Detection of human parechovirus in stool samples collected from children with acute gastroenteritis in Japan during 2007–2008
Abstract
Of 477 stool specimens, which had been screened for rotavirus, adenovirus, norovirus, sapovirus and astrovirus, collected from infants and children with acute gastroenteritis in pediatric clinics encompassing five localities (Sapporo, Tokyo, Maizuru, Osaka, and Saga) in Japan from July 2007 to June 2008, 247 negative samples (51.7%) were subjected to screening for human parechovirus. Human parechovirus (HPeV) was detected by RT-PCR using a primer pair to amplify 5′UTR region of its genome and was genotyped by sequencing of the VP1 gene. HPeV was detected in 20 of 247 specimens tested, and the detection rate was found to be 8.1%. Seventeen of the 20 strains that tested positive for HPeV were sequenced successfully the VP1 gene. The majority of the HPeV strains (n = 15) could be identified as HPeV1, and the remaining 2 strains could be typed as HPeV3. By phylogenetic and identical matrix analyses of HPeV VP1 sequences, HPeV1 should be divided into two lineages, and all of the Japanese studied HPeV1 strains belong to the lineage 2 accordingly. This is the first report of the circulation of HPeV, especially HPeV1 in Japan. J. Med. Virol. 83:331–336, 2011. © 2010 Wiley-Liss, Inc.
INTRODUCTION
Human parechoviruses (HPeVs) belong to the large family of Picornaviridae which is a highly diverse family of important pathogens of humans and animals. The HPeV genome is approximately 7.3 kb nucleotides in length and contains a large open reading frame (ORF) coding for single polyprotein. The polyprotein is cleaved post-translationally into three structural proteins (VP0, VP3, and VP1) and seven non-structural proteins (2A–2C and 3A–3D) [Hyypia et al., 1992; Stanway et al., 1994].
The previous findings reveal the genetic variability of HPeVs and the number of newly identified HPeV genotypes has been on the increase. Based on VP1 sequence comparisons, HPeVs have been classified into 14 genotypes (HPeV1–14) (http://www.picornaviridae. com/parechovirus/hpev/hpev.htm), of these, nine HPeV genotypes (HPeV1–8 and 14) have been published up to date [Ito et al., 2004; Abed and Boivin, 2005; Boivin et al., 2005; Benschop et al., 2006a,b; Al-Sunaidi et al., 2007; Watanabe et al., 2007; Baumgarte et al., 2008; Benschop et al., 2008; de Vries et al., 2008; Drexler et al., 2009; Li et al., 2009].
It is established well that rotaviruses, adenoviruses, astroviruses, and caliciviruses are the most important etiologic agents of acute gastroenteritis, which is a common cause of morbidity and mortality worldwide [Glass et al., 2001]. However, there remains a “diagnostic gap,” which has been attributed to less explored viral pathogens. The present study aimed to screen stool samples collected from Japanese children with acute gastroenteritis during 2007–2008 for HPeV infection, one of less explored viral pathogens which has been reported to be associated with diarrhea recently, and to characterize the detected HPeV strains.
MATERIALS AND METHODS
Clinical Specimens
Of 477 stool specimens, which had been screened for rotavirus, adenovirus, norovirus, sapovirus and astrovirus [Chanit et al., 2009], collected from infants and children aged from 2 months to 15 years who sought treatment of diarrhea at pediatric clinics encompassing five localities (Sapporo, Maizuru, Tokyo, Saga, and Osaka) in Japan from July 2007 to June 2008, 247 negative samples (51.7%) were subjected to screening for HPeV.
RNA Extraction and Reverse Transcription (RT)
The RNA genome of HPeV was extracted from 140 µl of 10% fecal suspension using the QIAamp viral RNA Mini kit (QIAGEN, Inc., GmbH Hilden, Germany) according to manufacturer's instructions. Then, for reverse transcription, 5 µl of the stored, extracted RNA was added to a reagent mixture consisting of 5× First Strand Buffer (Invitrogen, Carlsbad, CA), 10 mM dNTPs (Roche, Mannheim, Germany), 0.1 M DTT (Invitrogen), SuperScript Reverse Transcriptase III (200 U/µl) (Invitrogen), random primer (1 µg/µl) (hexa-deoxyribonucleotide mixture) (Takara, Shiga, Japan), RNase Inhibitor (33 U/µl) (Toyobo, Osaka, Japan), and distilled water. The total volume of reaction mixture was 15 µl. RT reaction was carried out at 50°C for 1 hr, followed by 95°C for 5 min and then held at 4°C. The cDNA was stored at −30°C until using for PCR reactions [Yan et al., 2004; Phan et al., 2005].
