Maternal ApoE genotype and risk of recurrent pregnancy loss: An updated systematic review and meta-analysis
Funding information: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
PROSPERO registration number: CRD42022333713.
Abstract
Background
In order to clarify the role of the maternal apolipoprotein E (ApoE) genotype and the risk of recurrent pregnancy loss (RPL), we herein performed an updated systematic review and meta-analysis to reevaluate the evidence on this association.
Methods
A comprehensive literature search was performed on PubMed, Web of Knowledge and the Cochrane library up to September 2022. Methodological study quality was assessed using the Newcastle–Ottawa Scale and the credibility of significant pooled odds ratios (ORs) was estimated by the false positive report probability and the Bayesian false discovery probability.
Results
Twelve studies published from 2009 to 2022 fulfilled the inclusion criteria. In the overall analysis, the ε4 allele was found to confer an increased risk of RPL compared to the ε3 allele (OR 1.60, 95% CI 1.00–2.55, p = 0.049) and women carrying the ApoE ε4 allele displayed a higher risk of RPL compared with those carrying the ε2 and ε3 alleles (OR 1.75, 95% CI 1.06–2.87, p = 0.028). Subgroup analysis based on subjects’ ethnicity revealed that these associations were restricted to the Asian population (ε4 allele vs. ε3 allele, OR 5.93, 95% CI 1.79–19.61, p = 0.004; ε4 allele carriers vs. carriers of ε2 and ε3 alleles, OR 8.42, 95% CI 1.47–48.12, p = 0.017). None of the associations detected were found to be noteworthy under false positive report probability or Bayesian false discovery probability at a prior probability of 0.001.
Conclusions
This updated meta-analysis highlights an association between maternal ApoE genotype and RPL risk in Asians, but not in Caucasians. Further case–control studies are warranted in women of Asian ancestry to exclude the possibility of false-positive findings.
1 INTRODUCTION
Recurrent pregnancy loss (RPL) is a relevant health problem in reproductive medicine, defined as two or more spontaneous abortions.1 According to which of the different definition for RPL is used,2, 3 the prevalence of women affected by RPL ranges from 1 to 5%.3-5 Recurrent pregnancy loss is recognized as a complex multifactorial condition that could be influenced by the interaction of several factors, such as chromosome abnormalities, uterine alterations, infections and endocrinological and autoimmune diseases,6, 7 as well as by genetic factors for thrombophilia susceptibility.8, 9 Among the candidate gene investigated for possible association with RPL, much interest has been focused during the past several years on the APOE gene which encodes for apolipoprotein E, a protein that plays a key role in the lipid metabolism and acts as a major regulator of steroid hormone function. The APOE gene is located on chromosome 9q13.2.2 and is characterized by three main alleles, namely ɛ2, ɛ4, and ɛ3, the former of which is the least widespread isoform while the latter is the most common one.10 These alleles are designated by two single nucleotide polymorphisms in exon 4 that originate cysteine–arginine interchanges at codons 112 (rs429358) and 158 (rs7412). Specifically, ɛ2 allele has cysteine at both positions (Cys 112, Cys 158) and ɛ4 has arginine at both positions (Arg 112, Arg 158) while ɛ3 allele has cysteine at 112 and arginine at 158 positions (Cys 112, Arg 158). The combination of these three alleles results in six different genotypes in humans (ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4 and ε4/ε4).11 The ApoE ε4 allele has been reported to correlate with higher levels of low-density lipoprotein cholesterol and with an increased risk of atherosclerosis, which in turn might contribute to the thrombogenesis that is believed to affect placental flow and fetal intrauterine growth.12 In addition, ApoE alleles are shown to be differentially involved in inflammatory response, platelet function, apoptosis and the modulation of oxidative stress.13 On the basis of this evidence, a number of studies have been carried out to assess the association of maternal ApoE genotype with the risk of RPL. While some studies have found that women carrying the Apo ε4 allele are at higher risk of RPL,14, 15 other studies reported no significant association of ApoE genotype with RPL.16, 17 The reasons for such contrasting results are unclear, but possible explanations might be the low statistical power of each single study and/or the different ethnic origin of patients enrolled. Given that these limitations can be partly overcome by combining primary studies using a meta-analytic approach,18 and that additional reports have been published after publication in 2014 of the most recent meta-analysis available on this topic,19 we conducted an updated systematic review and meta-analysis in order to quantitatively summarize all of the current available studies and provide further insights into the relationship between maternal ApoE genotype and susceptibility to recurrent pregnancy loss. In comparison with the previous meta-analysis,19 for the first time, subgroup analyses were performed based on ethnicity, and the false-positive report probability (FPRP) analysis20 and the Bayesian false discovery probability (BFDP) test,21 were applied to assess the noteworthiness of significant findings.
