Investigating an outbreak of acute viral hepatitis caused by hepatitis E virus variants in Karachi, South Pakistan
Abstract
Hepatitis E is a classic water-borne disease in developing countries. Detection of anti-HEV IgM and IgG antibodies, in addition to HEV RNA are useful epidemiological markers in diagnosis of hepatitis E. This study was conducted to investigate an outbreak of acute viral hepatitis in South-Pakistan. Anti-HEV IgM and IgG were assessed comparatively with serological kits manufactured by Abbott, Cosmic, TGH, and Wantai, selecting HEV RNA as reference assay. Molecular evolutionary analysis was performed by phylogeny and HEV spread time analysis by Bayesian Coalescent Theory approach. Of the 89 patients, 24 (26.9%) did not have acute hepatitis viral marker. Of the remaining 65 cases, 4 (6.1%) were positive for anti-HAV IgM, one (1.5%) for anti-HBc IgM, 2 (3%) for HCV, 53 (81.5%) for anti-HEV IgM, and 5 (7.7%) were hepatitis-negative. The Wantai test was 100% sensitive and specific followed by Cosmic (98.1% and 100%), TGH (98.1% and 97.2%) and Abbott (79.2% and 83.3%). Two HEV variant strains were detected by phylogeny responsible for this acute hepatitis outbreak. Estimates on demographic history of HEV showed that HEV in Pakistan has remained at a steady nonexpanding phase from around 1970 to the year 2005, in which it expanded explosively with the emergence of new HEV variants. In conclusion, the limited sensitivity of available assay (Abbott anti-HEV EIA) may be a concern in HEV diagnosis in Pakistan. This study cautions that the dissemination of the variant strains to other areas of Pakistan may lead to explosive HEV outbreaks. J. Med. Virol. 83:622–629, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Hepatitis E is an important public health concern in many developing countries particularly in Asia, Africa, and Latin America. Hepatitis E virus (HEV) has been responsible for a substantial proportion of sporadic (non-epidemic) cases of acute hepatitis as well as large water-borne epidemics related to poor hygiene and sanitation [Emerson and Purcell, 2003; Kar et al., 2008]. Although the disease is self-limiting and no chronic sequelae or carrier state has been documented in the general population, but it can acquire chronic/carrier state in immune compromised patients [Kamar et al., 2008].
HEV is a non-enveloped virus and its genome is a single-stranded, positive-sense RNA, which is capped and polyadenylated [Tam et al., 1991; Kabrane-Lazizi et al., 1999]. The genome is approximately 7.2 kb and contains three open reading frames (ORF1–3) [Tam et al., 1991]. ORF1 encodes non-structural proteins including the helicase and RNA-dependent RNA polymerase [Agrawal et al., 2001]. ORF2 and ORF3 overlap, and the ORF2 and ORF3 proteins are translated from a single bicistronic subgenomic RNA [Graff et al., 2006]. The ORF2 protein is the viral capsid protein, whilst the ORF3 protein is essential for virion egress from infected cells [Yamada et al., 2009].
HEV have been classified into four genotypes and regardless of the genotype of the virus or the country of origin, it appears to have one single serotype [Emerson and Purcell, 2003]. Most of the enzyme immunoassays (EIAs) for HEV infection are based on either recombinant HEV proteins or synthetic peptides. Serological detection of antibodies to the virus even of different origins is possible by relying on major epitopes derived from the ORFs of the virus and is the established procedure for the diagnosis of acute HEV infection [Seriwatana et al., 2002]. However, epidemiological studies require the detection of HEV RNA by PCR in addition to anti-HEV IgM and IgG antibodies, especially in outbreak investigations.
HEV along with hepatitis A virus (HAV) infections are the most common cause of viral hepatitis in Pakistan in the general population [Hamid et al., 1996, 2002; Rab et al., 1997]. HAV infection has been shown to cause severe illness in adult patients with underlying chronic liver disease (CLD). However, vaccination against HAV has reduced substantially this risk. HEV-induced acute hepatitis may also be fulminant [Hamid et al., 1996; Peron et al., 2007] and like HAV, HEV could be expected to cause a severe illness in patients with underlying CLD. Characteristically, in pregnant women, the illness is particularly severe and carries a high case fatality rate (15–25%) [Hamid et al., 1996; Bhatia et al., 2008].
