Methods for Brain Atrophy MR Quantification in Multiple Sclerosis: Application to the Multicenter INNI Dataset
Grant sponsor: This study was partially supported by Fondazione Italiana Sclerosi Multipla with a research fellowship (FISM 2019/BR/009) and a research grant (FISM2019/S/3), and financed or co-financed with the ‘5 per mille’ public funding.
Abstract
Background
Current therapeutic strategies in multiple sclerosis (MS) target neurodegeneration. However, the integration of atrophy measures into the clinical scenario is still an unmet need.
Purpose
To compare methods for whole-brain and gray matter (GM) atrophy measurements using the Italian Neuroimaging Network Initiative (INNI) dataset.
Study Type
Retrospective (data available from INNI).
Population
A total of 466 patients with relapsing–remitting MS (mean age = 37.3 ± 10 years, 323 women) and 279 healthy controls (HC; mean age = 38.2 ± 13 years, 164 women).
Field Strength/Sequence
A 3.0-T, T1-weighted (spin echo and gradient echo without gadolinium injection) and T2-weighted spin echo scans at baseline and after 1 year (170 MS, 48 HC).
Assessment
Structural Image Evaluation using Normalization of Atrophy (SIENA-X/XL; version 5.0.9), Statistical Parametric Mapping (SPM-v12); and Jim-v8 (Xinapse Systems, Colchester, UK) software were applied to all subjects.
Statistical Tests
In MS and HC, we evaluated the intraclass correlation coefficient (ICC) among FSL-SIENA(XL), SPM-v12, and Jim-v8 for cross-sectional whole-brain and GM tissue volumes and their longitudinal changes, the effect size according to the Cohen's d at baseline and the sample size requirement for whole-brain and GM atrophy progression at different power levels (lowest = 0.7, 0.05 alpha level). False discovery rate (Benjamini–Hochberg procedure) correction was applied. A P value <0.05 was considered statistically significant.
Results
SPM-v12 and Jim-v8 showed significant agreement for cross-sectional whole-brain (ICC = 0.93 for HC and ICC = 0.84 for MS) and GM volumes (ICC = 0.66 for HC and ICC = 0.90) and longitudinal assessment of GM atrophy (ICC = 0.35 for HC and ICC = 0.59 for MS), while no significant agreement was found in the comparisons between whole-brain and GM volumes for SIENA-X/XL and both SPM-v12 (P = 0.19 and P = 0.29, respectively) and Jim-v8 (P = 0.21 and P = 0.32, respectively). SPM-v12 and Jim-v8 showed the highest effect size for cross-sectional GM atrophy (Cohen's d = −0.63 and −0.61). Jim-v8 and SIENA(XL) showed the smallest sample size requirements for whole-brain (58) and GM atrophy (152), at 0.7 power level.
Data Conclusion
The findings obtained in this study should be considered when selecting the appropriate brain atrophy pipeline for MS studies.
Evidence Level
4.
Technical Efficacy
Stage 1.