2 MATERIALS AND METHODS
2.1 Search and inclusion/exclusion criteria
The present systematic review was reported according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines22 and the protocol published in the International Prospective Register of Systematic Reviews “PROSPERO” (registration number: CRD42022333713). The PubMed, Web of Knowledge and Cochrane Library databases were used up to 14 September 2022 to search primary studies using the Boolean combination of the following key terms: (abortion OR miscarriage OR pregnancy loss OR RPL OR fetal loss OR fetal death) AND (apolipoprotein OR ApoE OR Apo E) AND (polymorphism OR polymorphisms OR genetic OR genetics OR variant OR variants OR allele OR alleles).
Studies were included if they met the following criteria: (1) studies investigating the association between maternal ApoE gene genotype and the risk of recurrent pregnancy loss (i.e. two or more spontaneous abortions); (2) the use of a case–control study design; (3) cases with a history of two or more spontaneous miscarriages; (4) controls had to be women without history of RPL or, at most, one miscarriage; (5) analysis of rs429358 and rs7412 polymorphisms of ApoE gene; or (6) data on distribution of ApoE alleles or genotypes in both cases and controls. Studies were excluded if they met the following critera: (1) not human studies; (2) review articles, editorials, comments, meeting abstract; (3) obviously not related to the research topics; (4) duplication of previous publications; (5) not case–control studies; (6) not maternal ApoE alleles or genotypes; or (7) previous meta-analysis on the same research topics.
The retrieved studies were read in their entirety to assess their appropriateness for inclusion in the meta-analysis. There were no language restrictions for inclusion. Reference lists of eligible studies were also reviewed to identify additional published works not initially retrieved. All studies were independently analysed by two reviewers (S.C and F.A) and any discrepancies were resolved through a consensus discussion with a third reviewer (S.T.).
2.2 Data extraction
The following information was extracted from each included study: the first author’s last name, year of publication, study location, ethnic background, number of miscarriages in cases, definition of controls, total number of cases and control samples, distribution of maternal ApoE alleles and/or genotypes in both cases and controls. All studies were independently analyzed by two reviewers (F.A. and S.T.) and any discrepancies were resolved by consensus.
2.3 Study quality assessment
Methodological study quality was independently assessed by two authors (S.C. and S.T.) using the Newcastle–Ottawa scale (NOS) for case–control studies (available at https://www.ohri.ca//programs/clinical_epidemiology/oxford.Asp). Disagreements between reviewers were resolved by consensus. The NOS is composed of eight items categorized into three major components including selection, comparability and exposure. The NOS ranges from zero to nine stars, with a higher number of stars indicating a higher methodological quality. The methodological quality was determined according to the total score achieved by each study: low quality, 0–3; medium quality, 4–6; and high quality, 7–9.