A number of reports revealed different outbreaks of acute viral hepatitis in Pakistan associated primarily with HEV [Malik Iftikhar Ahmed, 1996; Rab et al., 1997]. In this study, the samples were collected from Karachi, the industrial capital and the largest metropolitan city in South-Pakistan. The aims of this study were to investigate: (1) the etiologies of acute viral hepatitis (2) serological diagnosis of HEV and (3) the demographic history of HEV in Pakistan.
MATERIALS AND METHODS
Patients
Between December 1, 2007, and July 31, 2008, the consecutive patients (n = 89) attending the outpatient and inpatient clinics of The Aga Khan University (AKU), who presented symptoms associated with acute liver disease, were included in the study. The most common symptoms included jaundice, fatigue, weakness, nausea, and a loss of appetite and in some cases bleeding, ascites and liver encephalopathy.
Serological Diagnosis
All patients were screened by serological tools for hepatitis A–D virus infections. HBsAg, anti-HCV, anti-HDV, and anti-HAV IgM were tested in each sample by Abbott enzyme immunoassays (EIAs) (Abbott Laboratories, Chicago, IL). Four different serological assays, that is, Abbott (Abbott Laboratories), Cosmic (Cosmic Viragent, Tokyo, Japan), TGH (Toshiba General Hospital, Tokyo, Japan) and Wantai (Wantai Co. Ltd, Beijing, China) were evaluated comparatively for the detection of anti-HEV. HEV RNA detection by reverse transcriptase PCR (RT-PCR) was selected as the reference assay to analyze the specificity and sensitivity of each test. To rule out the ambiguity in HEV RNA detection, samples were also tested for anti-HEV IgA, as this test is very sensitive in nature for detection of acute HEV infections and may be a choice marker for differentiating acute infections [Mitsui et al., 2005; Takahashi et al., 2005; Elkady et al., 2007]. A case of HEV infection was considered, if it is positive for both HEV RNA and anti-HEV IgA, where “early acute infection” was defined if a case is negative for anti-HEV IgG, while “acute infection” was defined if it is positive for anti-HEV IgG. The sensitivity of an anti-HEV IgM assay was defined as the percentage of anti-HEV IgM positivity from among the HEV cases showing the profiles of early acute or acute infection. The specificity of an anti-HEV IgM assay was defined as the percentage of negative samples among those negative to HEV RNA showing the profiles of only previous infection or not infected. Anti-HBc IgM in samples was tested by ELISA (Fujirebio, Inc., Tokyo, Japan) and anti-HIV using the Jinedia HIV-1/2 Mix PA kit (Bio-Rad, Fujirebio, Inc., Tokyo, Japan). Biochemical markers such as alanine-amino transferase (ALT), aspartate-aminotransferase (AST), alkaline phosphatase (ALP) and bilurubin levels were measured in all samples.
Informed consent was obtained at the time of blood sampling from each patient included in the study. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the ethical committee of the institution.
Detection of HEV RNA and HEV Genotyping
Total RNA was extracted from the serum samples using the SepaGene RV-R Nucleic acid extraction kit (Sanko Junyaku Co., Ltd, Tokyo, Japan) in accordance with the manufacturer's protocol. Viral RNA were reverse transcribed to complementary DNA using SuperScript II RNase H_Reverse Transcriptase (Invitrogen Corp., Carlsbad, CA) and random hexamer primer (Takara Shuzo Co. Ltd, Tokyo, Japan) as described previously [Ohno et al., 1997]. Screening of HEV RNA was carried out with a specific set of screening primers targeting a partial nucleotide sequence of ORF1 region of the HEV genome, as described previously [Takahashi et al., 2003]. The sequences spanning 326 nt in ORF1 and 247 nt in ORF2 of HEV RNA corresponding to nt 125–450 and nt 6050–6296 of the Nepali isolate; GenBank accession number AF015830 was obtained for HEV genotyping. A sequence of 868 nt in the RNA-dependent RNA polymerase region (corresponding to nt 3918–4785 of AF051830) was also obtained for HEV RNA positive cases for molecular evolutionary analysis.
Detection of HBV DNA
HBV DNA was extracted by a QIAamp DNA Blood Mini Kit (Qiagen, Inc., Hilden, Germany) from 100 ml of each HBsAg positive serum. Partial core and S regions were amplified in order to detect HBV DNA in the samples using the primers described previously [Sugauchi et al., 2001]. The detection limit for this study was 100 copies/ml [Tanaka et al., 2004].