Multiple sclerosis (MS) is no longer considered an exclusively inflammatory and demyelinating disease of the central nervous system, since neurodegeneration is also a major pathological hallmark.1 Currently, MRI is the most common diagnostic and research tool in MS, which is capable of noninvasively quantifying neurodegeneration especially through measures of atrophy.2
Using MRI, it has been demonstrated that gray matter (GM) atrophy is relevant in MS for explaining clinical disability, cognitive impairment, and clinical evolution of the disease.3-5 Moreover, the no evidence of disease activity (NEDA) status, which is defined by the absence of MRI activity (no appearance of new lesions on T2-weighted and gadolinium-enhancing lesions on T1-weighted sequences), relapses, and disability progression (included in NEDA-3), is upgraded to NEDA-4.6 This proposed measure includes whole-brain atrophy assessment (brain volume loss ≤0.4%), which reflects the neurodegenerative burden of the disease associated with disease progression and irreversible disability.6 Current MS therapeutic strategies target neurodegeneration and the promotion of neuroprotection.7-9
The improvement of image analysis techniques for the reliable estimation of whole-brain and GM atrophy to be used for individualized treatment decisions is still an open area of research.2 Specifically, these measures have still not found clinical application, because of time-consuming procedures and technical or disease-related challenges, due to high image quality requirements and the presence of MS lesions.2, 10, 11 In a previous study, the accuracy and precision of available methods for whole-brain and GM atrophy quantification have been compared by using a test–retest dataset and simulated brain MRI of MS patients, with the aim to provide guidelines for selecting suitable software applications for atrophy measurement.11 However, the existing automatic methods, as demonstrated also in other similar studies, are not reproducible enough to allow the monitoring of atrophy changes in individual patients.11-15 Moreover, the application to multicenter data and larger samples should also be evaluated to confirm previous results,11, 15 in an attempt to formulate updated guidelines for the selection of atrophy tools, with the aim of technical improvements and future clinical integration. Specifically, the majority of previous studies of atrophy quantification in MS (both global and regional) have enrolled only small numbers of patients (unlikely to be representative of the whole spectrum of the disease) and healthy controls (HC) acquired at a single center with the same MRI protocol.12, 14, 16-19
With the current study, we aimed to compare a set of common methods for whole-brain and GM atrophy measurements, taking advantage of the large multi-center dataset from the Italian Neuroimaging Network Initiative (INNI), to guide future users in the selection of an appropriate pipeline for MS studies, moving toward the use of brain atrophy measures in everyday clinical practice.
Materials and Methods
Ethics approval was received from the local Institutional Review Board at each Research Center, and written informed consent was obtained from all subjects.
Subjects
The INNI initiative has supported the creation of a repository where 3-T MRI, clinical, and neurophysiological data from MS patients and HC are collected from four Italian Research Centers in the MS field, with the main goal of improving the application of MRI to identifying novel MRI biomarkers for monitoring the disease course.20
We retrospectively studied 466 MS patients with a relapsing–remitting (RR) clinical phenotype21 and 279 HC collected by INNI from four centers identified here as A, B, C, and D. Inclusion/exclusion criteria for all subjects (patients and HC) were as follows: no contraindications for MRI, no history of alcohol or substance abuse, no neurologic diseases (other than MS in the patients), and no psychiatric diseases.
The Expanded Disability Status Scale (EDSS) scores have been also collected for MS patients in order to assess their clinical disability status and the distribution among the centers. Demographic and clinical characteristics of patients and HC at the baseline visit are reported in Table 1.
| HC | RRMS | P | |
|---|---|---|---|
| All centers | (n = 279) | (n = 466) | |
| Females/males | 164/115 | 323/143 | 0.003a |
| Mean age (SD) | 38 (13) | 37 (10) | 0.71b |
| Mean median disease duration (range) (years) | - | 9.5 8 (0–42) | - |
| Median EDSS (range) | - | 1.5 (0–4.5) | - |
| Median T2 LV (range) (mL) | 0.1 (0–0.5) | 3.9 (0.1–40.7) | <0.001b |
| Center A | (n = 122) | (n = 215) | |
| Females/males | 69/53 | 139/76 | 0.09a |
| Mean age (SD) | 36 (13) | 37 (11) | 0.47b |
| Mean median disease duration (range) (years) | - | 9.3 8.0 (2–30) | - |
| Median EDSS (range) | - | 2 (0–4) | - |
| Median T2 LV (range) (mL) | 0.1 (0–0.3) | 3.5 (0.5–33.8) | <0.001b |
| Center B | (n = 59) | (n = 57) | |
| Females/males | 38/21 | 40/17 | 0.50a |
| Mean age (SD) | 43 (13) | 39 (10) | 0.33b |
| Mean median disease duration (range) (years) | - | 11.8 9.5 (1–42) | - |
| Median EDSS (range) | - | 2 (1–4) | - |
| Median T2 LV (range) (mL) | 0.2 (0–0.5) | 3.1 (0.2–30.8) | <0.001b |
| Center C | (n = 72) | (n = 115) | |
| Females/males | 41/31 | 86/29 | 0.01a |
| Mean age (SD) | 36 (13) | 38 (9) | 0.04b |
| Mean median disease duration (range) (years) | - | 0.5 9.5 (0–27) | - |
| Median EDSS (range) | - | 2 (0–4) | - |
| Median T2 LV (range) (mL) | 0.1 (0–0.3) | 4.3 (0.4–32.9) | <0.001b |
| Center D | (n = 26) | (n = 79) | |
| Females/males | 17/9 | 58/21 | 0.43a |
| Mean age (SD) | 40 (9) | 38 (10) | 0.32b |
| Mean median disease duration (range) (years) | - | 7.9 6.5 (1–22) | - |
| Median EDSS (range) | - | 1.5 (0–4.5) | - |
| Median T2 LV (range) (mL) | 0.07 (0–0.2) | 2.9 (0.1–22.2) | <0.001b |
- SD = standard deviation; HC = healthy controls; RRMS = relapsing–remitting multiple sclerosis; EDSS = Expanded Disability Status Scale; LV = lesion volume.