2.4 Statistical analysis
The effect measure of interest was the odds ratio (OR), which was expressed as point estimate with 95% confidence interval (CI). Odds ratio estimates from each study were combined based on genetic contrasts of the ApoE allele (ε2 vs. ε3, ε4 vs. ε3, ε2 vs. ε4) and ApoE genotypes in different combinations. A Woolf–Haldane continuity correction of 0.5 was added in the two-by-two contingency table to generate a finite odds ratio when any zero cell occurred. The OR estimates were pooled using the random-effects model (DerSimonian–Laird method), which incorporates between-study heterogeneity and allows for a different effect in each study.23 Heterogeneity between studies was estimated by the chi-square–based Cochran's Q statistic (significant for p < 0.10) as well as the I2 index (range, 0–100%), which quantifies heterogeneity irrespective of the number of studies values. The random-effects model coincides with the fixed-effect model in the absence of heterogeneity (I2 = 0).24 Leave-one-out sensitivity analyses were conducted to assess robustness of the estimated effect sizes. In addition, subgroup analyses were performed based on the ethnic origin of subjects when relevant data were reported in at least three independent studies. Meta-analyses were performed with ProMeta version 2 software (INTERNOVI di Scarpellini Daniele s.a.s., Cesena FO, Italy), and the significance of pooled ORs was set at p < 0.05. Publication bias was evaluated graphically by drawing funnel plots and statistically analysed by means of Egger’s test using the ProMeta version 2 software. In case of statistical evidence of funnel plot asymmetry (Egger’s p-value <0.10), the “trim-and-fill” procedure was used to determine the stability of the results.25 The noteworthiness of significant pooled ORs was estimated by application of the Bayesian false positive report probability (FPRP)20 and the Bayesian false discovery probability (BFDP)21 tests. The FPRP and BFDP values were calculated at the prior probability of 0.001 that is expected for candidate single nucleotide polymorphisms.26 A significant pooled result (p < 0.05) with an FPRP value of <0.2, or with a BFDP value of <0.8 indicated a noteworthy association. The FPRP and BFDP calculations were done using the Excel spreadsheet provided by Wacholder et al. (2004)20 and Wakefield et al. (2007),21 respectively.
3 RESULTS
3.1 Study characteristics
A total of 267 studies were identified through a search of PubMed, Web of Knowledge and Cochrane library. After exclusion of 80 hits for duplication and additional 175 studies for not meeting the inclusion criteria, 12 articles published between 2009 and 2022 were included in the systematic review.14-17, 27-34 The detailed flowchart of the literature review process and reasons for study exclusion are shown in Figure 1. Overall, the present systematic review included 2,231 cases, ranging from a minimum of six in the study of Lambrinoudaki et al. (2010)28 to a maximum of 802 in the report of Gumus et al. (2018).31 The total number of control subjects was 2,241, ranging from a minimum of 3727 to a maximum of 1060.30 The case group consisted of women who had experienced at least two unexplained spontaneous miscarriages in all studies except in three reports,16, 31, 33 which included cases with at least three spontaneous miscarriages. Seven studies included women of Caucasian origin17, 28-33 and four studies enrolled women with Asian ancestry,14-16, 34 while in one study27 case–control women were of mixed ethnic origin. The distribution of ApoE alleles and genotypes from each included study is shown in Supplementary Table S1. Data on the distribution of ApoE alleles was not reported in one study,33 while ApoE genotypes distribution was not available in two studies.17, 34 With regard to the study quality analysis (Table 1), the NOS results had scores ranging from 3 to 6, with an average of 4.25, which indicated that the methodological quality of studies included was medium. Individual item scores for each study are presented in Supplementary Table S2. Other study characteristics, including definition of control population, mean maternal age of cases/controls and the method used to determine ApoE genotypes are shown in Table 1.
