Comparative evaluation of different DNA extraction methods for HPV genotyping by linear array and INNO-LiPA
Abstract
In order to investigate the influence of DNA extraction on two PCR-based HPV genotyping tests (Linear Array, Roche and INNO-LiPA Extra, Innogenetics), three different procedures were used to purify DNA from 28 cervico-vaginal samples tested previously by the Hybrid Capture 2: the AmpliLute Liquid Media Extraction kit (Roche), the QIAamp DNA Blood mini kit (QIAGEN), and the NucliSENS EasyMAG automated platform (bioMérieux). All HC2-positive samples were found positive by both assays, independently of the extract used. Type-specific concordance (i.e., identical HPV type-specific profile in all the extracts of the same sample) was observed in 55% and 75% of the cases testing samples by the Linear Array and the INNO-LiPA, respectively. Using the DNA extracted with the two manual methods the results were concordant in 75% of the cases both for the Linear Array and the INNO-LiPA. When comparing the Linear Array results obtained on either of the two manual extracts with those obtained following automated extraction, 65% of the samples showed type-specific concordance in both cases. The INNO-LiPA results were concordant in 80% of the cases comparing the AmpliLute versus the automated extract, while concordant results were observed in 90% of the cases when comparing the QIAGEN versus the automated extract. In conclusion, the Linear Array and INNO-LiPA results are affected by the method of DNA extraction. Consequently, different HPV type-specific profiles may be observed using different extracts of the same sample. The use of consistent protocols for DNA purification is a priority to guarantee intra-assay reproducibility over time. J. Med. Virol. 83:1042–1047, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Human Papillomaviruses (HPVs) are the etiological agents of cervical carcinoma, the second most common malignancy among women after breast cancer [Munoz et al., 2006; zur Hausen, 2009]. Virtually all cases of cervical carcinoma worldwide are HPV-positive, although only a limited number of HPV genotypes among the more than 100 types identified so far seem to have a role in the development of cervical cancer. In particular, 12 genotypes have been classified as “carcinogenic” or “high-risk” [Munoz et al., 2006; Bouvard et al., 2009], with HPV 16 representing the most prevalent type, being present in 61% of all cervical cancer cases [de Sanjose et al., 2010].
Cervical cytology screening, which represents one of the best examples of the efficacy of secondary prevention for cancer, resulted in a reduction in cervical cancer morbidity and mortality [Kitchener et al., 2006]. Furthermore, the relatively recent introduction of HPV molecular tests has added a powerful tool to cervical cancer prevention programs [Wheeler, 2007]. These tests now represent an integral part of the management of women with equivocal cytology and have major clinical relevance as an adjunct to cytology [Snijders et al., 2003; Wright et al., 2007]. A number of assays are available for the molecular diagnosis of HPV infections [Molijn et al., 2005]. Some tests only provide a positive versus negative result, with no individual identification of the genotypes (e.g., Hybrid Capture 2, HC2), while others allow simultaneous HPV detection and genotyping (e.g., the Linear Array HPV Genotyping Test and the INNO-LiPA). Importantly, since the oncogenic potential of the different HPV types, even in the high-risk group, varies greatly and the risk of developing high-grade cervical lesions and cancer depends on the infecting genotypes [Munoz et al., 2003; Munoz et al., 2006], the accurate assessment of the type-profile is a major step toward a reliable evaluation of cancer risk. Although the clinical value of HPV genotyping tests is still debated, these tests are, however, useful for assessing the prevalence of specific genotypes in population-based studies and might assume an increasing importance for determining a possible variation in the prevalence of non-vaccine HPV types in the HPV vaccine era, in order to monitor type-specific viral persistence [Kjaer et al., 2010], to plan a correct management of the patient, and to predict treatment effectiveness in cervical cancer patients [Nagai et al., 2004; Nobeyama et al., 2004].
