Characterization of HIV-1 from patients with virological failure to a boosted protease inhibitor regimen
Abstract
The use of highly active antiretroviral treatment (HAART) regimens with unboosted protease inhibitors (PIs) has resulted in a high level of virological failure primarily due to the development of resistant virus. Current boosted PI regimens combine successfully low-dose ritonavir (r) with a second PI. The aim of the study was to estimate the proportion of patients, in a population based setting, who develop virological failure on a PI/r regimen. Through The Danish HIV Cohort Study 1,007 patients who received PI/r based treatment between 1995 and 2008 were identified. Twenty-three (2.3%) experienced virological failure, of whom 19 (83%) started PI/r treatment before 2001. Patients from Copenhagen (n = 19) were selected to study the development of protease (PR) and gag cleavage site (CS) mutations during PI/r treatment and PI plasma levels at the time of virological failure. Three patients (16%) developed major PI resistance mutations. Mutations in the p7/p1 and p1/p6 gag CS only developed in patients with major or minor mutations in PR. Drug concentrations were low or undetectable in 10 out of the 19 patients. In total PR resistance mutations and low drug levels could account for 12 (63%) of the failure cases. In conclusion, virological failure to PI/r is a low and decreasing problem primarily caused by low plasma drug levels and to a lesser extent major PR mutations. Gag CS mutations did not contribute significantly to resistance development and virological failure. J. Med. Virol. 83:377–383, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
During maturation of the human immunodeficiency virus type 1 (HIV-1) viral particle the protease (PR) enzyme cleaves the gag polyprotein into functional proteins essential to the infectious virus [Loeb et al., 1989; Katz and Skalka, 1994; Mammano et al., 1998]. The gag polyprotein contains five specific protease cleavage sites (CS), p24/p17, p17/p2, p2/p7, p7/p1, and p1/p6 with different affinity for the enzyme. These are cleaved at specific rates and order to obtain the mature infectious viral particle. Protease inhibitors (PIs) block the cleavage activity of PR by binding to the active site of the enzyme and thereby hinder the maturation of the virus [Ashorn et al., 1990; McQuade et al., 1990].
Highly active antiretroviral treatment (HAART) regimens with unboosted PIs have shown a high level of virological failure primarily due to the development of resistant virus. Resistance towards PIs arises when specific major resistance mutations appear in the PR gene, causing a conformational change in the enzyme's active site. This lowers the affinity to the PIs and allows for cleavage to occur. Minor mutations develop secondarily to restore loss of viral fitness and replication capacity induced by the major mutations [Gulnik et al., 1995; Borman et al., 1996; Croteau et al., 1997; Nijhuis et al., 1999; Mammano et al., 2000].
Current boosted PI regimens combine low-dose ritonavir with a second high-dose PI (PI/r). This decreases the degradation rate by the cytochrome P450 enzyme in the liver and increases blood and intracellular levels of the PIs [Moyle and Back, 2001] which has proven to decrease the risk of virological failure and resistance development [Cooper et al., 2003; Lima et al., 2008].
A previous study [Ananworanich et al., 2006] found a low prevalence (3.8%) of virological failure in patients on PI/r regimens. von Wyl et al. 2007 found that only 30% of PI/r failure patients harbored resistance mutations and Ananworanich et al. 2006 found no resistance mutations in nine patients with PI/r failure. The major reasons for failure on PI/r regimens in the absence of resistance have not been determined, though theories such as altered absorption and metabolism of the PIs in patients failing PI/r have been suggested as an explanation for the low plasma concentrations and subsequent virological failure seen in some patients [Zazzi, 2007].
Since gag is the natural substrate for PR, mutations in or around the gag cleavage sites have been linked to PI resistance development. Some studies assign a compensatory fitness role to gag CS mutations [Doyon et al., 1996; Zhang et al., 1997; Bally et al., 2000], whereas others found resistance development based exclusively on mutations in gag [Prabu-Jeyabalan et al., 2004; Verheyen et al., 2006; Nijhuis et al., 2007]. Some studies have looked in gag outside the CS and found non-CS mutations leading to improved viral replication [Gatanaga et al., 2002; Myint et al., 2004; Aoki et al., 2009]. Ghosn et al. 2009 suggested that the emergence of certain gag CS mutations could predict virological failure. Though the exact role of gag CS and non-CS mutations is yet to be determined, it is widely accepted that specific mutations such as A431V and I437V in p7/p1 and R452S in p1/p6 can contribute to continued viral replication in the presence of PIs [Zhang et al., 1997; Myint et al., 2004; Verheyen et al., 2006; Malet et al., 2007; Nijhuis et al., 2007].