- a Pearson's chi-square test.
- b Mann–Whitney test.
- Bold is for significant values.
Of this population, 170 MS patients and 48 HC underwent a follow-up re-evaluation 1 year after the baseline. At follow-up, all patients had been relapse-free and steroid-free for at least 1 month before the MRI acquisition.
At baseline, the group of patients with the follow-up visit were age- and sex-matched, as well as clinically matched according to the EDSS score compared to those who had the baseline visit only (median = 1.5, range = 0–4, P = 0.1 compared to patients with baseline visit only).
MRI Acquisitions
Baseline and follow-up brain MRI scans were obtained from each participating Center using 3.0-T scanners. Three-dimensional (3D) T1-weighted (without gadolinium injection) and T2-weighted scans were acquired at each Center using a local protocol and the same MRI scanner at the follow-up acquisitions. We describe the principal pulse sequence parameters in Table 2, although these are more extensively reported elsewhere.22 Although protocols were not standardized, the 3D T1-weighted sequence resulted in similar resolution and coverage among centers.22
| Center A | Center B | Center C | Center D | |||||
|---|---|---|---|---|---|---|---|---|
| Scanner | Philips Intera | GE signa HDxt | Siemens Verio | Philips Achieva | ||||
| Coil | 8-channel head | 8-channel head | 12-channel head | 32-channel head | ||||
| Sequence | Dual-echo | T1w FFE | FLAIR | T1w IR–FSPGR | Dual-echo | T1w MPRAGE | Dual-echo | T1w TFE |
| Coil | 8-channel head coil | 8-channel head coil | 12-channel head coil | 32-channel head coil | ||||
| Plane | Axial | Axial | Axial | Sagittal | Axial | Sagittal | Axial | Axial |
| Voxel (mm3) | 1 × 1 × 3 | 1 × 1 × 1 | 1 × 1 × 3 | 1 × 1 × 1.2 | 1 × 1 × 4 | 1 × 1 × 1 | 1 × 1 × 3 | 1 × 1 × 1 |
| FOV (mm2) | 243 × 243 | 230 × 230 | 256 × 256 | 256 × 256 | 220 × 220 | 256 × 256 | 240 × 240 | 256 × 256 |
| TR (msec) | 2910 | 25 | 9002 | 6.988 | 5310 | 1900 | 4000 | 10 |
| TE (msec) | 16–80 | 4.6 | 120 | 2.85 | 10–103 | 2.9 | 15–100 | 3.9 |
| TI (msec) | – | – | 2500 | 650 | – | 900 | - | 900 |
| FA (deg) | 90 | 30 | 90 | 8 | 150 | 9 | 90 | 8 |
- GE = General Electrics; T1w = T1-weighted; FLAIR = Fluid Attenuated Inversion Recovery; T2w = T2-weighted; FOV = field of view; TR = repetition time; TE = echo time; TI = inversion time; FA = flip angle; TFE = transient field echo; IR = inversion recovery; FSPGR = fast spoiled gradient echo; MPRAGE = magnetization prepared rapid gradient echo; FFE = fast field echo.