PCR-based methods are used widely in clinical and research laboratories. The Linear Array HPV Genotyping Test (Roche, Milan, Italy) and the INNO-LiPA (Innogenetics, Pomezia, Italy) has received the CE mark certification, are registered for in vitro diagnostic use in Europe and, among all the available methods, have the most extensive clinical validation. The Linear Array, which is based on the amplification of a 450 bp sequence within the L1 region using PGMY primers, identifies 37 anogenital HPV genotypes. The INNO-LiPA HPV Genotyping Extra is based on the use of SPF10 primers for the amplification of a 65 bp fragment within L1, and is designed for the identification of 28 different genotypes. The Linear Array is validated only with a specific, ad hoc developed method: cervical samples have to be collected in PreservCyt and only the AmpliLute Liquid Media Extraction kit (Roche) is validated for sample preparation. In contrast, in the case of the INNO-LiPA, a variety of media can be used for collection of cervical material (e.g., water, PBS, and PreservCyt) and DNA extraction can be carried out using proteinase K or commercial kits.
Several studies have shown that the Linear Array and the INNO-LiPA often lead to different genotyping results, because of the different primer sets, reaction conditions, sensitivity, specificity, plus a number of other parameters [van Hamont et al., 2006]. However, not many studies have focused on the influence of the DNA extraction method on the genotyping results [Dunn et al., 2007].
The aim of the present study was to investigate the influence of the method used for DNA extraction from cervico-vaginal samples on HPV genotyping results obtained by the Linear Array and the INNO-LiPA. Three different procedures, two manual and one automated, were used: the Roche-recommended AmpliLute Liquid Media Extraction kit (Roche), the QIAamp DNA Blood mini kit (QIAGEN, Milan, Italy) and the NucliSENS EasyMAG automated platform (bioMérieux, Florence, Italy). The QIAamp DNA Blood mini kit was employed since it represents one of the most common manual extraction methods in clinical and research laboratories. Finally, an automated extractor was included in the present investigation as automated extraction instruments are now widely adopted in many laboratories, because they are highly flexible, allow the simultaneous processing of many different samples and they reduce the hand-on time of the technicians.
MATERIALS AND METHODS
Samples
Cervico-vaginal samples were obtained from women attending the Pathology Department of the Regina Elena Cancer Institute (Rome, Italy). Specimens, collected in PreservCyt (Cytyc Corp., Rome, Italy) using a cytobrush (Cytyc Corp.) and an Ayre spatula (Cytyc Corp.), were tested by the HC2 high-risk HPV DNA test (QIAGEN) according to the manufacturer's instructions. Twenty HC2-positive samples (i.e., specimen RLU/cut-off value > 1) and eight HC2-negative samples (i.e., specimen RLU/cut-off value < 1) were selected.
DNA Extraction
The three methods of extraction used are explained in Figure 1. The specimen preparation with the AmpliLute Liquid Media Extraction kit (Roche) was performed following the manufacturer's instruction. Briefly, cervico-vaginal samples were vortexed and 250 µl aliquots of each specimen were used for the extraction. The final elution step was performed using 120 µl of the Roche-provided elution buffer (AVE), obtaining “extract A.” The “DNA purification from blood or body fluids spin protocol” of the QIAamp DNA Blood mini kit (QIAGEN) was used to purify the DNA from 1 ml of the cervico-vaginal specimens. Hundred microliter of the QIAGEN-provided elution buffer (AVE) were used for the final elution step, obtaining “extract B.” For the extraction with the NucliSENS EasyMAG automated platform (bioMérieux), the lysis step was performed by the instrument (on-board protocol) and the “Specific B” protocol was then used to extract total nucleic acids from 500 µl of the cervico-vaginal samples. The elution step was carried out with 110 µl of NucliSENS Elution buffer (bioMérieux), obtaining “extract C.” All extracts were stored at −20°C prior to processing.