The purpose of the present study was to estimate the proportion of patients, in a population based setting, who develop virological failure on a PI/r based regimen. A group of patients was selected for molecular analysis to study the development of PR and gag mutations during PI/r treatment as well as the plasma concentration levels of the PIs at the time of virological failure.
PATIENTS AND METHODS
Setting
The estimated HIV-1 prevalence among adults in Denmark is 0.07% [Lohse et al., 2005]. About 60% of the patients are infected with subtype B virus [Jorgensen et al., 2003]. Medical care, including antiretroviral treatment (ARV), is tax-paid and provided free-of-charge to all residents of Denmark infected with HIV-1. During the study's follow-up period, national criteria for initiating HAART were HIV-1-related disease, acute HIV-1 infection, pregnancy, CD4 cell count <300 cells/µl, and, until 2001, plasma HIV-1-RNA >100,000 copies/ml.
Study Population
Patients were identified through The Danish HIV Cohort Study, described earlier [Lohse et al., 2005]. In brief, this cohort encompasses all patients infected with HIV-1 seen at Danish HIV treatment clinics since 1st of January 1995. The Danish HIV Cohort Study is approved by the Danish Data Protection Agency (J-no: 2001-41-0624).
All patients who, as of December 31st 2008, had received treatment with a PI/r regimen including Saquinavir, Indinavir, Atazanavir, or Lopinavir and with no prior failure to unboosted PI were identified from the cohort. Patients with previous virological failure to a non-PI containing regimen were included in the study population.
The patients with virological failure while compliant with a boosted PI regimen were identified from the study population. Virological failure was defined as plasma HIV-1-RNA >400 copies/ml in two consecutive measurements minimum 4 weeks apart after at least 3 months of compliance to a continuous PI/r regimen or after HIV-1-RNA had been <20 copies/ml. Patient files of the failure patients were scrutinized to evaluate compliance. For almost all patients with virological failure or rebound the reasons for this had been discussed with the patient. Patients who had stopped treatment or who reported regularly missing treatment doses were excluded. Information on the degree of compliance (i.e., percentage of missed doses) had not been collected systematically.
For the molecular analysis of the pol and gag genes, all failure patients seen at the two HIV treating centers in Copenhagen, managing 2/3 of the Danish HIV patients, were selected. Plasma samples obtained at or after the time of failure, and baseline samples obtained prior to any PI treatment were analyzed. Failure samples taken more than 2 months after the failure criteria had been fulfilled were only included if the patient received the same PI/r treatment at failure and sample date.
The study was approved by The Danish National Committee on Biomedical Research Ethics (H-D-2009-017). Informed consent was obtained from each patient.
PCR and Sequencing
Sequences of the pol region were extracted from the Danish HIV Sequence Database housed at the Department of Virology, State Serum Institute, which includes sequences from resistance tests performed on samples from Danish HIV patients from 2000 to 2008. The database currently includes approximately 5,000 sequences from 1,200 patients. The Danish HIV Sequence Database is approved by the Danish Data Protection Agency (J-no: 2006-41-6709).
If the sequence was not available from the database, RNA from stored plasma samples was extracted using QiaAmp viral RNA mini kit (Qiagen, Hilden, Germany). The pol gene including the PR and reverse transcriptase (RT) regions was amplified and sequenced using a previously described in house nested RT-PCR method amplifying 1,302 bp [Madsen et al., 2007].
Amplification of the complete gag gene was done using primers JA152 [Leitner et al., 1995] and XM1_mod [modified from Gatanaga et al., 2002] for both cDNA (65°C, 10 min; 42°C, 5 min; 42°C, 1 h, 99°C, 3 min) and first round of PCR (94°C, 10 min; 94°C, 30 sec; 65°C, 30 sec; 72°C, 1 min) 40 cycles (72°C, 10 min). For the second round of PCR (94°C, 30 sec; 65°C, 30 sec; 72°C, 1 min) 30 cycles (72°C, 10 min) primers LF3 [Gatanaga et al., 2002] and TVM1_R 5′-CCTCCAATTCCCCCTATCATTTTTG-3′ (in-house) were used, generating a 1,723 bp PCR-fragment including all five PR cleavage sites.