| First authorref | Year | Country (ethnicity) | Total cases | Miscarriage, (n) | Total controls | Definition of controls | Mean maternal age, cases/controls | ApoE genotyping method | NOS score |
|---|---|---|---|---|---|---|---|---|---|
| Goodman C.27 | 2009 | USA (Mix) | 69 | ≥2 | 37 | Women with at least two live births and not more than one spontaneous abortion | 34.7/37.5 | PCR-reverse blot hybridization | 3 |
| Agarwal M.16 | 2010 | North India (Asian) | 200 | ≥3 | 200 | Healthy parous women with at least two live births, with no history of abortions | 28.4 ± 5.9/31.9 ± 7.3 | PCR-RFLP | 6 |
| Lambrinoudaki I.28 | 2010 | Greece (Caucasian) | 6 | ≥2 | 62 | Women with no history of pregnancy loss | – /− | Multiple PCR/reverse blot hybridization | 4 |
| Ozdemir O.29 | 2012 | Turkey (Caucasian) | 543 | ≥2 | 113 | Women with no history of pregnancy loss and at least one normal pregnancy | 27.8 ± 2.1/28.9 ± 2.2 | Multiple PCR/reverse blot hybridization and Real-Time PCR | 5 |
| Rynekrova J.30 | 2012 | Czech Republic (Caucasian) | 76 | ≥2 | 1060 | Women with at least two healthy children and no history of abortions | 15–44/25–65 | PCR-RFLP | 4 |
| Asgari M.14 | 2013 | Iran (Asian) | 81 | ≥2 | 81 | With two healthy children and no history of RPL | 27.7/30 | PCR-RFLP | 4 |
| Korkmazer E.17 | 2013 | Turkey (Caucasian) | 45 | ≥2 | 45 | With at least one live birth and no history of spontaneous abortion | 33 ± 0.78/33.5 ± 0.7 | Real-Time PCR | 5 |
| Poursadegh Zonouzi A.15 | 2014 | Iran (Asian) | 100 | ≥2 | 100 | With no history of pregnancy loss | 31.96 ± 4.76/29.64 ± 3.63 | PCR-RFLP | 4 |
| Gumus E.31 | 2018 | Turkey (Caucasian) | 802 | ≥3 | 244 | Women with at most one abortion in three consecutive pregnancies | 28.17 ± 4.07/27.96 ± 3.15 | Real-Time PCR | 4 |
| El Achi H.32 | 2018 | Lebanon (Caucasian) | 70 | ≥2 | 160 | Population-based control women | 32.2/− | PCR-reverse blot hybridization | 3 |
| Turienzo A.33 | 2020 | Spain (Caucasian) | 89 | ≥3 | 89 | Women with no history of pregnancy loss | −/− | Real-Time PCR | 4 |
| Younis M.34 | 2022 | Saudi Arabia (Asian) | 150 | ≥2 | 50 | With at least two live births and no history of miscarriages | 31 ± 5.1/32 ± 5.04 | PCR-reverse blot hybridization | 5 |
- NOS, Newcastle–Ottawa Scale; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; RPL, recurrent pregnancy loss.
3.2 Quantitative data synthesis
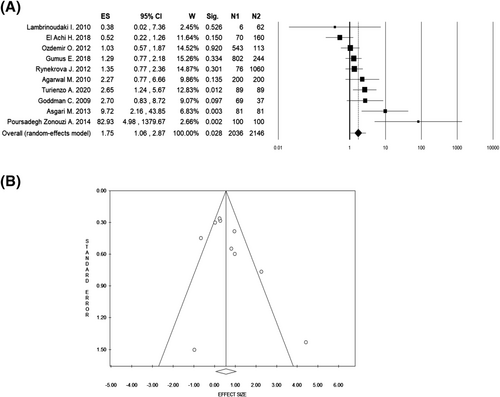
The summary of random-effect meta-analyses is shown in Table 2. In the overall analysis of 10 studies, including 2036 cases and 2146 controls, women carrying the ε4 allele (i.e. ApoE ε2/ε4, ε3/ε4 and ε4/ε4) were found to be at higher risk of RPL (OR 1.75, 95% CI 1.06–2.87, p = 0.028, Figure 2A) compared with those carrying other ApoE alleles (i.e. ε2/ε2, ε2/ε3 and ε3/ε3), in the presence of significant heterogeneity among studies (I2 = 64.9%, p = 0.002). To evaluate the robustness of the overall pooled estimate, we performed a leave-one-out sensitivity analysis by iteratively removing one study at a time and recalculating the summary OR. The results of this leave-one-out meta-analysis (Supplementary Figure S1) showed that the pooled OR ranged from 1.51 (95% CI 0.95–2.39, p = 0.080) when the study of Asgari et al. (2020)14 was omitted, to 2.00 (95% CI 1.22–3.27, p = 0.006) when the study of El Achi et al (2018)32 was excluded from the analysis (Figure 3), suggesting a lack of robustness of the overall pooled results. With regard to the other genetic contrasts of ApoE alleles or genotypes (Table 2), a borderline significant association was found for the comparison of the ε4 allele vs. the ε3 allele in the overall pooled analysis (OR1.60, 95% CI 1.00–2.55, p = 0.049, Figure 3A) in the presence of significant heterogeneity among studies (I2 = 64.7%, p = 0.002). No significant associations emerged in the remaining overall pooled results.