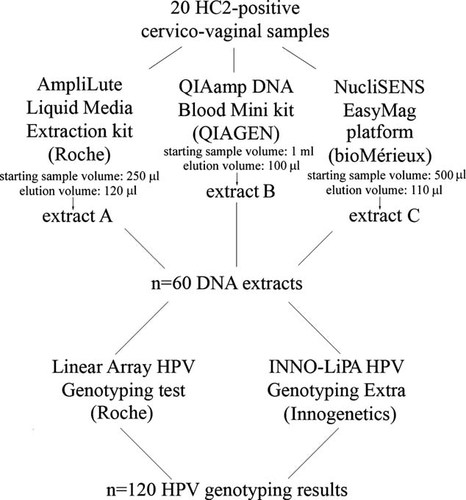
Diagram of the study flow.
HPV Genotyping Tests
HPV amplification, amplicons hybridization, and detection were performed in parallel on each extract following the Roche and Innogenetics instructions for the Linear Array and the INNO-LiPA, respectively. The volume of each extract used to perform the PCR was calculated in order to correspond to the same volume of the original cervico-vaginal sample and, whenever required, adjusted to 50 µl for the Linear Array and 10 µl for the INNO-LiPA. The Gold-plated 96-Well GeneAmp® PCR System 9700 (Applied Biosystems, Milan, Italy) was used for both the Linear Array and INNO-LiPA amplification steps. All hybridization steps up to color development were carried out with a Profiblot T48 instrument (Tecan, Männedorf, Switzerland) for samples tested by the Linear Array and in an Auto-LiPA (Innogenetics) for samples analyzed by the INNO-LiPA.
The Linear Array results were interpreted visually. The Linear Array testing algorithm, which is unable to discriminate HPV 52 infection in case of HPVs 33, 35, or 58 infections, was used. HPV 52 infection was not ascertained by HPV 52-specific PCR. The LiRAS® for LiPA HPV software was used for the interpretation of the INNO-LiPA results. Visual interpretation was adopted to confirm the automatic reading.
Results were considered valid if successful amplification of the human DNA control, β-globin for the Linear Array and a fragment of HLA-DPB1 gene for the INNO-LiPA, was observed.
For the Linear Array and INNO-LiPA comparison, results were defined as concordant when identical assay-common genotypes were detected by both assays, compatible when one or more assay-common genotypes were not detected by either of the assays, and discordant when no similarities in the assay-common genotypes were found between the two assays. When comparing the genotyping results obtained by either assay on the three different extracts of the same cervico-vaginal sample, type-specific concordance was defined as an identical HPV type-specific profile in the extracts compared.
RESULTS
For the present study, three different nucleic acids extracts were obtained from 20 HC2-positive and 8 HC2-negative cervico-vaginal samples using commercially available methods, as explained in Materials and Methods Section. Subsequently, all extracts were tested both by the Linear Array and the INNO-LiPA (Fig. 1). Successful amplification of the human DNA control was obtained for all the extracts in both assays, proving the efficiency of each extraction technique employed and the quality of the sample, that is, inadequate specimen processing and the presence of inhibitors can be excluded.
Among the eight HC2-negative samples examined, seven were consistently negative both by the Linear Array and the INNO-LiPA, independently of the extract used. Therefore, these samples were not useful for comparing the HPV type-specific profiles obtained on different extracts. One sample was negative by the Linear Array and positive for HPV 74 by the INNO-LiPA in all the three extracts. HPV 74 is not included in neither the HC2 nor in the Linear Array. Thus, this sample was not taken into consideration in our analysis.
Results obtained by the Linear Array and INNO-LiPA on HC2-positive samples were compared considering only the assay-common genotypes. The number of concordant, compatible, and discordant results obtained using the same extract for both assays are shown in Table I.
| Extract | No. of samples (%) | ||
|---|---|---|---|
| Concordant | Compatible | Discordant | |
| A | 8/20 (40) | 11/20 (55) | 1/20 (5) |
| B | 7/20 (35) | 12/20 (60) | 1/20 (5) |
| C | 8/20 (40) | 12/20 (60) | 0/20 (0) |
- See Materials and Methods Section for definition of concordant, compatible, and discordant results.