The gag sequences were obtained using BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) with primers LF3, TVM1_R, TVM_seq1 5′-AAGACACCAAGGAAGC-3′, TVM_seq2 5′-TTCCTAGGGGCCCTGCAATT-3′, TVM_seq3 5′-ACTTTTACCCATGCATT-3′, and TVM_seq4 5′-ACATTAGAAGAAATGATG-3′ (all in-house).
The population based sequences were run on an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems).
Sequence Analysis
Sequences (Genbank accession numbers: AM933271–AM933291; AM933522–AM933542) were analyzed using SeqScape v2.5 (Applied Biosystems) and Bionumerics v5.0 (Applied Maths, Sint-Martens-Latem, Belgium) and aligned using Clustal W in Geneious v4.7 (Biomatters Ltd., Auckland, New Zealand). Phylogenetic neighbor-joining tree analyses for subtyping and quality control were carried out using Geneious v4.7 (Biomatters Ltd.). Sequences from the Danish HIV Sequence Database and subtype specific references from the Los Alamos Sequence Database (http://www.hiv.lanl.gov/) were included as reference sequences.
Resistance associated mutations in pol according to the Stanford HIV Drug Resistance Database [Liu and Shafer, 2006; last updated June 10, 2008] were identified. Full gag sequences from pre-treatment and failure samples were aligned pairwise in order to detect mutations in the five CS regions that had developed during treatment. CS regions were defined by five amino acids (aa) on each side of the CS.
Plasma Concentrations
PI plasma concentrations from the time of virological failure were measured using a validated method based on high performance liquid chromatography [Justesen et al., 2003].
RESULTS
1,007 patients in The Danish HIV Cohort Study database had received ritonavir boosted regimens with Saquinavir, Indinavir, Lopinavir, or Atazanavir and had no prior failure to unboosted PI. Among these 1,007 patients 36 had experienced virological failure. For 13 of these incompliance was reported in the patient files. The remaining 23 patients (2.3%) had thus experienced virological failure according to the definition described in the “Methods” section.
Characteristics of the study population as well as the 23 patients experiencing virological failure are given in Table I. Virological failure was experienced by 2.7% of males (19 out of 707) versus 1.3% of females (4 out of 300). In total 521 patients started boosted PI after year 2000 of whom 4 (0.8%) experienced virological failure. This incidence was higher for the years 1995–2000 where 3.9% (19 out of 486) starting PI/r developed virological failure. For patients with a first-line regimen including PI/r 3.4% (18 out of 528) developed virological failure compared to 1.0% (5 out of 479) of the patients starting a non-PI/r regimen. The failure rates were similar in the groups of patients starting any treatment before and after 1995 (2.3% vs. 2.2%). The incidence rates of failure for each specific PI/r regimen were: boosted Saquinavir: 10.8/1,000 person years of follow up (PYR) (95% confidence interval 6.4–18.2) (total 1,297 PYR), boosted Indinavir 15.8/1,000 PYR (95% confidence interval 7.1–35.2) (total 380 PYR), boosted Lopinavir 1.4/1,000 PYR (95% confidence interval 0.2–9.7) (total 731 PYR), and boosted Atazanavir 4.5/1,000 PYR (95% confidence interval 1.1–17.8) (total 449 PYR).
