles strains (M185) of genotype C2, were also included. Sequences analyzed in this study are in bold. Significant bootstrap values (>80) are indicated.
The most closely related measles virus to the Hoedts virus was the genotype C2 reference strain and nucleotide divergence was 2.2% and 1.1% for N and H gene, respectively. Phylogenetic analysis based on the C-terminal part of the N gene showed previously that it belonged to genotype C2 [Souraud et al., 2009]. These results were confirmed in this study with the H gene (Fig. 2). The sequence comparison of the C-terminal part of the N gene with other measles virus sequences available in Genbank showed that Hoedts strain is most close to the measles strain isolated in Canada in 1984 (Monteral.CAN/14.84-AF410973) [Tipples et al., 2004] with 98% identity and to the genotype C2 reference strain (Maryland.USA/77), isolated in the USA in 1977 [Rota et al., 1994].
Sequence Variation in the Coding Regions of the Genome of the SSPE Strains
The sequences of the complete coding regions of the genome of both SSPE strains (Laine and Hoedts) were sequenced. They were compared to each other, to measles strains available on Genbank and also with a wild-type measles strain of the same genotype or a closely related genotype.
Sequence comparison of both SSPE strains showed that, the M gene started by a threonine (Fig. 3, Table III) instead of a methionine usually found as the start amino acids of proteins. Premature stop codons were identified in the F gene (Fig. 4) whereas the H gene was elongated in both SSPE strains. Although the majority of mutations found were specific to each strain, common mutations could be identified (Table III). In the F protein, a mutation I446T was identified. Sequence alignment on Genbank showed that this mutation is present in only 4 of the 100 sequences analyzed. In the M gene, a mutation V101A was found whereas in the H protein sequence, mutations R7Q and Y12H were observed in this study. Along the L gene, mutations Y723C and D1887N appear to be specific to both SSPE strains.

Amino acid sequence alignment of the M protein. Amino acid identity are given as dot and an amino acid indicates disagreement with Edmonston strain. The star (*) indicates the stop codon. The question mark (?) were added for the purpose of alignment to obtain equal sequence length for all the sequences. Amino acid specific to SSPE strains (Laine and Hoedts) are indicated.
| Gene | Amino acid position | Edmonston strain (genotype A) | Laine strain (SSPE) | M185 strain (genotype C2) | Hoedts strain (SSPE) |
|---|---|---|---|---|---|
| P/V | 225 | G | E | G | E |
| 300 | stop | Q | stop | stop | |
| M | 1 | M | T | M | T |
| 5 | Y | H | Y | H | |
| 65 | L | P | L | P | |
| 97 | L | P | L | P | |
| 101 | V | A | V | A | |
| 135 | F | L | F | L | |
| 165 | L | P | L | L | |
| 170 | Y | H | Y | H | |
| 180 | F | L | F | L | |
| 232 | Y | H | Y | H | |
| 248 | F | S | F | S | |
| 291 | L | P | L | P | |
| 303 | V | A | V | A | |
| 335 | stop | Q | stop | stop | |
| 350 | - | stop | - | - | |
| F | 94 | M | M | M | V |
| 449 | I | T | I | T | |
| 535 | L | P | L | stop | |
| 546 | Y | stop | Y | - | |
| H | 7 | R | Q | R | Q |
| 12 | Y | H | Y | H | |
| 618 | stop | Q | stop | W | |
| 622 | - | ? | - | stop | |
| L | 723 | Y | C | Y | C |
| 1887 | D | N | D | N |
- Important mutations specific to only one of the two SSPE strains are indicated in bold. The question mark (?) represented unanalyzed amino acid.
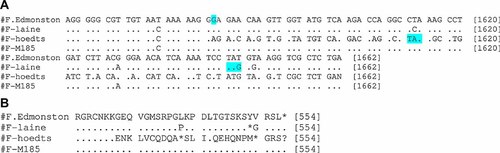
Alignment of nucleotide sequence (A) and the related amino acid sequence (B) of the F gene (C-terminal partial sequence). Nucleotide and amino acids identities are given as dot and a nucleotide or an amino acid indicates disagreement with Edmontston strain. The star (*) indicates the stop codon. The letter N in the nucleotide sequence and the question mark (?) in amino acid sequence were added for the purpose of alignment. The nucleotide G deleted in the Hoedts sequence is indicated and the premature stop codon of SSPE strains (Laine and Hoedts) are also indicated.