All HC2-positive samples were positive by both assays, independently of the extract used to perform the PCR. As far as the Linear Array genotyping results are concerned, type-specific concordance in all the three extracts was observed in 11/20 cases (55%), while INNO-LiPA results were concordant in 15/20 cases (75%, Table II). When comparing the results of the HPV genotyping tests performed on the DNA extracted with the two manual methods (extracts A and B), 15/20 cases (75%) showed type-specific concordance both by the Linear Array and the INNO-LiPA. When comparing the Linear Array results obtained using either of the two manually extracted DNA (extract A or B) with those obtained on the DNA isolated by the automated extractor (extract C), 13/20 samples (65%) showed type-specific concordance in both cases. On the other hand, the INNO-LiPA results were concordant in 16/20 cases (80%) when extract A (AmpliLute) was compared with extract C (automated extract), while concordant results were observed in 18/20 cases (90%) when comparing extract B (QIAGEN) with extract C. The sample size was too limited to perform any statistical analysis to determine if the observed differences were statistically significant.
| No. of concordant samples (%) | ||||
|---|---|---|---|---|
| HPV test | A vs. B vs. C | A vs. B | A vs. C | B vs. C |
| Linear Array | 11/20 (55) | 15/20 (75) | 13/20 (65) | 13/20 (65) |
| INNO-LiPA | 15/20 (75) | 15/20 (75) | 16/20 (80) | 18/20 (90) |
- A, AmpliLute extract (Roche); B, QIAamp DNA Blood Mini kit extract (QIAGEN); C, NucliSENS EasyMAG extract (bioMérieux).
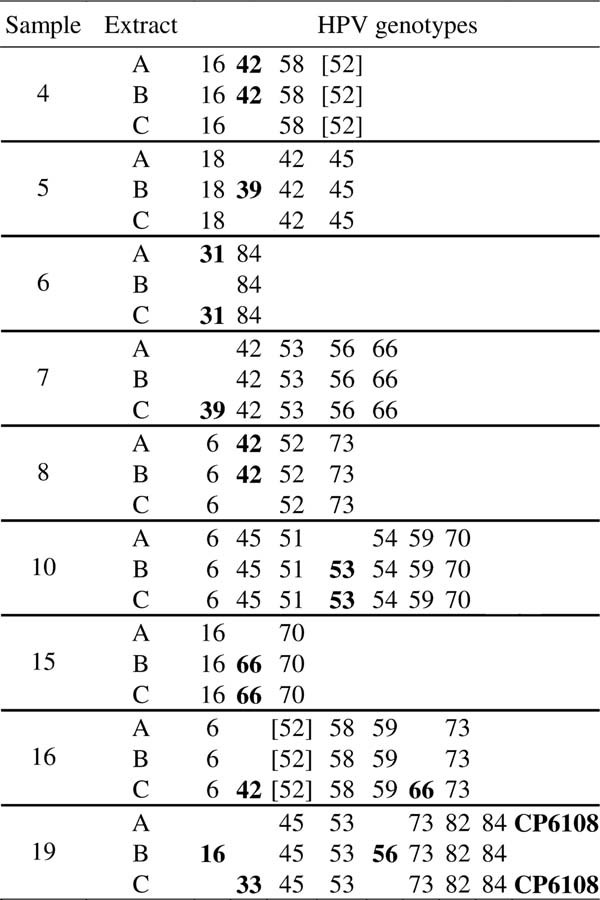
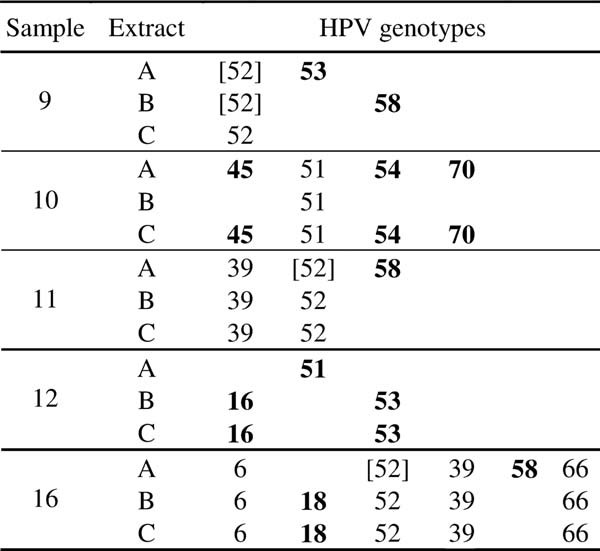
For all the nine samples with a discordant Linear Array type-specific profile and for four out of the five samples with a discordant INNO-LiPA type-specific profile, there was at least one HPV type in common in all the three extracts (Tables III and IV). More specifically, the lack of type-specific concordance in the Linear Array results concerned nine HPVs, two low-risk (HPVs 42 and CP6108), five high-risk (HPVs 16, 31, 33, 39, and 56) and two possible high-risk types (HPVs 53 and 66), as shown in Table III. In the case of the five samples with a discordant INNO-LiPA type-specific profile, lack of concordance was observed for five high-risk (HPVs 16, 18, 45, 51, and 58), two possible high-risk (HPVs 53 and 70), and one low-risk HPV types (HPV 54), as shown in Table IV.
DISCUSSION
