Genotypes and viral load of hepatitis C virus among persons attending a voluntary counseling and testing center in Ethiopia
Abstract
The prevalence of different genotypes of hepatitis C virus (HCV) in Ethiopia is not known. HCV genotypes influence the response to therapy with alpha-interferon alone or in combination with ribavirin. A cross sectional study was conducted on attendees of voluntary counseling and testing center. Serum samples from 1,954 (734 HIV positive and 1,220 HIV negative) individuals were screened for HCV antibody. Active HCV infection was confirmed by quantitative PCR in 18 of the 71 samples with anti-HCV antibodies. The HCV viral load ranged from 39,650 to 9,878,341 IU/ml (median 1,589,631 IU/ml) with no significant difference [χ2(17) = 18.00, P = 0.389] between persons positive or negative for HIV. The viral load of HCV was, however, higher in older study subjects (r = 0.80, P = 0.000). HCV genotypes were determined using the VERSANT HCV Genotype Assay (LiPA) and sequence analysis of the NS5b region of the HCV genome. Diverse HCV genotypes were found including genotypes 1, 2, 4, and 5. There was no difference in the distribution regarding the HIV status. As in other parts of the world, genotyping of HCV must be considered whenever HCV is incriminated as a cause of hepatitis. J. Med. Virol. 83:776–782, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Hepatitis C Virus (HCV) infection leads to chronic infection in approximately 80% of the persons infected [Amarapurkar, 2000] affecting about 170 million of the world's population [WHO, 1999]. Chronic hepatitis C eventually leads to the development of hepatocellular carcinoma (HCC) in 0.4–2.5% of infected persons [Colombo, 1999]. Different studies have shown that HCV genotypes influence the response to therapy with alpha-interferon alone or in combination with ribavirin [Zein et al., 1996; McHutchinson et al., 1998]. The prevalence of the genotypes and subtypes of HCV vary significantly in different parts of the world. Genotype 1 is the most prevalent in most parts of the world [Simmonds, 1995]. In Central and Northern Europe and the United States genotype 1 (subtypes 1a and 1b) is more prevalent [Feucht et al., 1997; Alter et al., 1999], while HCV subtypes 2a, 2b, 3a, 4a, and others occur less frequently. Type 2 with its subtypes is known to be one of the dominant types in Asia and is also found commonly in the Mediterranean countries and northern Europe. Subtype 4a is predominant in the Middle East and North and Central Africa [Simmonds et al., 1993; McOmish et al., 1994; Xu et al., 1994]. Genotypes 5 and 6 are confined mainly to South Africa and South-East Asia, respectively [McOmish et al., 1994; Xu et al., 1994; Maertens and Stuyver, 1997].
Co-infection with HIV and HCV is emerging as a major cause of morbidity and mortality [Jones et al., 2005]. Among the estimated 40 million persons infected with HIV worldwide, an estimated 4–5 million are infected chronically with HCV [Alter, 2006]. Ethiopia is among the countries most affected by HIV/AIDS and a total of 1,320,000 people (45% male and 55% female) were living with HIV/AIDS in 2005 with a national prevalence rate of 3.5% (10.5% in urban and 1.9% in rural areas) [FMOH/HAPCO, 2006]. In one study, prevalence of HCV antibody of higher rate between HIV-positive compared to HIV-negative individuals was reported from a community based survey in Addis Ababa, Ethiopia (4.5% vs. 0.8%, respectively, P < 0.001, n = 4,593) [Ayele et al., 2002]. The presence of HCV increases the frequency of hepatotoxicity with antiretroviral therapy, and may also influence the choice of antiretroviral drug, with drugs which are potentially hepatotoxic such as ritonavir and nevirapine [Soriano et al., 2002]. Co-infection of HIV with HCV is reported to cause a more rapid and more severe histopathological course [Eyster et al., 1993] and increases HCV replication with high HCV RNA levels [Di Martino et al., 2001; Matthews-Greer et al., 2001]. Persons co-infected with HIV have been observed to be less likely to clear HCV spontaneously than those without HIV infection [Thomas et al., 1996].