Polymerase Chain Reaction (PCR) for Detection of HPeV
After adding 2 µl of cDNA into 23 µl of the reagent mixture containing 5× Taq DNA polymerase buffer (Promega, Madison, WI), dNTPs (10 mM), primers (20 µM), Taq DNA polymerase (5 U/µl) (Promega), and distilled water, screening PCR was conducted using primers ev22(+) and ev22(−) to amplify a 270-bp PCR product of 5′UTR region [Joki-Korpela and Hyypia, 1998] (Table I). The PCR protocol was 95°C for 5 min, followed by 35 cycles of 95°C for 30 sec, 50°C for 30 sec, 72°C for 1 min, and a final extension at 72°C for 7 min.
| Primer | Gene | Sequence 5′–3′ | Sense | Position* | Amplicon | References |
|---|---|---|---|---|---|---|
| ev22(+) | 5′TR | CYCACACAGCCATCCTC | + | 312–328 | 270 | Joki-Korpela and Hyypia [1998] |
| ev22(−) | 5′TR | TRCGGGTACCTTCTGGG | − | 581–565 | ||
| VP1-parEchoF1 | VP1 | CCAAAATTCRTGGGGTTC | + | 2332–349 | 760 | Benschop et al. [2006b] |
| VP1-parEchoR1 | VP1 | AAACCYCTRTCTAAATAWGC | − | 3090–071 | ||
| Cap-parEcho-F | VP1 | TCHACWTGGATGMGRAARAC | + | 2162–181 | 1076 | This study |
| Cap-parEcho-R | VP1 | TCYARYTCACAYTCYTCYTC | − | 3237–218 |
- Y stands for C or T, R: G or A, W: A or T, H: A, C, or T, and M means C or A. Sequence position (*) is based on the full genome sequence of the prototype HPeV1 strain, Harris strain, with the accession number of L02971.
Genotyping by VP1 Sequencing and Primer Designation
At first, to amplify the VP1 gene, the previously described primers developed by Benschop et al. [2006b] were used. Because of failure in obtaining PCR products of most HPeV-positive samples except for three, two new primers were designed for the first PCR. Then, for the nested PCR, it was performed using the inner primer pair described by Benschop et al. [2006b].
For primer designation, to obtain the full length of the VP1 gene, alignment of full genome sequences of reference strains of eight known HPeV genotypes available in GenBank databases was performed using Clustal X software to find the conserve regions and the two new primers were designed outside the VP1 region. Oligonucleotide sequences of the newly developed primers and their positions were described in Table I.
The first PCR was done using the new designed primers and the thermal cycle program was as follows: 5 min at 95°C, followed by 35 cycles of 95°C for 1 min, 52°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 7 min. The nested PCR was conducted using the known primer pair: VP1-parEchoF1 and VP1-parEchoR1 [Benschop et al., 2006b] at annealing temperature of 48°C to generate a 760-bp product (Table I). Analysis of the amplification products was performed by 1.5% agarose gel electrophoresis, and DNA bands were visualized by SYBR Safe (Invitrogen, Tokyo, Japan) staining under ultraviolet light.
Sequence and Phylogenetic Analysis
The PCR amplicons of the VP1 gene were purified and sequenced in both directions using the BigDye Terminator Cycle Sequencing kit (Perkin Elmer-Applied Biosystems, Inc., Foster City, CA). The primers for amplification of VP1 gene were used as sequencing primers. The sequence data were collected by an ABI Prism 310 Genetic Analyzer (Perkin Elmer-Applied Biosystems, Inc.).
The comparison analysis of the VP1 gene was conducted between the obtained HPeV strains and other reference HPeV strains of 9 known genotypes available in GenBank database. The sequence data and the phylogenesis were analyzed using BioEdit v7.0.5. A parsimony analysis was conducted using MEGA version 3.1 to determine the evolutionary relationship among studied sequences [Kumar et al., 2004]. The method was performed using close-neighbor interchange with a random option and with 500 bootstrap repetitions.