Image Analyses
Standardized preprocessing was systematically performed on all INNI MRI data, including a procedure for quality control described in detail elsewhere.22 Briefly, steps of such a procedure included: conversion from DICOM to NIFTI image format; check and monitoring of head positioning; evaluation of image distortions and signal inhomogeneities; evaluation of the presence of artifacts and decision about inclusion/exclusion from the final dataset; for patients with MS, check (and editing, if needed) of T2 lesion masks sent from peripheral sites; co-registration of T2 lesion masks on high-resolution T1-weighted scans, and lesion refilling. During quality control of the longitudinal data, 8 HC and 17 MS patients were excluded for the follow-up scans due to the presence of image artifacts, poor repositioning, or insufficient coverage of the brain. Thus, 279 HC and 466 MS patients were included in the cross-sectional analysis; while 40 HC and 153 MS patient were finally included in the longitudinal analysis.
All sagittal acquisitions were reoriented to the axial plane, and for the 3D T1-weighted images the portion of the neck extending below the cerebellum was cropped. Focal T2-hyperintense white matter (WM) lesions had already been manually identified according to standardized procedures23 and segmented by an expert neurologist (with more than 5 years of experience in MRI) from each participating Center. Lesion volumes were automatically extracted as the number of non-zero voxels in the binary masks of WM lesions multiplied by the voxel size.
In our study, baseline T1-weighted images after applying the lesion-filling technique implemented within the Functional MRI of the Brain (FMRIB) Software Library (FSL; version 5.0.9; https://fsl.fmrib.ox.ac.uk/fsl/fslwiki),24 in order to account for the presence of black holes, were used as input to the toolbox for cross-sectional whole-brain and GM tissue volumes extraction. For longitudinal atrophy quantification, both baseline and follow-up T1-weighted images (after lesion filling) were used as an input to all compared pipelines, in order to assess changes in whole-brain and GM atrophy over time. These cross-sectional and longitudinal (for those with follow-up) analyses were performed for each subject enrolled for this study (both MS and HC).
- SIENA(X)/SIENA-XL, FSL version 5.0.9 (SIENA-XL has been provided by developers upon reasonable request), University of Oxford, UK.25, 27
- SPM toolbox version 12, Matlab (Release 2012a, MathWorks, University College London, UK).28 For SPM-v12 longitudinal atrophy assessment, we need to point out that the pipeline was built in-house using a combination of the longitudinal pairwise registration (a tool already implemented in SPM) and the Jacobian integration technique (see Supplemental Material S1).
- Brain Atrophy Toolbox, Jim version 8 (Xinapse Systems, Colchester, UK).26
An automatic procedure is implemented within all these tools, which are extensively described in the Supplemental Material S1 for each software compared. Thus, after the previously described prerocessing steps on T1-weighted input images, we ran the automatic installed tools with default/recommended parameters.
The methods implemented within each automatic pipeline are extensively described in the Supplemental Material S1. The methods compared have computer processing times ranging from a minimum of 15 minutes for SPM-v12 and 20–30 minutes for Jim-v8 cross-sectional pipelines, to a maximum of 50–60 minutes for SIENAX on a computer with an Intel Xeon 12 core processor (Intel, Santa Clara, California, USA), 16 GB RAM and a Quadro K600 GPU (NVIDIA, Santa Clara), running CentOS Linux 7.