In Ethiopia, there are no previous reports on HCV genotypes, HCV viral load and the association with persons infected with HIV. The aim of the present study was to determine the genotypes of HCV and the viral load of HCV among both HIV positive and negative individuals. It was conducted in apparently healthy persons visiting a voluntary counseling and testing center. Attendees were either ignorant of their HIV status, or they were naïve for HAART.
MATERIALS AND METHODS
The Faculty research and publication committee of the Medical Faculty of Addis Ababa University, AHRI/ALERT ethical review committee, the Ethics committee of Leipzig University, Germany, approved the study protocol. The study commenced after the National Ethics Review Committee of Ethiopia approved it.
Study Design and Sample Collection
A cross-sectional study was carried out from June to September 2006 with participants who were informed and consented to the study who were visiting a regional voluntary counseling and testing center located at Zewditu Hospital of Addis Ababa, Ethiopia. The rationale for selecting study participants from the voluntary counseling and testing center attendees is simply because it was convenient to obtain a large number of persons knowing their HIV status. Sampling was therefore stratified into each of HIV positive and HIV negative groups. The mean age of the study participants (n = 1,954) is 29.1 years (95% CI = 28.7–29.5 years, SE = 0.20), 32.3 years (95% CI = 31.7–32.9 years, SE = 0.32) for the HIV positive study participants (n = 734) and 27.1 years (95% CI = 26.6–27.6 years, SE = 0.25) for the HIV negative study participants (n = 1,220). Female participants were 1,117 (57.2%) and male participants were 837 (42.8%). Venous blood samples of 5–10 ml were obtained from each of the study participants. Part of the whole blood sample was used for HIV screening and the remaining blood sample was allowed to clot and then serum was separated within 4 hr by centrifugation at 2,500g for 10 min, and stored in aliquots at −80°C until tested for HCV.
HIV Screening
Tests for HIV was done at the voluntary counseling and testing center according to the national guidelines for HIV testing (Rapid HIV Test Algorithm) which includes a combination of three tests namely, Determine® HIV-1/2 (Abbott Laboratories, Abbott Park, IL), Capillus™ HIV-1/HIV-2 and Uni-Gold™ (both IDA Business Park, Bray, Co., Wicklow, Ireland).
Whole blood samples were tested initially by the Determine HIV-1/2 test and those found positive were subjected to the Capillus test kit. Samples positive by both tests were considered to be positive for anti-HIV-1/2 antibodies. Samples with discrepant test results were subjected to the third test kit, Uni-Gold. Samples which were positive on Uni-Gold test kit were considered to contain antibodies against HIV-1/2. All tests were done according to the recommendations of the respective manufacturer of the kits.
HCV Screening
The Murex anti-HCV [Version 4.0] (Abbott-Murex Diagnostics, Murex Biotech S.A. (Pty) Ltd., Kyalami Boulevard, Republic of South Africa), a third generation enzyme linked immunosorbent assay (ELISA), was used to screen all HIV positive and HIV negative serum samples for antibodies against HCV.
HCV Viral Load
Viral load was determined in all samples found positive by initial testing by the Murex anti-HCV test. Viral load was performed using the Abbott Real Time HCV assay (Abbott Molecular Inc., Des Plaines, IL). The various steps of the procedure were carried out according to the manufacturer's instructions in three different rooms, one each for sample preparation (RNA extraction), reagent preparation (mixing up of activation reagent, oligonucleotide reagent and thermostable rTth DNA Polymerase), and amplification and detection of target on an automated Abbott m2000rt instrument. A volume of 500 µl of serum was used for extraction of nucleic acid to reach a lower detection limit of 12 IU/ml of HCV RNA.
HCV Genotype Determination
Line probe assay (LiPA)
HCV RNA positive samples were subjected to genotyping based on 5′ non-coding sequences of the HCV genome using VERSANT® HCV Genotype Assay (LiPA) 1.0 (Bayer HealthCare LLC, Tarrytown, NY) according to the manufacturer's instructions.