Sequencing and Phylogenetic Analysis
The amplicons obtained were sequenced directly with Prism Big Dye (Applied Biosystems, Foster City, CA) in an ABI 3100 DNA automated sequencer. The sequences for phylogenetic analysis were retrieved from DDBJ/EMBL/GenBank. Alignments were performed using CLUSTALW (http://clustalw.ddbj.nig.ac.jp/top-e. html) and neighbor-joining trees were constructed with 6-Parametric method and bootstrapped 1,000 times to confirm the reliability of the phylogenetic tree [Shin et al., 2008].
The nucleotide sequence data reported in this paper appear in the DDBJ/EMBL/GenBank nucleotide sequence database with the accession numbers AB513496–AB513616.
Molecular Evolutionary Analyses
A reconstructed tree was built on the ORF1 sequence of 868 nucleotides of partial RNA polymerase region (RDRP) of HEV genome by Mega using Pakistan isolates and all the available reference sequences retrieved from databases (DDBJ/EMBL/GenBank). To estimate virus effective population sizes through time, a coalescent-based approach was used which is implemented in a Bayesian Markov Chain Monte Carlo (MCMC) inference framework in the program BEAST [Lemey et al., 2009]. All possible combinations of the relaxed [Drummond et al., 2006] and strict molecular clock models with Bayesian skyline [Drummond et al., 2005], constant, exponential, and logistic growth coalescent models were analyzed. The Bayes Factors (BF) were estimated for each pair of models, as implemented in Tracer v1.4 and suggested previously [Suchard et al., 2001]. MCMC sampling was performed for at least 1 × 107 generations, sampling a tree every 1,000 generations. HKY model of nucleotide substitution was used with gamma distribution, five rate categories and a substitution rate of 0.84 × 10−3 as described by Tanaka et al. [2006]. The program Tracer (http://tree.bio.ed.ac.uk) was used to check for convergence and to determine whether appropriate mixing of the posterior target distribution had been achieved (effective sample size >100).
RESULTS
Etiology of Acute Hepatitis
The time lag between the start of symptoms of acute infection and the collection of samples in this study was 6.9 ± 4.7 days. Of the 89 patients, 24 (26.9%) had chronic HBV infection with no detectable acute viral marker. Of the remaining 65 cases, 4 (6.1%) were positive for anti-HAV IgM, 1 (1.5%) was positive for anti-HBc IgM, 2 (3%) were positive for HCV, 53 (81.5%) were positive for HEV RNA, and 5 (7.7%) were hepatitis-negative (Table IA). Serological assays (Anti-HEV IgM and IgG) of Abbott, Cosmic, TGH, and Wantai were evaluated comparatively selecting HEV RNA detection by PCR as a reference. The Wantai test was 100% sensitive and specific followed by Cosmic (98.1% and 100%), TGH (98.1% and 97.2%) and Abbott (79.2% and 83.3%) (Table IB). The results of anti-HEV IgA testing for subset of samples (n = 17) with non-specific or not-sensitive IgM results were in complete agreement with the results of HEV RNA detection by PCR (data not shown). There were two cases (3%) with early acute infection, which were detectable by all kits except Abbott. Five patients (7.7%) were negative for all hepatitis viral markers. Underlying HBV infection with detectable HBV DNA was observed in 10 (15.4%) of acute hepatitis E patients. The baseline and clinical features of these 65 patients with acute liver disease are summarized in Table IA.