| PI/r | Success | Failure | % | |
|---|---|---|---|---|
| Sex | ||||
| Male | 707 | 688 | 19 | 2.7 |
| Female | 300 | 296 | 4 | 1.3 |
| Origin | ||||
| Denmark | 719 | 703 | 16 | 2.2 |
| Europe | 46 | 44 | 2 | 4.3 |
| Africa | 148 | 144 | 4 | 2.7 |
| Other/unknown | 94 | 93 | 1 | 1.1 |
| Mode of transmission | ||||
| Homosexual | 427 | 419 | 8 | 1.9 |
| Heterosexual | 397 | 383 | 14 | 3.5 |
| Other/unknown | 183 | 182 | 1 | 0.5 |
| Start PI/r (year) | ||||
| 1996 | 33 | 31 | 2 | 6.1 |
| 1997 | 21 | 83 | 8 | 8.8 |
| 1998 | 74 | 71 | 3 | 4.1 |
| 1999 | 149 | 146 | 3 | 2.0 |
| 2000 | 139 | 136 | 3 | 2.2 |
| 2001 | 89 | 88 | 1 | 1.1 |
| 2002 | 48 | 47 | 1 | 2.1 |
| 2003 | 68 | 68 | 0 | 0.0 |
| 2004 | 87 | 86 | 1 | 1.1 |
| 2005 | 99 | 98 | 1 | 1.0 |
| 2006 | 70 | 70 | 0 | 0.0 |
| 2007 | 46 | 46 | 0 | 0.0 |
| 2008 | 14 | 14 | 0 | 0.0 |
| First-line treatment | ||||
| PI/r | 528 | 510 | 18 | 3.4 |
| Not PI/r | 479 | 474 | 5 | 1.0 |
| Start of any treatment | ||||
| Before 1995 | 365 | 357 | 8 | 2.2 |
| After 1995 | 642 | 627 | 15 | 2.3 |
| Total | 1,007 | 984 | 23 | 2.3 |
Nineteen patients attending either of the two clinics in Copenhagen experienced virological failure on a PI/r based treatment. These were selected for sequence analyses of the pol and gag regions at baseline and at time of virological failure (Table II). For two patients (patients 16 and 17), it was not possible to obtain gag sequences from the failure sample due to the restricted amount of stored material.
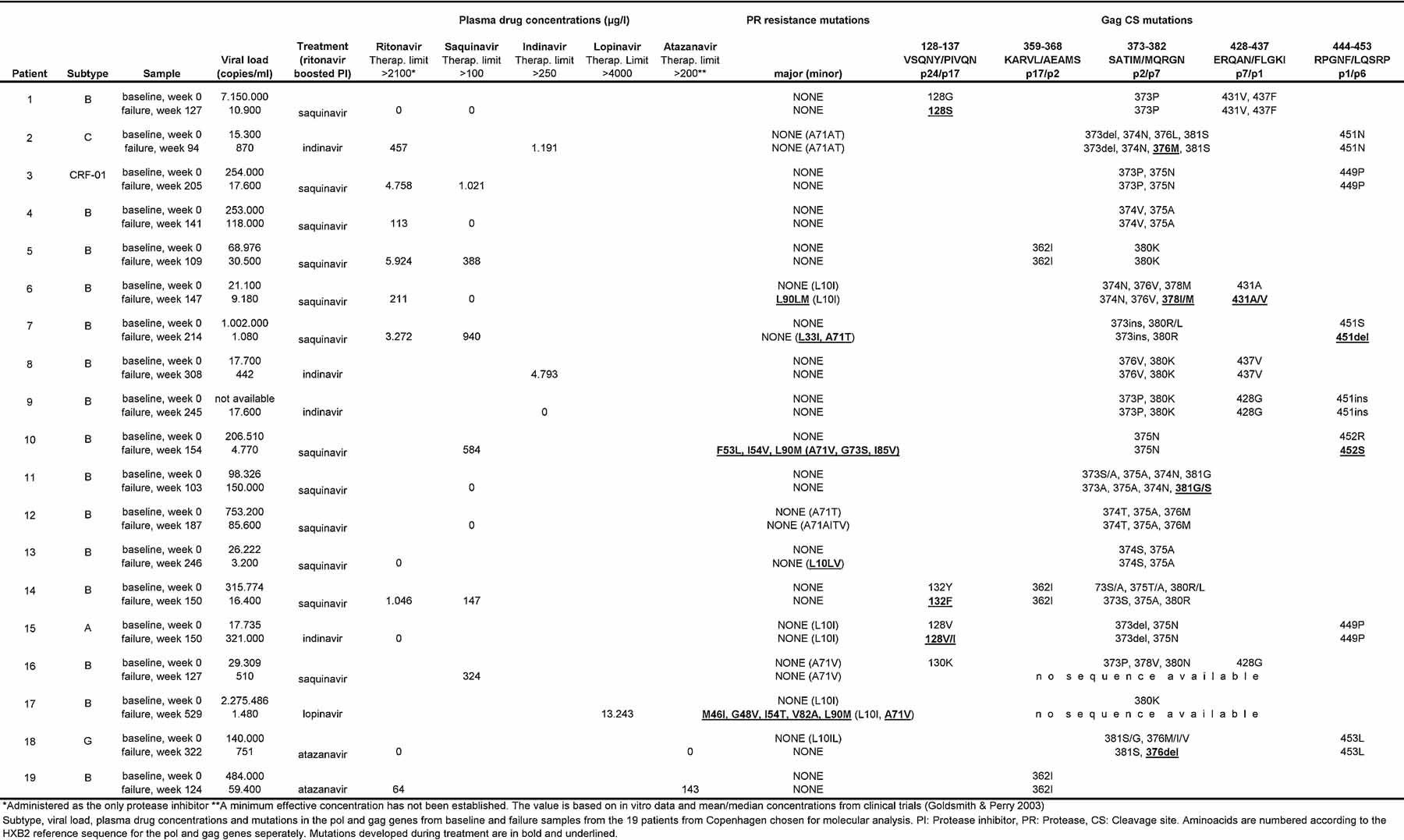
- *Administered as the only protease inhibitor.