Due to the fact that the Laine strain could not be assigned to any know MV genotype, the most closely related wild-type Edmonston strain, was used for sequence analysis of N, P, M, F, H, and L genes. The results showed that the amino acid sequence divergence was 3.6%, 3.9%, 9.5%, 1.8%, 3.8%, and 1.2% for N, P, M, F, H, and L of the Laine and Edmonston strains, respectively. Furthermore, a T-C mutation was identified in the P/V gene which results in the replacement of the stop codon of the V protein by a glutamine (Q 300). Therefore, the predicted V protein of the Laine strain may have one more amino acid than the Edmonston strain. In the M gene, the start codon (methionine) was altered to threonine due to a T-C change in the gene. A newly generated termination codon was identified at position amino acid 350 (Table III, Fig. 3). Hence, the predicted M protein of the Laine strain may have 15 amino acids more than one of the Edmonston strain (335 amino acids). Another important observation in the Laine M protein sequence was that 66% of the mutations were L-P due to T-C mutation in the M gene. In the F protein sequence, a T-G mutation at nucleotide position 7095 results in an earlier termination codon at amino acid position 546 (Table III). This generates a predicted F protein which may be four amino acids shorter than the F protein of the Edmonston strain (Fig. 4). Sequence analysis of the H gene revealed that there were no termination codons. The attempts to amplify the intergenic region between H and L failed. In the N protein sequence, the Laine strain differs from Edmonston strain by 3.6%. The L protein sequence seems to be more stable than other proteins sequences, with only 1.2% mutations.
The entire coding sequence of the Hoedts was also analyzed. As the sequences of the complete genome of the reference strain for genotype C2 (Maryland.USA/77) was not available on Genbank, the genome of the strain M185 [Alla et al., 2006] was sequenced and used as the wild-type strain for comparison. The results showed that the amino acid sequence divergence was 1.6%, 2.3%, 6.5%, 2.6%, 1.4%, and 0.9% for N, P, M, F, H, and L, respectively. The sequence analysis showed that as in Laine strain, the start codon of the predicted M protein of the Hoedts strain was altered to a threonine (Fig. 3, Table III). In the sequence of the F protein, a deletion of a G nucleotide was detected at position 7031 which generate a reading frame shift that resulted in a premature termination codon at amino acid position 535 (Fig. 4). Taken together, these two evens lead to a predicted altered F protein in the Hoedts strain which may be 11 amino acids shorter than the one of the wild-type strain of the same genotype, the M185 strain. In addition, a mutation M94V was found in the F protein sequence. The analysis of the H sequence revealed that the termination code is located at position 622 instead of 618 as in M185 strain (Table III). Therefore, the H protein of the Hoedts strain may have four amino acids more than M185 strain. In the N protein, there was only one mutation (S427N) in Hoedts strain. In contrast, the P protein contained 12 amino acids changes. The L protein had only 0.8% mutations.
DISCUSSION
SSPE is rare slowly progressive neurological disorder caused by the persistent infection of human brain by a defective measles virus. Only wild-type measles virus sequences have been found in SSPE cases [Rima and Duprex, 2005]. It is estimated that about 1 out of 100,000 individuals infected by the measles virus will develop SSPE. This study provides for the first time molecular data on SSPE cases from an historical and a contemporaneous strains. These data are the basis that will help for a better understanding of measles strain circulation in France.
Phylogenetic analyses from SSPE cases usually indicate the genotype circulating in the geographic area where the patient contracted the primary measles infection [Rima et al., 1997]. This characteristic have been used to identify the source of virus strains causing SSPE [Miki et al., 2002; Forcic et al., 2004; Bellini et al., 2005; Mahadevan et al., 2008].
The Laine strain, suspected to have circulated in France between the 1940s and 1960s was found to be close to an untyped SSPE strain reported previously to be circulating in the UK in 1956 [Jin et al., 2002]. However, the Laine strain, could not be assigned a genotype. These findings suggested that the Laine strain is an untyped historic strain probably circulating in France while Edmonston strain was circulating in the USA (1954).
The high nucleotide identity, 97% and 98% between Hoedts strain and measles strain circulating in the USA (Maryland.USA/77) and in Montreal in 1984 (Montreal.CAN/14.84), respectively, confirmed the presumption that the patient would have been infected in the 1980s. Furthermore, the genotype C2 was first detected in Europe in the 1970s where it was considered to be an indigenous genotype and had recently been exported to the USA and Canada [Riddell et al., 2005]. Thus the genotype C2 found is coherent with temporal and geographical distribution of measles virus.