HCV sequencing
Samples were also subjected to direct sequencing of PCR amplicons from the HCV NS5b region. Extracted RNA (5 µl, spared from the extraction for Real Time PCR) was used for reverse transcription (45 min, 50°C, 4 µl of 5× buffer, 4 µl of RNase free water, 0.5 µl of RNase inhibitor (40 units/µl), 2.5 µl of dNTPs (10 mM), 3 µl (10 pmol/µl) of the reverse primer Asn1121 5′GCNGARTAYCTVGTCATAGCCTC3′ (positions 8281–8303, accession no. M62321) and 1 µl of AMV-RT (10 units/µl) in a final volume of 20 µl). The resulting cDNA was used as template in the ensuing PCR (50 µl aliquots containing 5 µl cDNA, 2 µl dNTP (10 mM), 3.5 µl MgCl2 (50 mM), 5 µl of 10X PCR buffer and 1 µl (10 pmol/µl) of each of the forward primer Sn755 5′TATGAYACCCGCTGYTTTGACTC3′ (position 7915–7937) and reverse primer (described above), and 0.4 µl of Taq polymerase (5 units/µl). The PCR amplification was carried out with a touchdown approach as described elsewhere with slight modifications [Morice et al., 2001]. The amplification consisted of initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 30 sec, primer annealing at 60°C for 20 sec and extension at 72°C for 1 min. During the first 5 cycles, the annealing temperature was 60°C, reduced by 0.3°C per cycle for 30 cycles (touchdown), followed by 5 cycles at 50°C, and a final extension at 72°C for 5 min). Amplification was monitored by 2% agarose gel electrophoresis for the production of 389 bp. The amplified products were cleaned by the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI) according to the manufacturer's instructions. Purified amplicons were subjected to direct sequencing of both strands with the PCR primers using the ABI Prism BigDye® Terminator v1.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA). Analysis of each sequence was made by HCV-BLAST (http://hcv.lanl.gov/content/hcv-db/BASIC_BLAST/basic_blast.html) to define the HCV genotype by the closest matching sequences.
Statistical Analysis
Data entered in Microsoft Office Excel 2003 were transferred to STATA version 8.0. Data were organized and summarized in descriptive statistics and the viral load and distribution of HCV genotypes among the HIV-positive and HIV negative samples were observed. Chi square (χ2) test was carried out to see if there was any association of assessed variables. P-value of less than 0.05 was considered statistically significant.
RESULTS
HIV Screening
All individuals attending the voluntary counseling and testing center were notified about their HIV status on the same day of their visit. Out of the 1,954 enrolled study subjects, 1,220 were negative and 734 positive for HIV.
HCV Screening
Screening of the 1,954 samples by the Murex anti-HCV ELISA resulted in 71 (3.63%) positive samples (50 in HIV positive and 21 in HIV negative persons).
HCV Viral Load and Genotype Determination
HCV RNA extraction and quantitation was performed in the 71 Murex anti-HCV positive samples using the Abbott Real Time HCV Assay that target the 5′ UTR of the genome. HCV RNA target was detected and quantified only in 18 of these samples. However, HCV RNA was not detected in another 56 anti-HCV negative but HIV positive samples.
In 15 of the total 18 HCV PCR positive samples, genotypes were determined by the VERSANT HCV Genotype Assay (LiPA). The HCV genotype could not be determined with this method in three samples. The genotypes determined by LiPA are 1b, 2a/c, 4, 4c/d, and 5a. Except the two LiPA determined 5a subtypes, the other thirteen including the three LiPA indeterminant subtypes were amplified by RT-PCR for the synthesis of cDNA targeting the NS5b region. However, two strains of 4c/d and one strain of genotype 4 as determined by LiPA were not sequenced because of insufficient sample. The subsequent amplification of the cDNA in the 13 samples resulted in an amplicon synthesis of 389 bp that was verified by 2% agarose gel electrophoresis. After sequencing of the three LiPA undetermined samples, HCV subtype 2c was determined in one sample and subtype 4r, according to the provisional HCV genotype classification [Simmonds et al., 2005], was confirmed in the other two samples. In sequencing of the NS5b region on the LiPA determined subtypes, genotype 1b was subtyped as 1c; all the five 2a/c strains to subtype 2c, and four strains of 4c/d to subtype 4d (Table I). The comparison of the distribution of these HCV genotypes in both HIV positive and negative individuals showed no statistically significant difference (χ2(6) = 9.234, P = 0.161) (Table II).