- **A minimum effective concentration has not been established. The value is based on in vitro data and mean/median concentrations from clinical trials [Goldsmith and Perry, 2003].
- Subtype, viral load, plasma drug concentrations and mutations in the pol and gag genes from baseline and failure samples from the 19 patients from Copenhagen chosen for molecular analysis. PI: Protease inhibitor, PR: Protease, CS: Cleavage site. Aminoacids are numbered according to the HXB2 reference sequence for the pol and gag genes seperately. Mutations developed during treatment are in bold and underlined.
The median time between the baseline and failure samples was 44 months (range 24–124 months). Fifteen patients were infected with subtype B virus, the remaining four with subtypes A, CRF-01, C, and G, respectively (Table II).
No patients had major PI resistance mutations at baseline. Patients 6, 10 and 17 developed major PI resistance mutations during PI/r treatment, in all three cases including L90M. Two had received boosted Saquinavir, and one had received boosted Lopinavir. Patients 7 and 13 developed only minor PI mutations. For the remaining four patients with minor PI mutations at failure (2, 12, 15, and 16), these were already present at baseline. From ten patients no major or minor PI resistance mutations could be detected at time of virological failure.
Fifteen patients had mutations related to RT inhibitors (data not shown). In ten patients the mutations developed during the study period and in the remaining five the mutations were already present at baseline.
Gag sequences were analyzed from 19 baseline samples and 17 failure samples. All sequences deviated from the HXB2 reference sequence in one or more positions at the five cleavage sites. Subtype non-B specimens exhibited more nucleotide differences from the subtype B reference than the subtype B sequences. These deviations were typically maintained from baseline to failure and are likely to be subtype specific polymorphisms.
Nine patients developed aa mutations in one or more CS between baseline and failure.
Most mutations and polymorphisms were observed at the p2/p7 cleavage site. The most conserved site was p17/p2 with no development of mutations and only one polymorph position (position 362). Only patients with major or minor mutations in PR developed mutations at the p7/p1 and p1/p6 CS during failure.
Several insertions and deletions relative to the HXB2 reference were observed throughout the gene especially in the p1 and p6 regions (data not shown). All except one were identical at baseline and failure. This was a four aa insertion in p24 in the failure sample of patient 12 (data not shown).
Drug concentrations were below target trough value in ten out of the 19 patients (Patients 1, 4, 6, 9, 11, 12, 13, 15, 18, and 19, see Table II). Saquinavir or Indinavir were not detectable in 6 out of the 14 patients in whom it was measured. In two of these patients ritonavir was detectable. The remaining eight patients had Saquinavir/Indinavir drug concentrations in a range above the therapeutic limit. For two patients ritonavir was the only drug measured, and in both cases it was undetectable, suggesting the additional PI to be present at low concentrations as well. Lopinavir was detected in one patient receiving this PI, but Atazanavir was below the therapeutic limit in the two patients receiving it. For nine patients, drug concentrations were above target trough value. In these patients the median viral load (VL) was 1,480 copies/ml (442–30,500) at the time of virological failure. For the three patients with major resistance mutations, one had plasma concentrations below target trough value and two had values above.