| Features | Total | AHA | AHB | AHE | HCV | NABCDE |
|---|---|---|---|---|---|---|
| Age (years) | 30.9 ± 14.5 | 26.5 ± 11.4 | 19 | 29.4 ± 11.3 | 32.6 ± 9.4 | 31.8 ± 23 |
| Gender (male) | 43 | 2 | 0 | 38 | 2 | 3 |
| ALT, IU/L (mean ± SD) | 1,630.6 ± 1,687.5 | 1,677.7 ± 870.5 | 2,140 | 2,146.4 ± 1,570.4 | 819.3 ± 683.5 | 2,301.8 ± 3,171.8 |
| AST, IU/L (mean ± SD) | 1,955 ± 1,986.3 | 2,087.3 ± 977.6 | 2,140 | 1,893.4 ± 1,900.3 | 651.6 ± 531 | 2,645 ± 3,609.5 |
| ALP, IU/L (mean ± SD) | 171.8 ± 106.4 | 150 ± 60.3 | 133 | 160.9 ± 70.4 | 106.3 ± 17 | 333.8 ± 270 |
| GGT, IU/L (mean ± SD) | 119.8 ± 160.1 | 112 ± 58 | 182 | 103.9 ± 95 | 55 ± 17.4 | 431.8 ± 531.9 |
| Total Bil. mg/dl (mean ± SD) | 11.4 ± 14.1 | 9.3 ± 3.2 | 3 | 11.3 ± 10.2 | 12.5 ± 12.8 | 3.6 ± 3 |
| Anti-HAV IgM, n (%) | 4 (6.1) | 4 (100) | 0 | 0 | 0 | 0 |
| Anti-HEV IgM, n (%) | 53 (84.1) | 0 | 0 | 53 (100) | 0 | 0 |
| Anti-HEV IgG, n (%) | 52 (82.5) | 1 (25) | 0 | 51 (96.2) | 1 (50) | 5 (100) |
| HEV RNA, n (%) | 53 (84.1) | 0 | 0 | 53 (100) | 0 | 0 |
| Anti-HBc IgM, n (%) | 1 (1.5) | 0 | 1 (1.5) | 0 | 0 | 0 |
| HBV DNA, n (%) | 11 (17.4) | 0 | 1 (100) | 10 (18.9) | 0 | 0 |
| Total | 65 | 4 | 1 | 53 | 2 | 5 |
| Tests | Sensitivity% (+/tested) | Specificity% (−/tested) |
|---|---|---|
| Abbott | 79.2 (42/53) | 83.3 (30/36) |
| Cosmic | 98.1 (52/53) | 100 (36/36) |
| TGH | 98.1 (52/53) | 97.2 (35/36) |
| Wantai | 100 (53/53) | 100 (36/36) |
Phylogenetic Analysis
In order to analyze the genomic heterogeneity of HEV, phylogenetic trees were constructed in two partial ORF1 parts and in ORF2 of the HEV genome against all sequences available in the database. All sequences corresponded to genotype 1 clustering in two groups referred from now “variant 1” and “variant 2.” Phylogenetic trees based on alignment of the ORF1 sequences suggested that the strains were related more closely to the strains from Nepal and India. The topology of the tree obtained either the 5′ terminal end of ORF1 or in RNA dependent RNA polymerase (RDRP) region of ORF1 was similar to that obtained in the 3′terminal sequence of the ORF2 (Fig. 1A–C). Phylogenetic tree in RDRP region distinctly separates the both clusters with high bootstrap values (Fig. 1C). The consistent high bootstrap values within the clusters describe the homology of the strains with each other, a characteristic of epidemic.
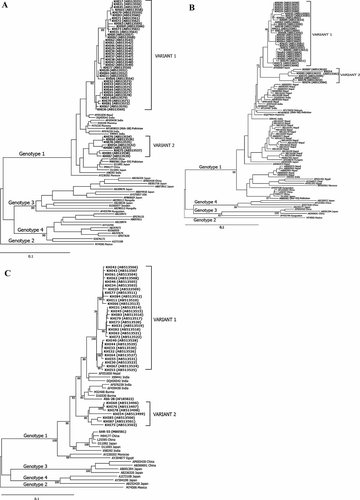
A–C: Phylogenetic trees constructed (A: ORF1 region 326nt) (B: ORF2, 247nt) (C: ORF1-RDRP region, 868 nt) of HEV genome. Pakistan isolates are in bold letters, aligned with all the available reference sequences retrieved from databases (DDBJ/EMBL/Gene Bank). Variant strains isolated in this study are represented in bold letters showing isolate name (database reference number). The numbers in the tree indicate bootstrap reliability by the interior branch test. Exceptional strains are indicated according to their area of origin.
Sequence Analysis in 5′Terminus of ORF1, RDRP Region of ORF1, and 3′Terminus of ORF2
The comparison of the ORF1 and ORF2 nucleotide sequences obtained in this study with all previously published strains indicated that the HEV variant 1 was more closely related to Nepalese strain (96–97% identity), followed by Burma (94–96% identity), Indian (93–95% identity), and previous Pakistani strains (90–95% identity; Table II). The HEV variant 2 of this study indicated an almost similar degree of identity (94–95%) with Nepalese, Burma, Indian, and previous Pakistani strains in ORF1 but differed substantially in ORF2 with only 89–91% of identity. These sequences were divergent significantly from Mexican, Chinese, and Japanese strains of HEV genotype 2, 3, 4 strains, respectively (80–86% identity). Interestingly, the amino acid sequence comparisons did not show any significant difference from other Asian strains (96–100% identity) (Table II). Cumulatively these results indicate that the new variant strains although have much nucleotide variations in different regions of HEV genome from previous Asian strains but these nucleotide variations are more or less synonymous ones.