| Sample code | Age of study participants | HCV viral load (IU/ml) | Genotypes identified by | Genotype interpretation | |
|---|---|---|---|---|---|
| LiPA (5′ UTR) | NS5b region sequencing | ||||
| 07/12629 | 22 | 59,056 | 4 | Not donea | 4 |
| 07/12650 | 28 | 1,173,793 | 2a/c | 2c | 2c |
| 07/12594 | 30 | 1,499,523 | 4c/d | 4d | 4d |
| 07/12604 | 30 | 300,612 | 1b | 1c | 1c |
| 07/12605 | 30 | 237,338 | 4c/d | Not donea | 4c/d |
| 07/12635 | 31 | 3,991,650 | 2a/c | 2c | 2c |
| 07/12603 | 32 | 39,650 | 5a | Not doneb | 5a |
| 07/12609 | 32 | 1,848,682 | 4c/d | 4d | 4d |
| 07/12613 | 34 | 939,024 | Untypable | 2c | 2c |
| 07/12638 | 38 | 3,655,729 | 2a/c | 2c | 2c |
| 07/12647 | 38 | 2,277,182 | Untypable | 4r | 4r |
| 07/12606 | 39 | 95,507 | 4c/d | Not donea | 4c/d |
| 07/12586 | 40 | 1,004,719 | 5a | Not doneb | 5a |
| 07/12636 | 45 | 1,679,738 | 4c/d | 4d | 4d |
| 07/12626 | 50 | 3,393,666 | 4c/d | 4d | 4d |
| 07/12655 | 55 | 7,770,254 | 2a/c | 2c | 2c |
| 07/12651 | 60 | 3,534,191 | Untypable | 4r | 4rc |
| 07/12625 | 70 | 9,878,341 | 2a/c | 2c | 2c |
- a NS5b sequencing could not be performed due to the insufficient sample material left.
- b Amplification of the NS5b region was left undone as studies showed concordant results with LiPA analysis on this subtype [Cantaloube et al., 2006].
- c Only provisionally classified (4r is formerly known as 4a).
| HIV status | HCV Genotype | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| 1c | 2c | 4 | 4c/d | 4d | 4r | 5a | ||
| Negative | 0 | 4 | 0 | 0 | 1 | 2 | 0 | 7 |
| Positive | 1 | 2 | 1 | 2 | 3 | 0 | 2 | 11 |
| Total | 1 | 6 | 1 | 2 | 4 | 2 | 2 | 18 |
- Pearson χ2(6) = 9.234, P = 0.161.
The HCV viral load in the samples as determined by a real time PCR assay revealed a quantity ranging from 39,650 to 9,878,341 IU/ml (median 1,589,631 IU/ml) with no significant difference [χ2(17) = 18.00, P = 0.389] between HIV negative groups that ranged from 1,173,793 to 7,770,254 IU/ml (median 3,534,191 IU/ml) and HIV positive groups that ranges from 39,650 to 9,878,341 IU/ml (median 939,204 IU/ml). The mean HCV viral load of the different HCV genotypes and the frequency of occurrence of each genotype are shown in Table III. HCV viral load is also not significantly associated with the type of HCV genotypes encountered (P = 0.323). HCV viral load was, however, higher in older study subjects (r = 0.80, P = 0.000) (Fig. 1). The relative prevalence of each genotype was observed to be 50% for HCV genotype 4 (4, 4c/d, 4d, 4r), 33% for genotype 2c, 11.11% for genotype 5a and 5.56% for genotype 1c (Table III).