Statistical Analysis
Statistical analysis was performed using the R software package (version 3.1.1; The R Project for Statistical Computing; https://www.r-project.org/). Demographic data and T2 lesion volumes were compared between groups using the χ2 Pearson test for categorical variables and the Mann–Whitney U or t-tests for continuous variables. We evaluated the intraclass correlation coefficient (ICC), to assess the strength of the agreement among the different software packages based on whole-brain and GM atrophy measures (both cross sectional and longitudinal). The ICC was also evaluated separately for each center for cross-sectional whole-brain and GM segmentations in order to check for possible bias due to the different MRI acquisitions at each Center and/or MRI sequences. Between-group differences (center-adjusted) in whole-brain and GM cross-sectional volumes for the different software packages were expressed as effect size, calculated according to the Cohen's d definition.29
The sample size requirements at three different power levels (lowest = 0.7) for detecting a significant difference in the rates of whole-brain and GM atrophy progression between HC and MS at the 0.05 alpha level were estimated for each software package. False discovery rate (Benjamini–Hochberg procedure) correction was applied. A P value <0.05 was considered statistically significant.
Results
Demographic and Clinical Features
Table 1 summarizes the main demographic and clinical characteristics of the subject groups at the baseline visit. Considering all datasets together, we found that the ratio of females to males included was significantly different between MS patients and HC. Separately, for the majority of the datasets considered, no significant differences in age or sex were found between the two groups (P values ranging from 0.32 to 0.50), except for center C, where patients were significantly older than HC. Center C showed a significant sex difference between MS patients and HC.
Disease duration was significantly different between datasets from centers B and C, lesion load was significantly different between patients of centers C and D, and EDSS scores were significantly different between several dataset pairs: A and D, B and C, B and D, and C and D.
At follow-up (mean follow-up interval = 0.98 ± 0.24 years for MS, 1.05 ± 0.21 years for HC; P = 0.07), median EDSS was 1.5 (range = 0–4.5, P = 0.1 vs. baseline) and 12 MS patients had worsened clinically (EDSS score increase ≥1.5 when baseline EDSS was 0, ≥1.0 when EDSS at baseline was <6.0, and ≥0.5 when EDSS at baseline was ≥6.0).
Assessment of Cross-Sectional Atrophy Measures
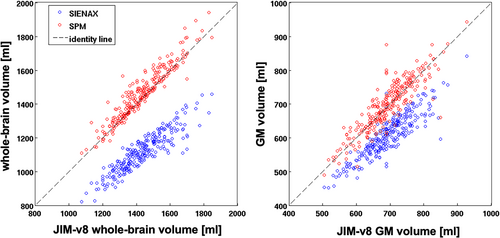
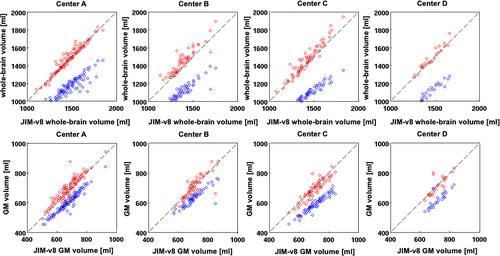
For the cross-sectional assessment, we found a significant agreement between whole-brain volumes obtained by SPM-v12 and Jim-v8 for both HC (ICC = 0.93) and MS patients (ICC = 0.84), while no significant agreements were found in the comparisons between SIENAX and both SPM-v12 (P = 0.19) and Jim-v8 (P = 0.21) (Fig. 1), with a systematic shift of SIENAX whole-brain volumes over lower values. Similarly, for GM cross-sectional atrophy, we found a significant agreement between the GM volumes obtained by SPM-v12 and Jim-v8 for both HC (ICC = 0.66) and MS patients (ICC = 0.90), while no significant agreement was found for the comparisons among SIENAX and both SPM-v12 (P = 0.29) and Jim-v8 (P = 0.32) (Fig. 1). Considering each Center separately (Fig. 2), SPM-v12 and Jim-v8 showed again the highest agreements both for whole-brain and GM volumes for HC and MS patients, as shown in Table 3. No significant agreements were found in the comparisons between SIENAX and both SPM-v12 and Jim-v8 atrophy results for each Center individually and for both whole-brain (center A: P = 0.18 and P = 0.21; center B: P = 0.16 and P = 0.32; center C: P = 0.09 and P = 0.12; center D: P = 0.25 and P = 0.43) and GM volume (center A: P = 0.21 and P = 0.22; center B: P = 0.11 and P = 0.33; center C: P = 0.15 and P = 0.13; center D: P = 0.36 and P = 0.53).