It is widely known that biased hypermutations are a hallmark of SSPE measles virus. Mutations in the F protein of SSPE strains have been previously described [Cattaneo et al., 1988, 1989; Billeter et al., 1994]. So far these mutations resulted in three different type of F protein: (i) a F protein with an elongated carboxy-terminus tail [Ning et al., 2002], (ii) a F protein with a shortened carboxy-terminus [Cattaneo et al., 1989; Billeter et al., 1994; Ning et al., 2002], (iii) a F protein with unchanged length despite many amino acids changes [Ning et al., 2002; Ayata et al., 2007, 2010]. In this study, the cytoplasmic tail of the F protein, predicted from sequences analysis of the gene, is altered in both SSPE strains and presented a short tail pattern. However, the extent and mode of alteration was different in each strain. In Laine strain, a premature stop codon was introduced by a point mutation leading to a stop codon at amino acid position 546 while in Hoedts strain, a deletion of a G nucleotide at position 7031 was responsible for a reading frame shift which, subsequently results in a premature stop codon. These two different mechanism are similar to those reported previously [Cattaneo et al., 1988, 1989; Ning et al., 2002]. It was reported recently that the F gene of SSPE viruses is a major determinant of neurovirolence, [Ayata et al., 2010]. In the same report, it was suggested that mutation T461I was sufficient to transform a non-neuropathogenic wild-type measles virus into lethal virus. In this study, a different mutation I446T was found in the same region of the F protein of both SSPE strains. This mutation seems to be very rare as it was found in only 4% of sequences available in Genbank. It might therefore be interesting to study the role of that specific mutation in the propagation of SSPE strains in the brain. A mutation M94V was observed in the F protein sequence of the Hoedts strain. It was reported that the amino acid at position 94, located in the putative heptad repeat C (HRC) domain of the F protein plays an important role in the fusogenicity and glycoprotein interaction of measles virus [Plemper and Compans, 2003]. This mutation has been reported for another SSPE strain (Osaka2), isolated in Japan [Ayata et al., 2007].
The results of sequence analysis of the M gene complies with the published data, that is, biased hypermutations were found in the two SSPE strains. The initial codon was substituted by a threonine, raising the question of the functionality of the resulting M protein. It might be interesting to analyze the P/M intergenic region to explore whether there is an early start codon for M protein. According to Ayata et al. [2002] mutation in the P3′ untranslated region can cause increased read-through at the P/M junction and directly affects M gene expression. A late stop codon was found in the Laine strain, suggesting that the M protein is elongated. Similar results were reported previously [Jin et al., 2002; Forcic et al., 2004]. However, in contrast to the present study, most of published studies reported truncated M proteins. It has been reported that the V101A mutation, found in the predicted M Protein of both SSPE strains, is sufficient to generate a functionally defective virus assembly [Runkler et al., 2007]. Among mutations found in the M protein sequence of the Laine strain, one was L165P which is known to impair the ability of measles virus to produce cell-free progeny virus [Jiang et al., 2009]. Biased and even point mutations in the M gene are known to render the M protein insoluble, non-functional and therefore, impair the ability of the measles virus to produce cell-free progeny virus [Sheppard et al., 1986; Jiang et al., 2009].
Compared to the M gene, the H gene of both SSPE strains were less mutated. However, in both SSPE strains, the predicted H protein was longer than those of the wild-type strains. In fact, the stop codon could not be found in the Laine stain, suggesting that the predicted H protein in this strain is elongated. It might therefore be interesting to amplify the intergenic region between H and L genes to ascertain whether the termination codon for H protein exists or not. Mutations were found throughout the H protein of both SSPE strains, however, two of them (R7Q) and (Y12H), were shared by both SSPE strains and were reported previously in SSPE strains isolated in the United Kingdom in the 1950s [Jin et al., 2002]. The study of their biological role should give more insight onto the pathogenesis of SSPE.
In the present investigation, the complete sequences of the N, P, F, M, H, and L gene of two SSPE strains were analyzed. The genotypes of measles virus identified in SSPE cases provided information about the circulation of measles strains in France in the 1940–1960s and 1980s. Sequences analyses results showed that the N, P, and L genes had no exceptional mutations. In contrast, striking alterations where observed in the sequence of M protein which has an altered start codon, in H protein where elongated C-terminal tail was found and in the F protein which was partially deleted. Detailed virological and immunological studies will be necessary to explore the biological impacts of these mutations for a better comprehension of SSPE pathogenesis.
Acknowledgements
We are grateful to Dr. F. L. Cosset, Inserm U758 (Lyon, France) for his support, Dr. A. Alla, National Institute of Hygiene (Rabat, Morocco) for measles strain M185.