| Genotypes | Number of observations | Viral load (IU/ml) | ||
|---|---|---|---|---|
| Absolute | Relative % | Mean | Range (min–max) | |
| 1c | 1 | 5.56 | 300,612 | |
| 2c | 6 | 33.33 | 4,568,132 | 939,024–9,878,341 |
| 4 | 1 | 5.56 | 59, 056 | |
| 4c/d | 2 | 11.11 | 166,423 | 95,507–237,338 |
| 4d | 4 | 22.22 | 2,105,402 | 1,499,523–3,393,666 |
| 4r | 2 | 11.11 | 2,905,687 | 2,277,182–3,534,191 |
| 5a | 2 | 11.11 | 522,185 | 39,650–1,004,719 |
| Total | 18 | 100.00 | 2,409,925 | 95,507–9,878,341 |
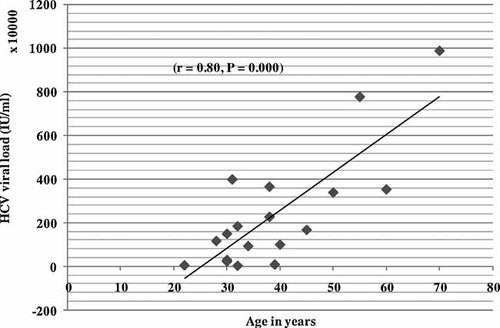
HCV viral load versus age.
DISCUSSION
In this study, the 71/1,954 (3.6%) Murex anti-HCV positive samples were subjected to a quantitative PCR. Only 18 (25.4%) samples were found to be HCV RNA positive. High false positive test results in ELISA systems are encountered commonly in low-prevalence settings such as healthy blood donors [Sakugawa et al., 1995; Gretch, 1997] and as a cross-reaction to several infections such as malaria, schistomiasis, respiratory viruses and autoimmune diseases [Aceti et al., 1990; Bar-Shany et al., 1996; Agha et al., 2006]. However, patients who have HCV antibody but negative to HCV RNA on repeated PCR testing are generally considered to have cleared or resolved the infection [Chou et al., 2004]. Another random sample of 56 sera from 684 HIV positive but anti-HCV antibody-negative samples were tested for HCV RNA to seek acute hepatitis C. Despite the high sensitivity of the assay used none of these sera were tested HCV RNA positive. This result is in contrast to previously published observations that reported almost 6% HIV infected and HCV seronegative people with detectable HCV RNA using the standard commercial assays [Bonacini et al., 2001; George et al., 2002]. This figure could rise to 20% using the more sensitive whole blood HCV RNA methods. This could be attributed to the intermittent viraemia or lower concentrations of HCV RNA which are near the limits of detection of the RT-PCR method utilized [George et al., 2002]. The HCV prevalence is not significantly different in HIV positive and HIV negative persons probably because the study population was among persons attending a voluntary counseling and testing center and not selected at random from the general population.
To our knowledge, this is the first report on the HCV genotypes in Ethiopia. The genotypes encountered in the 18 HCV RNA positive samples were 1b, 2a/c, 2c, 4, 4c/d, 4r, and 5a as determined by LiPA. Since subtyping is not important for routine clinical purposes, this LiPA results may be satisfactory in clinical settings. However, NS5b region sequence analysis yielding more accurate results will be a major improvement for epidemiological studies and medicolegal investigations by removing classification errors that occur using 5′ UTR analysis [Cantaloube et al., 2006]. It has been shown in several studies that LiPA misclassifies a significant percentage of type 1a isolates as type 1b [Chen and Weck, 2002; Tamalet et al., 2003]. The poor performance of LiPA in respect of subtyping of genotype 1 has been reported previously [Chen and Weck, 2002; Tamalet et al., 2003]. Furthermore, the LiPA does not discriminate between subtypes 2a and 2c, and also fails to correctly subtype samples containing genotype 4 [Tamalet et al., 2003; Cantaloube et al., 2006; Ross et al., 2007]. The “gold standard” for genotyping is direct sequencing of the NS5b or E1 region followed by sequence alignment with reference sequences and phylogenetic analysis [Simmonds and Smith, 1999]. NS5b sequencing revealed LiPA misclassification of the 1c genotype in this study as 1b. According to the consensus proposals for unified system of nomenclature of HCV genotypes, 1b, 2a, 2c, and 5a are classified as confirmed designations of HCV genotypes whereas 4c, 4d, and 4r are classified as provisional designations [Simmonds et al., 2005]. The prevalence of the different HCV genotypes was similarly distributed among the HIV negative and positive individuals (χ2(6) = 9.234, P = 0.161). There are very limited studies on the distribution of HCV genotypes in HCV mono-infected and HIV co-infected patients. In one study, a larger proportion of genotype 1 infections was recorded both in HIV positive and HIV negative hemophiliacs. The subtype distribution among genotype 1 infected patients was different, with a significantly increased frequency of subtype 1a over 1b among HIV positive patients [Picchio et al., 1997].