| ICC | Jim-v8 SIENAX | Jim-v8 SPM-v12 | SIENAX SPM-v12 | ||||
|---|---|---|---|---|---|---|---|
| Whole-brain | GM | Whole-brain | GM | Whole-brain | GM | ||
| HC | Center A | 0.25 | 0.52 | 0.78* | 0.70* | 0.23 | 0.54 |
| Center B | 0.22 | 0.48 | 0.83* | 0.86* | 0.17 | 0.51 | |
| Center C | 0.19 | 0.32 | 0.92* | 0.87* | 0.16 | 0.37 | |
| Center D | 0.12 | 0.29 | 0.93* | 0.81* | 0.14 | 0.40 | |
| MS | Center A | 0.23 | 0.47 | 0.97* | 0.76* | 0.21 | 0.50 |
| Center B | 0.17 | 0.56 | 0.80* | 0.90* | 0.13 | 0.59 | |
| Center C | 0.14 | 0.33 | 0.86* | 0.82* | 0.11 | 0.45 | |
| Center D | 0.18 | 0.39 | 0.94* | 0.62* | 0.16 | 0.58 | |
- Abbreviations: ICC=Intra-class correlation coefficient; GM = gray matter; HC = healthy controls; MS = multiple sclerosis; SPM = Statistical Parametric Mapping.
- * P < 0.05.
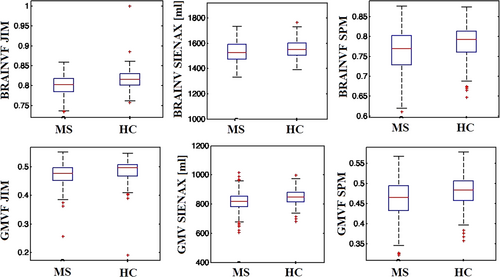
Regarding the difference between MS patients and HC (Fig. 3), all software tools showed low effect sizes (Cohen's d = −0.40, −0.32, −0.3, for SPM-v12, Jim-v8, and SIENAX, respectively) for whole-brain atrophy quantification, while SPM-v12 and Jim-v8 showed the highest effect sizes for cross-sectional GM atrophy (Cohen's d = −0.63 and −0.61, respectively) compared to SIENAX (Cohen's d = −0.53). All software applications showed a significant difference for both whole-brain and GM volume results between MS patients and HC.
Assessment of Longitudinal Atrophy Measures
For longitudinal whole-brain atrophy assessment, SPM-v12 and SIENA showed significant agreement for both HC (ICC = 0.62) and MS patients (ICC = 0.43), while no significant agreement was found in the comparisons between Jim-v8 and both SPM-v12 and SIENA whole-brain volume changes (all ICCs < 0.1, P = 0.54 and P = 0.37, respectively). For longitudinal GM atrophy quantification, SPM-v12 and Jim-v8 showed a significant agreement for MS patients (ICC = 0.59), while a lower value of agreement was found for GM volumes (ICC = 0.35) in HC, as well as between SIENA-XL and SPM-v12 (ICC = 0.4 for MS; ICC = 0.26, P = 0.05 for HC). No significant agreement was found for longitudinal GM atrophy quantification between SIENA-XL and Jim-v8 results (ICC = 0.12, P = 0.63).
Jim-v8 and SIENA-XL showed the smallest and comparable sample size requirements for both whole-brain and GM longitudinal atrophy assessment at the different power levels. Table 4 shows the sample size requirements for each software package at each power level.
| Power level | 70% | 80% | 90% | |||
|---|---|---|---|---|---|---|
| Sample size | ||||||
| Whole-brain | GM | Whole-brain | GM | Whole-brain | GM | |
| SIENA/SIENA-XL | 62 | 152 | 79 | 193 | 105 | 258 |
| SPM-v12 | 384 | 549 | 488 | 699 | 653 | 935 |
| Jim-v8 | 58 | 163 | 73 | 207 | 98 | 277 |
- GM = gray matter; SPM = Statistical Parametric Mapping.