The present study compared the HPV genotyping results obtained by the Linear Array HPV Genotyping Test and the INNO-LiPA HPV Genotyping Extra performed on three different nucleic acids extracts of the same cervico-vaginal samples.
The comparison between the Linear Array and the Inno-LiPA results on HC2-positive samples showed that most, if not all, the genotyping results were either concordant or compatible, as already demonstrated in previous studies performed on much larger sets of samples [van Hamont et al., 2006; Castle et al., 2008]. The data also confirmed that the Linear Array is able to detect more multiple infections and a greater number of HPV types per multiple infections (data not shown) [Castle et al., 2008]. The differences observed between the two methods may be due to the fact that these assays differ in several ways, such as the length of the amplicons and the amplicon detection. Neither the detailed genotyping results obtained for each sample nor a detailed discussion concerning the direct comparison between the two assays were considered in this study, since studies aimed at comparing results obtained by these two methods have been reported extensively.
The variability in the HPV type-specific profiles obtained both by the Linear Array and the INNO-LiPA on different extracts of the same samples suggests that both assays are affected by the DNA isolation method used. These data are consistent with the results of a previous study that showed the influence of the extraction protocol on the Linear Array performance [Dunn et al., 2007]. However, the present study seems to indicate that the INNO-LiPA is less affected than the Linear Array by the DNA purification technique used.
Interestingly, the inclusivity level of the Linear Array for most of the nine HPV types discordant among the several extracts is medium to high (900–30,000 copies/ml), that is, this assay detects consistently these genotypes across replicates and different runs only at high concentration, with the exception of HPVs 16, 53, and 66 for which the 95% positive hit rate concentration level predicted by Probit analysis is 195, 256, and 250 copies/ml, respectively, as reported in the Roche Linear Array test manual. The most frequent discordant HPV type among the three extracts when tested by Linear Array was HPV 42, which has an inclusivity level of 30,000 copies/ml. In fact, in three cases the Linear Array failed to detect HPV 42 in all the extracts of the same sample (Table III). On the contrary, only in one case HPV 16 was not detected in all the extracts of the same sample, while in five cases this genotype was found in all the three extracts (Table III and data not shown).