The nucleotide sequences of the reference HPeV strains described in this study have been deposited in GenBank under accession numbers: HPeV1: Harris (L02971), 652281 (FJ373120), BNI-R09/03 (EU024632), BNI-R32/03 (EU024636), BNI-R15/03 (EU024633), BNI-788St (EF051629), 677033 (FJ373136), 69960AE (AM933170), A229-05 (AB300968), A234-05 (AB300969), A708-99 (AB300935), BNI-R04/03 (EU024631), A65-05 (AB300963), A222-05 (AB300967), BNI-R21/03 (EU 024634), 652780 (FJ373127), 650648 (FJ373108), A191-05 (AB300966), A527-99 (AB300928), BNI-90/03 (EU024630), BNI-R30/03 (EU024635); HPeV2: Williamson (AJ005695); HPeV3: Can82853-01 (AJ889918), 677146 (FJ373162), A415-01 (AB300945), A308/99 (AB084913), 651689 (FJ373153); HPeV4: Fuk2001-282 (AB433630), NII370-93 (AB434673), T75-4077 (AM 235750), 653046 (FJ373170), K251176-02 (DQ315670); HPeV5: CT86-6760 (AF055846), T92-15 (AM235749), 676618 (FJ373175); HPeV6: 2005-823 (EU077518), NII 561-2000 (AB252582), BNI-67/03 (EU024629), 650045 (FJ373178); HPeV7: PAK5045 (EU556224); HPeV8: BR/217/2006 (EU716175); HPeV14: 451564 (FJ373179). For the studied Japanese strains, they are FJ648741-FJ648754, GQ149452, GQ203502, and GQ203503.
RESULTS
Of the 247 samples tested, 20 were positive for HPeV and the detection rate of HPeV was 8.1%. Of these, 17 strains were amplified and sequenced of the full-length VP1 capsid sequences. All 20 patients whose stool specimens showed positive for HPeV were infants and children aged from 5 to 51 months with the mean and median ages of 14.8 and 14 months, respectively. A half of the patients were les than 18 months of age. Besides diarrhea, fever and vomiting were found in 30% and 15% of the patients, respectively. Coughing and coryza were present in 10% and 5% of the patients, respectively. No dehydration and neurological symptoms were noted. For seasonal pattern of HPeV infection, in this study, HPeV was detected nearly the year round, except for April, July, and December, with the peak of incidence in February (data not shown).
Figure 1 shows the phylogenetic tree constructed from 624 bases of partial VP1 segment of reference HPeV strains and 17 Japanese strains found in this study. As shown in Figure 1, the majority of the Japanese HPeV strains (n = 15) could be identified as HPeV1, and the remaining two strains could be typed as HPeV3. The figure shows clearly that HPeV1 strains clustered into two separate branches. The prototype strains of HPeV1, the Harris strain, clustered into a branch (designed as lineage (1) together with a strain detected recently in the Netherlands, the 652281 strain. Fifteen studied Japanese HPeV1 strains were clustered into the remaining branch (designed as lineage (2) consisting of the majority of contemporary HPeV1 strains.
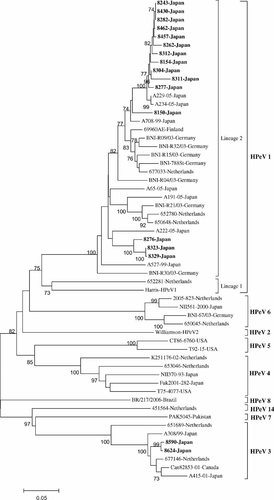
Phylogenetic tree constructed from the 624 nucleotides of the VP1 gene of the strains studied and reference HPeV strains with 500 bootstrap repetitions. Percentage bootstrap values above 70% are shown at the branch nodes. The studied HPeV strains are in boldface type.
Identical matrix analysis of VP1 amino acid sequences of the studied Japanese strains and global reference strains was performed. For HPeV1 strains, amino acid identities among the 15 Japanese strains were greater than or equal to 91.5%. High amino acid similarities were noted between the strains studied and reference strains of the same lineage 2 and ranged from 91.0% to 99.5%; while those between the strains studied and the lineage 1 strains were from 83.6% to 90.1%. Between the HPeV1 strains studied and those of other genotypes, amino acid similarities were less than 80% and varied from 55.7% (with genotype 3) to 79.6% (with genotype 6) (data not shown).
For the studied HPeV3 strains, they clustered closely to the Japanese HPeV3 prototype strain A308/99 and had 95.8% amino acid similarity to this strain. In comparison to the strain 651689 detected recently in the Netherlands and clustered separately from the HPeV3 prototype strain (Fig. 1), the studied Japanese strains showed a mean amino acid similarity of 95.3%.