Discussion
In this work, we aimed to compare three of the available methods for whole-brain and GM atrophy measurements on a multicenter dataset, in order to guide the selection of a suitable atrophy processing pipeline for large MS studies. Using the INNI dataset, we found good agreement between SPM-v12 and Jim-v8, which also better separated GM atrophy distribution between MS patients and HC in comparison to the third software package (SIENAX). However, the newly proposed SIENA(XL) and Jim-v8 required the smallest sample size for longitudinal atrophy quantification.
The high agreements found between SPM-v12 and Jim-v8 for cross-sectional whole-brain and GM atrophy quantification may be explained by the fact that the implementation of both methods was similar. These pipelines started with the same International Consortium from Brain Mapping (ICBM) atlas30 as a prior tissue type probability map, which was registered to the T1-weighted image. Then, the T1-weighted image was automatically segmented into GM, WM, and cerebrospinal fluid, giving spatially normalized volumes (see Supplemental Material S1 for a detailed description).31 On the other hand, with SIENAX, voxels within the brain were classified based on image intensities using a hidden Markov random field model, and tissue volumes were normalized using a scaling factor accounting for head size.32
The ICC reliability indices between the atrophy results for each software package, estimated separately for each center, were as expected when considering all centers together: the comparisons for the different software packages did not change for any specific center. Thus, pooling T1-weighted images with similar quality from different MRI acquisition centers did not affect or introduce a bias into the degree of agreement between the software packages. In this study, even though the acquisition was not standardized, the 3D T1-weighted anatomical brain images from the INNI database, provided as input to the atrophy pipelines, were similar between the centers especially in terms of resolution and field of view (no relevant effect on lesion filling). Thus, our findings here may endorse the importance of large multicenter quantitative studies of brain tissue atrophy in MS with high-resolution T1-weighted images, even if they are acquired without using highly standardized acquisition protocols. However, a high degree of standardization of the different MRI acquisition protocols at the different centers and data harmonization procedures are strongly recommended if the most reliable results are to be obtained.10, 33, 34
For analyses at group level, both whole-brain and GM atrophy measures obtained by all software packages facilitated differentiation between HC and MS patients. However, it is important not to combine measures obtained using different pipelines within the same analysis, even for large studies due to their different levels of accuracy and precision in estimating brain atrophy.11 Moreover, the different software packages did not show similar strengths in the assessment of between-group differences (effect sizes), especially for GM cross-sectional atrophy quantification. This is likely because GM volume is smaller than the whole-brain volume, and therefore subject to greater measurement errors as a percentage of any real change in volume.2 Moreover, the measurement of GM atrophy in MS could be heavily affected by disease-specific technical challenges (eg presence of T1-hypointense MS lesions), more so than for whole-brain volume quantification.35
In contrast to the cross-sectional results, when SPM-v12 and SIENA/SIENA-XL were applied longitudinally, they showed a significant agreement for whole-brain and GM atrophy values. In this case, both pipelines were initialized in a similar way: they performed a longitudinal pairwise registration to align the baseline and follow-up 3D T1-weighted scans in a half-way space and produced a result consisting of a direct estimation of whole-brain volume change in this space (see Supplemental Material S1 for methodological details). The longitudinal pipeline from Jim-v8 also performed a half-way registration between baseline and follow-up images and used the Jacobian determinants of the deformation field, as in SPM-v12 longitudinal pipeline, but only when estimating GM volume change.26 Thus, Jim-v8 showed significant agreement with SPM-v12 only for GM atrophy quantification.
In the majority of cases, we found that the agreement between analysis methods obtained for MS patients was higher than for HC, for both cross-sectional and longitudinal atrophy. This may be due to the fact that we expected smaller cross-sectional brain volume variations between subjects, and smaller longitudinal changes in atrophy in HC than in patients with MS. Thus, not all pipelines may be sensitive enough to capture these smaller variations, leading to lower levels of agreement in HC.