In this study, a dominance of genotype 4 (50%, nine cases) is found. At present it is unknown whether this genotype originated in Ethiopia or was introduced from other regions such as the Middle East to where there is frequent travel of people to this geographical location from Ethiopia and vice versa. HCV genotypes of 4, 4e, and 4c/4d were identified recently in patients with hepatosplenic schistosomiasis attending at the National Centre for Gastrointestinal and Liver Disease in Khartoum, Sudan [Mudawi et al., 2007]. Genotype 2c was the second most common group (6 cases) with a frequency of 34%. Type 2 with its subtypes is known to be one of the dominant types in Asia and can also be observed to a large extent in the Mediterranean countries and Northern Europe [McOmish et al., 1994]. HCV types 1b, 2b, and 2k are also reported from Madagascar [Razafindratsimandresy et al., 2007], and types 1, 2, and 4 were found in West Africa [Pasquier et al., 2005; Plamondon et al., 2007]. Therefore, phylogenetic analysis of the different isolates encountered in the present study should reveal their origin. The diverse HCV genotypes encountered in this study indicates the need for thorough evaluation of HCV in case of clinical hepatitis presentation as it determines the indication, the duration of treatment, the dose of ribavirin and the virological monitoring procedure [Hadziyannis et al., 2004].
In this study, the HCV viral load was not significantly different in the HCV mono-infected and HIV/HCV co-infected persons (χ2(17) = 18.0, P = 0.389). In previous studies, HIV positive patients co-infected with genotype 1b had the highest mean HCV viral load [Picchio et al., 1997]. In this study, one female HIV infected individual was observed to be co-infected with genotype 1c that showed a relatively low viral load (300,612 IU/ml) when compared to the mean (698,136 IU/ml) and median (1,589,631 IU/ml) viral load of the overall genotypes encountered. In another recent study the mean HCV RNA level was higher in the genotype 1 group than in the non-genotype 1 groups, in both the HIV co-infected and HIV negative groups. Although the HCV genotype was not associated with differences in HIV viral load, an increased risk for progression to AIDS-related mortality and a significantly lower mean CD4+ count and percentage level was observed in the individuals in the genotype 1 group compared with those in the non-genotype 1 group [Yoo et al., 2005]. Another cohort study indicated that HIV/HCV co-infected patients display similar ALT profiles as HCV mono-infected patient, but a larger proportion of detectable serum HCV RNA [Bonacini et al., 2001]. No difference in HCV genotype distribution or viral load was detected in this study comparing HCV mono-infected and HCV/HIV co-infected individuals. Older study subjects were observed to have higher viral load (r = 0.80 and P = 0.000) as compared to the younger study participants which might indicate increasing viral load over time in patients infected chronically. A survey conducted among anti-HCV positive participants in the United States showed that more persons who were 40 years of age or older (89.6%) found to have HCV RNA positive results than did persons who were 6–39 years of age (60.2%) (P = 0.023) [Armstrong et al., 2006].
In conclusion, this report on HCV genotypes in Ethiopia showed a relatively diversified type of HCV genotypes. Larger studies representing the whole country would reveal the predominant HCV genotypes in Ethiopia and phylogenetic analysis their origin. The limitations of this study are that it was not community based; the protocol was not designed to define the role of HCV on HIV infection or vice versa on a cohort of HIV infected and uninfected groups; and did not address prevalence of HCV among clinical cases of hepatitis. Future studies should therefore address these limitations.