The alignment of deduced amino acid sequences of the strains studied and global HPeV reference strains of HPeVs genotypes 1-8 and 14 revealed that the arginine–glycine–aspartic acid (RGD) motif, which is considered to be critical for human parechovirus 1 entry, was present in all the studied HPeV1 strains. This RGD motif was not noted in the 2 studied HPeV3 strains, and among reference strains of HPeV3, HPeV7, HPeV8, and HPeV14 as well (data not shown).
DISCUSSION
To date, a variable spectrum of symptoms caused by HPeVs has been described. The common symptoms are similar to that caused by some enteroviruses, including mostly enteritis with diarrhea, and respiratory disease [Joki-Korpela and Hyypia, 1998; Stanway and Hyypia, 1999; Stanway et al., 2000; Benschop et al., 2006b; Baumgarte et al., 2008]. Other symptoms such as rash, fever, flaccid transient paralysis, diseases such as meningoencephalitis, encephalomyelitis, myocarditis, myositis, lymphadenopathy, and syndromes such as Reye's syndrome, hemolytic uremic syndrome have been reported also [Maller et al., 1967; Russell and Bell, 1970; Grist et al., 1978; O'Regan et al., 1980; Figueroa et al., 1989; Koskiniemi et al., 1989; Ehrnst and Eriksson, 1993, 1996; Legay et al., 2002; Ito et al., 2004; Boivin et al., 2005; Watanabe et al., 2007]. In the present study, the described clinical symptoms of the 20 patients whose fecal specimens were positive for HPeV do not seem to be different from those of other diarrheal diseases.
This is the first report on detection of HPeV in a large number of fecal samples collected from Japanese infants and children with acute gastroenteritis. In this study, a high detection rate of HPeV in the tested samples (8.1%) was found. However, it is not represent the percentage of HPeV-infected cases in all patients with acute gastroenteritis (477 cases) during 1-year period, because only 247 samples which had been known to be negative for rotavirus, adenovirus, norovirus, sapovirus, and astrovirus were screened for HPeV. As a result, possible mixed infection of enteric viruses was excluded from analysis. The fact that this study did not include mixed infection of enteric viruses is a limitation of the study. Nevertheless, a proportion of 4.2% (20/477) of HPeV mono-infection was noted among the Japanese infants and children with acute gastroenteritis.
In this study, 702-base full length of the VP1 gene of the studied Japanese strains was obtained successfully. However, some of VP1 sequences of reference strains, especially the strain 451564 of the new genotype HPeV14 [Benschop et al., 2008], available in GenBank databases were partial VP1 sequences. Consequently, the phylogenetic tree was constructed based on 624 bases. The majority of the strains studied (15 strains) were identified as HPeV1, and two strains could be genotyped as HPeV3. In addition, the phylogenetic tree shows that HPeV1 strains clustered into two separate branches, one branch comprises of a limited number of HPeV1 strains including the prototype Harris strain, and the another branch consists of contemporary strains of HPeV1. Accordingly, incorporated with low amino acid identities between HPeV1 strains of the two branches, HPeV1 strains should be divided into two lineages as described in Results Section.
There have been four genotypes of HPeVs found in Japan up to date. They are HPeV1, HPeV3, HPeV4, and HPeV6 [Takao et al., 2001; Ito et al., 2004; Watanabe et al., 2007; Wakatsuki et al., 2008]. By this study, two different types of HPeVs, HPeV1 and HPeV3, were identified in Japanese infants and children with acute gastroenteritis, and the majority of the strains studied belonged to HPeV1. As a result, among the four HPeVs detected in Japan, HPeV1 has a largest number of strains reported and isolated mainly from children with acute gastroenteritis.
In a recent study, a high detection rate of HPeVs (14.6%) was also found in stool samples which were negative for rotavirus, adenovirus, norovirus, sapovirus, and astrovirus, collected from Thai children with acute gastroenteritis [Pham et al., in press]. In that study, four genotypes of HPeV, HPeV1–4, were identified with a predominance of HPeV1. Taken together with previous findings, these results suggest that HPeVs might be possible causative agents of acute gastroenteritis among the studied patients, and HPeV1 is the predominant genotype associated with acute gastroenteritis.
In conclusion, 15 HPeV1 and two HPeV3 strains were found in children with acute gastroenteritis in Japan during 2007–2008. This is the first report of the circulation of HPeV, especially HPeV1 in Japan. The study suggests that HPeV1 should be divided into two lineages.
Acknowledgements
We are grateful to Dr. Miyabi Ito (Department of Microbiology, Aichi Prefecture Institute of Public Health, Japan) for advice for this study.