For the use in a clinical setting, atrophy assessment should be both highly sensitive and precise. In this study, we found that the Jim-v8 and SIENA(XL) longitudinal pipelines, for both whole-brain and GM atrophy quantification, required comparable and the smallest sample sizes at all power levels compared to the third software, making these tools appealing for application in MS studies. This was due to the lower difference for SPM-v12 between the means of the distribution of atrophy rates for HC and MS patients relative to the standard deviation found for HC, in comparison to the other two software packages (0.2 for whole-brain assessments against 0.47 and 0.45 for Jim-v8 and SIENA, respectively). For SIENA whole-brain atrophy assessment, our findings on sample size requirements may be in line with previous literature.36, 37 The requirement of a small sample size is also desirable for the creation of normative data for atrophy measures, to be used in individualized medicine when evaluating treatment effects.38 However, for SPM-v12 longitudinal atrophy assessment, we need to point out that the pipeline was built in-house using a combination of the longitudinal pairwise registration (a tool already implemented in SPM) and the Jacobian integration technique. Thus, longitudinal atrophy assessment is not part of an officially released processing pipeline.
From empirical evaluations obtained with the use of these tools, while having access to a free license (as in the case of SIENA(X)) is desirable, the ease of integration into the clinical routine (which is more straightforward for Jim-v8) and the ease of use (as in the case of both Jim-v8 and SPM-v12) are additional important considerations when selecting an atrophy processing pipeline. Especially for nonexpert users, the possibility to have a simple graphical user interface to guide the entire atrophy procedure is desirable, with respect to the use of a pipeline from the command-line interface. However, in research settings, the versatility of use and the free license are often desirable even at the expense of a less user-friendly toolbox.
Limitations
One limitation of this study was that it could not provide information about the accuracy and precision of the atrophy assessment methods investigated, due to the lack of a test–retest dataset or a comparison with ground truth, as it has been achieved in a previous study on a smaller cohort.11 However, the analyses performed in this study focused on the application of currently available techniques to a large dataset, in order to compare their performances in detecting whole-brain and GM atrophy on cross-sectional and longitudinal multicenter MRI data. Moreover, we enrolled only the RR MS phenotype to focus on the software comparison, limiting the heterogeneity due to the disease. Further investigations should include also progressive forms of the disease. Finally, brain alterations due to the normal aging could affect atrophy measures. In this study, we tried to overcome this issue by including a group of age-matched HC for the cross-sectional comparisons and we considered a limited follow-up period of 1 year. Furthermore, the SIENA-XL toolbox is a recently published longitudinal atrophy method not publicly available that has been included in this study under a permission of the developers. Currently, this is a limitation for the reproducibility of this study that would need to be overcome in the future when the software will be freely available.
Even if the choice not to include Freesurfer in the comparison could be seen as a limitation, in this study we decided to focus on two main aspects: 1) to restrict the comparison to those software that resulted as well-performing from our previous study on a single center and smaller sample size11; 2) to avoid including methods that are not specifically implemented and optimized for a volumetric quantification of brain tissues (as Freesurfer). Moreover, the inclusion of other atrophy measures as cortical thickness did not allow a direct comparison among the results of the different pipelines evaluated and goes beyond the purpose of this study: to focus on tools that would give an easy estimation of both whole-brain and gray matter (volumetric) atrophy measures for a possible future introduction in the clinical scenario.
Conclusion
Using the INNI dataset, we compared the performance of different atrophy tools on a large sample of patients acquired by multiple centers. We found good agreement between SPM-v12 and Jim-v8 atrophy results, while Jim-v8 and SIENA(XL) may provide the smallest estimated sample size for longitudinal atrophy quantification. The findings obtained in this study should be considered when selecting the appropriate brain atrophy pipeline for MS studies.
Acknowledgment
Open access funding provided by BIBLIOSAN.