| HEV strain | Genotype | Percentage identity nt. (a.a) | |||
|---|---|---|---|---|---|
| ORF1, 326 nt | ORF2, 247 nt | ||||
| Variant 1 | Variant 2 | Variant 1 | Variant 2 | ||
| Pakistan, M80581 | 1 | 92 (100) | 95 (99) | 90 (100) | 90 (96) |
| Pakistan, AF185822 | 1 | 93 (100) | 95 (98) | 95 (100) | 90 (96) |
| India, X99441 | 1 | 93 (98) | 94 (96) | 95 (100) | 89 (96) |
| Nepal, AF051830 | 1 | 96 (100) | 95 (98) | 97 (100) | 91 (96) |
| Burma, D10330 | 1 | 94 (99) | 95 (97) | 96 (100) | 91 (96) |
| Mexico, M74506 | 2 | 80 (94) | 83 (93) | 86 (85) | 85 (88) |
| China, AP003430 | 3 | 81 (91) | 82 (88) | 85 (98) | 87 (94) |
| Japan, AJ272108 | 4 | 81 (92) | 80 (90) | 82 (96) | 80 (93) |
Historical Analysis of HEV Population
Evolutionary analysis was performed in BEAST under a range of molecular clock and coalescent model combinations [Magiorkinis et al., 2009; Pybus et al., 2009], using the sequences in the most conserved RDRP region of HEV genome. In each case, the best fitting model was the relaxed molecular clock model plus the Bayesian skyline demographic model. The Bayesian skyline plot summarizes the spread and epidemic growth of HEV in Pakistan (Fig. 2). It shows clearly that HEV genotype 1 was in a steady nonexpanding phase from around 1970 (the lower 95% credible interval of the tMRCA) to the year 2005, in which it expanded explosively with the emergence of new HEV variant strain. The growth phase is preceded by a reduction in the effective number of infections (Fig. 2). However, this reduction is not significant given the size of the estimated confidence limits.
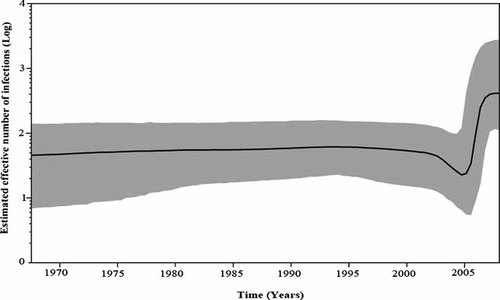
Hepatitis E population dynamics in Pakistan based on relaxed-clock analysis of ORF1-RDRP region. The dark line in Bayesian skyline plot shows the estimated effective population size through time. The gray area represents the 95% highest posterior density confidence intervals for this estimate.
DISCUSSION
The IgM class of antibody to the virus is considered as the specific marker for differentiating the acute from the convalescent phase of an infection [Ma et al., 2009]. Consecutive patients (n = 89) presenting with symptoms of acute liver diseases were tested to differentiate HAV, HBV, and HEV acute infections. Of the 89 patients, 24 (26.9%) did not have detectable acute viral marker however were detected with chronic HBV infection. HEV appeared to be the major etiological agent detected positive in 81.5% of the remaining 65 cases with acute viral hepatitis.
Although it has been understood that HEV outbreaks in developing nations are waterborne and occur in areas with poor sanitation, the only available and rapid methods in most of these countries is serological testing for HEV (anti-HEV IgM/IgG). Reliable serological tests are thus indispensible in such situations and the periodic evaluation of already available assays with new ones is therefore necessary not only for the management of hepatitis E but also for studying the epidemiology of disease. Given the findings that the viremia in acute HEV infection may prolong for a period of ≥2 weeks from the onset of illness [Clayson et al., 1995; Myint et al., 2006] and in the absence of reference serological assay, HEV RT-PCR was selected as the reference assay for HEV detection in this study. Among the four EIAs for detecting IgM anti-HEV, the Wantai assay showed 100% precision. The Cosmic assay showed 99.7% concordance with the Wantai IgM assay, while Abbott IgM assay was the least suitable assay in detection of these HEV variants. The difference in the detection among these assays may be attributed to the difference in the strain-specific antigenic domains included in these tests [Ma et al., 2009]. However, the difference in detections among these tests was observed for variant-1 only. To rule out the ambiguity in detection of HEV RNA, anti-HEV IgA was tested in the subset of samples with non-specific or not-sensitive results. IgA is the class of antibody, which is elicited during the acute phase of viral infections including HEV [Elkady et al., 2007], which was found to be detectable for a longer time of infection even at the stage when HEV RNA is no longer detectable [Mitsui et al., 2005; Takahashi et al., 2005]. Results of anti-HEV IgA showed 100% concordance with HEV RNA detection. One of the limitations of this study was that the performance of all serological assays was assessed for symptomatic infections. Cases of asymptomatic HEV infection in subclinical forms of infection exist and they are thought to exceed icteric infections in HEV outbreak [Clayson et al., 1995; Myint et al., 2006]. Further studies in the anti-HEV testing therefore could be vital in investigations to identify all infected persons rather than just those with clinical disease.