DISCUSSION
In the present study, 23 (2.3%) of Danish HIV-1 patients on PI/r treatment in the period from 1995 to 2008 developed virological failure.
The patients included in this study received Indinavir, Saquinavir, Lopinavir, or Atazanavir as part of the PI/r regimen. Virological failure during Lopinavir/ritonavir treatment without the development of PI mutations is well described [Hicks et al., 2004]. In the current study comparing previous PR regimens the failure incidence rates were higher in those treated with Saquinavir and Indinavir than in those treated with Lopinavir and Atazanavir (10.8/1,000 PYR and 15.8/1,000 PYR vs. 1.4/1,000 PYR and 4.5/1,000 PYR respectively). These rates were not corrected for calendar year, backbone treatment and reasons of choice of treatment regimen.
Out of the 19 patients chosen for molecular analyses only three (16%) developed major PI resistance mutations in PR explaining the viral drug escape. Three patients developed minor mutations in the PR gene, but no major mutations were detected, that could account for the virological failure observed. For the remaining patients sequencing of PR revealed no development of resistance mutations, and the virological failure must, thus, be explained by other factors. These findings are in concordance with findings from other studies, were 0–30% of virological failures to PI/r were explained by PR mutations [Ananworanich et al., 2006; von Wyl et al., 2007].
Previous studies have shown low PI concentrations in patients failing PI/r regimens and high intra- and inter-patient variability [Masquelier et al., 2005; Best et al., 2007; Zazzi, 2007]. In accordance with these observations the present study found that drug concentrations were below target trough value in 10 out of the 19 patients. Only one patient with measured plasma concentrations below target trough value developed major mutations. In the remaining nine patients with low PI concentrations the detected VL was caused by replicating wild type virus. Patients who are incompliant, but take their medication prior to sampling, will appear with measurable plasma drug concentrations, but high VL and wild type virus due to the delayed viral response. This could explain the observed pattern in these patients.
Samples were unlikely to have been collected immediately before a dose, so it is possible that some of these patients may in fact have had too low trough levels. Drug concentrations were only measured for one time point for each patient. Sequential measurements might have given a more complete picture of the actual situation. The drug concentrations below target trough level are likely to be explained by poor compliance even though this was considered when selecting the study population. Information on compliance was not collected prospectively but evaluated looking through patient files and no data for successful compliance were collected. Even for patients with no compliance issues registered these might have been contributing to the virological failure observed. Drug levels equal to zero makes altered absorption or metabolism unlikely explanations.
Baseline and failure gag sequences were analyzed for seventeen patients. Gag mutation development from baseline to failure at the p7/p1 and p1/p6 CS was only seen together with the development of major or minor PR mutations. In none of the 19 failure cases analyzed in this study it could be confirmed that these gag mutations had given rise to resistance development and virological failure on their own. Mutations in p24/p17 and p2/p7 were observed independently of PR mutations at positions 128, 132, 376, and 381. These positions have not previously been linked to PR resistance development.
Variations from the reference sequence in p7/p1 and p1/p6 were observed at positions 428, 431, 437, 449, 451, and 452 including the 431V, 437V, and 452S variants described in previous studies in relation to PI resistance development [Zhang et al., 1997; Myint et al., 2004; Verheyen et al., 2006; Malet et al., 2007; Nijhuis et al., 2007]. These variants were present at baseline as well as at the time of virological failure. They were therefore most likely present during the period of viral suppression observed in all patients according to the criteria and did thus not prevent viral suppression. Previous studies have shown that certain CS mutations might contribute directly to PI resistance by improving cleavage efficiency by wild type PR [Prabu-Jeyabalan et al., 2004; Verheyen et al., 2006; Nijhuis et al., 2007]. In the present study it could not be confirmed that this was of any significance for the limited amounts of virological failure cases observed.
In conclusion, virological failure is infrequent in patients on a PI/r based regimen. The proportion of patients failing PI/r treatment has decreased during the years 1995–2008. PR resistance mutations and low drug levels could account for 12 (63%) of the failure cases observed. The analyses of gag sequences did not confirm a significant role of gag CS mutations in resistance development among the limited amount of patients with virological failure to PI/r. The use of regimens evaluated in the present study has decreased over the years, and are only used for a minority of present day patients in Denmark.