These data indicate that the reproducibility of the results obtained on different extracts is lower for HPV types with a higher inclusivity level. Although, the reliable detection of both low-risk and high-risk genotypes is important to assess the prevalence of individual genotypes in epidemiological studies, failure in detecting low-risk types, such as HPV 42, might not have a major clinical implication. On the contrary, failure in detecting high-risk HPVs, such as HPVs 16 and 33, is significant because of the clinical relevance of infections by these high-risk types. In fact, HPVs 16 and 33 are among the five high-risk types which are found most frequently in high-grade cervical lesions and cervical cancer cases [Bosch et al., 2008; de Sanjose et al., 2010] and HPV 16-positive atypical squamous cells of undetermined significance (ASCUS)/low-grade squamous intraepithelial lesions (LSIL) have an increased risk to progress to high-grade lesions [Castle et al., 2005].
In the case of the INNO-LiPA, it was more difficult to interpret the discordant cases. In fact, while in the Linear Array each hybridization band corresponds to one genotype (with the exception of HPV 33, 35, and 58 that require the presence of a type-specific band together with a cross-reactive band), by the INNO-LiPA samples are scored positive for certain genotypes only when a combination of two or more hybridization lines is observed. Thus, discordant type-specific profiles were due to the absence of one or more lines that form the specific hybridization pattern of a certain genotype.
DNA concentration in the several extracts was not evaluated in the current study. However, it is plausible that the overall amount of DNA recovered from each extraction procedure and then used for PCR was different. Nevertheless, it is also possible that the observed differences in the genotyping results depend on a differential recovery of different HPV genotypes achieved using different extraction methods. The preferential amplification of certain types, present at higher concentration, may play an important role in determining non-identical genotyping results for all the extracts of the same sample. Dunn et al. [2007] have shown that even small variations in the extraction protocol, for instance in the centrifuge speed used to process the PreservCyt cervico-vaginal samples, determine a difference in the Linear Array performance.
The Linear Array results showed higher concordance when the two manual extraction protocols were compared. Roche-recommended protocol of extraction (A) is based on a two-step lysis followed by DNA purification on a QIAGEN vacuum manifold. The QIAGEN protocol (B) is based on a four-step procedure (lyse, bind, wash, and elute), which is similar to the Roche protocol. The automated nucleic acids extraction method (C) also includes similar steps, but it is based on the use of magnetic silica particles instead of silica columns and entails multiple washes. It is possible that the effectiveness of the two manual methods in yielding DNA samples suitable for amplification was comparable in terms of quantity and quality of the DNA sample, while the automatic procedure might have had an overall different performance.
Published data indicate that the use of an automated DNA extraction system prior to amplification may increase the detection of microbiological agents from several clinical samples when compared with manual techniques, possibly because of a higher DNA yield or fewer inhibitors [Loens et al., 2007; Pillet et al., 2009]. The present results, although obtained from a limited number of patients, also suggest a superior performance of the automated procedure (C) compared to the Roche-recommended method (A). In fact, considering the seven samples with discordant Linear Array results for extracts A and C, in five cases the Linear Array detected a higher number of HPV types in extract C (Table III). On the other hand, when comparing the automated extraction (C) with the QIAGEN method (B), extra genotypes were detected in extracts C and B in an equal number of cases.
Possible variability introduced by the use of different volumes of the original cervico-vaginal samples for DNA extraction and other possible, random sources of variations need to be taken into account. In spite of these limits, the present study confirms previous results showing that changes in the DNA extraction method modify HPV genotyping tests performance [Dunn et al., 2007]. Thus, the consistent use of the same DNA isolation protocol is recommended, in order to not have an extraction-dependent variability.
In conclusion, these preliminary data suggest that DNA extraction is a critical step for HPV genotyping tests, although a larger sample size is needed in order to evaluate thoroughly how different DNA extraction protocols affect these assays and to obtain a statistical validation of the results.