HEV strains belong to a single serotype and display considerable genetic diversity according to the time and place of isolation [Lu et al., 2006]. In order to gain insight into the genetic variability and mode of evolution of HEV in Pakistan, the HEV strains were sequenced in two parts of ORF1 (5′end of ORF1 and RDRP region) and the hyper-variable region of the ORF2. The data were consistent for all three parts, showing two independent and distinct phylogenetic clusters of HEV within genotype 1. These new strains were related more closely to the strains from Nepal, India, and Burma rather than previous Pakistan strains [Bryan et al., 2002; Shrestha et al., 2004; He, 2006], indicating that different epidemiological strains may be circulating in different regions of Pakistan. The other explanation may be that HEV has been in continuous spread in Pakistan and it had continued evolution in infected individuals in Pakistan, leading to the observed genomic variability. The fact that no significant amino acid substitutions were observed indicates that genomic mutations of HEV may occur naturally in infected individuals without significant immunological pressure from the host and that selective forces that do not allow amino acid substitutions may be involved in observed pattern of divergence [Shrestha et al., 2004].
To estimate the virus affecting the population through time, a coalescent-based approach was used which is implemented in a Bayesian Markov Chain Monte Carlo (MCMC) inference framework in the program BEAST [Drummond et al., 2006; Lemey et al., 2009; Pybus et al., 2009]. BEAST incorporates uncertainty in the phylogeny by integrating across tree topologies to estimate relative genetic diversity, an indicator of effective population size under a neutral evolutionary process. Estimates show that HEV genotype 1 was in a steady nonexpanding phase in Pakistan, from around 1970 (the lower 95% credible interval of the tMRCA) to the year 2005, in which it spread explosively with the emergence of new HEV variant strains. The growth phase is preceded by a dip in the effective number of infections. This could be due to a combination of epidemiological processes-stochastic or spatial effects that result in delay of exponential growth of emerging population due to reduced mean growth rates [Lande, 1998; May et al., 2001; Pybus et al., 2003]. However, this dip is not significant given the size of the estimated confidence limits. HEV has been reported in Pakistan as early as the 1950s and 1960s to the early 1990s during which HEV was found to be single main cause of at least 70% of acute hepatitis cases mainly affecting the adult population [reviewed by Malik Iftikhar Ahmed, 1996]. In 1994, a massive outbreak of HEV (December 1993–March 1994) occurred in Islamabad, the capital of Pakistan [Iqbal et al., 1989]. The epidemic was also reported in Lahore in December of the same year, when 283 cases were admitted to the army hospital at the same time [Malik Iftikhar Ahmed, 1996].
The epidemic history of these newly emerged Pakistan strains of HEV in the year 2005 is evidenced by the gastroenteritis outbreak which occurred in Karachi, Pakistan in summer (June–September) 2005 (http://www.dawn.com/2005/05/26/local10.htm), associated with sewage-contaminated water supply. Tens of thousands of people were affected by this gastroenteritis outbreak, which masked apparently the HEV outbreak and the newly emerged strain remained undetected and continued to spread in the community in an unrecognized form causing acute hepatitis.
In South Asia both HAV and HEV are considered as the most common causes of acute hepatitis. However, HEV has been the leading etiological agent responsible for large-scale epidemics in Pakistan [Malik Iftikhar Ahmed, 1996; Rab et al., 1997]. In this study, HEV variants were found responsible for acute hepatitis outbreak in Karachi, South-Pakistan. Limited sensitivity of available serological assay in Pakistan (Abbott anti-HEV EIA) may be a concern in diagnosing early acute infections and outbreak investigations. This study cautions that HEV variant strains have great epidemic potential and their dissemination to other areas in the country may lead to explosive HEV outbreaks.