| Group or subgroup | Number of studies | Cases/controls, n | Test of association | Test of heterogeneity | Egger's p-value | FPRP value at prior probability of 0.001 | BFDP 0.001 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-Value | I2 (%) | p-Value | Power* | OR = 1.5 | |||||
| ε2 vs. ε3 | ||||||||||
| Overall | 11 | 3977/3995 | 1.07 (0.78–1.47) | 0.680 | 39.7 | 0.084 | 0.421 | |||
| Caucasian | 6 | 2886/3079 | 1.17 (0.79–1.73) | 0.429 | 34.9 | 0.175 | ||||
| Asian | 4 | 971/846 | 1.07 (0.56–2.06) | 0.838 | 55.9 | 0.078 | ||||
| ε4 vs. ε3 | ||||||||||
| Overall | 11 | 3992/4006 | 1.60 (1.00–2.55) | 0.049 | 64.7 | 0.002 | 0.107 | 0.393 | 0.992 | 0.998 |
| Caucasian | 6 | 2864/3139 | 1.07 (0.81–1.41) | 0.629 | 3.1 | 0.397 | ||||
| Asian | 4 | 998/800 | 5.93 (1.79–19.6) | 0.004 | 62.2 | 0.047 | 0.012 | 0.997 | 0.995 | |
| ε2 vs. ε4 | ||||||||||
| Overall | 10 | 597/579 | 0.57 (0.32–1.02) | 0.060 | 61.2 | 0.006 | 0.001 | |||
| Caucasian | 5 | 418/502 | 1.13 (0.77–1.67) | 0.518 | 0 | 0.767 | ||||
| Asian | 4 | 153/66 | 0.18 (0.07–0.48) | <0.001 | 28.6 | 0.241 | 0.004 | 0.993 | 0.983 | |
| ε2 allele carriers vs. carriers of ε3 and ε4 alleles | ||||||||||
| Overall | 9 | 1947/2057 | 1.10 (0.75–1.62) | 0.621 | 47.6 | 0.054 | 0.496 | |||
| Caucasian | 5 | 1497/1639 | 1.31 (0.88–1.95) | 0.180 | 26.4 | 0.245 | ||||
| Asian | 3 | 381/381 | 1.09 (0.43–2.78) | 0.863 | 68.8 | 0.040 | ||||
| ε3 allele carriers vs. carriers of ε2 and ε4 alleles | ||||||||||
| Overall | 9 | 1947/2057 | 0.60 (0.32–1.12) | 0.109 | 1.4 | 0.422 | 0.017 | |||
| Caucasian | 5 | 1497/1639 | 0.57 (0.22–1.51) | 0.257 | 0 | 0.721 | ||||
| Asian | 3 | 381/381 | 0.22 (0.03–1.74) | 0.151 | 62.6 | 0.069 | ||||
| ε4 allele carriers vs. carriers of ε2 and ε3 alleles | ||||||||||
| Overall | 10 | 2036/2146 | 1.75 (1.06–2.87) | 0.028 | 64.9 | 0.002 | 0.143 | 0.271 | 0.990 | 0.997 |
| Caucasian | 6 | 1586/1728 | 1.21 (0.82–1.79) | 0.336 | 41.7 | 0.127 | ||||
| Asian | 3 | 381/381 | 8.42 (1.47–48.12) | 0.017 | 69.9 | 0.036 | 0.026 | 0.998 | 0.998 | |
| ε2/ε2 vs. other ApoE genotypes | ||||||||||
| Overall | 9 | 1947/2057 | 1.16 (0.54–2.49) | 0.696 | 0 | 0.947 | 0.824 | |||
| Caucasian | 5 | 1497/1639 | 1.43 (0.34–6.04) | 0.627 | 0 | 0.637 | ||||
| Asian | 3 | 381/381 | 1.12 (0.45–2.80) | 0.815 | 0 | 0.997 | ||||
| ε2/ε3 vs. other ApoE genotypes | ||||||||||
| Overall | 9 | 1947/2057 | 1.03 (0.73–1.44) | 0.884 | 27.8 | 0.197 | 0.199 | |||
| Caucasian | 5 | 1497/1639 | 1.26 (0.86–1.84) | 0.242 | 19.4 | 0.291 | ||||
| Asian | 3 | 381/381 | 0.76 (0.42–1.38) | 0.363 | 14.9 | 0.309 | ||||
| ε2/ε4 vs. other ApoE genotypes | ||||||||||
| Overall | 9 | 1947/2057 | 1.90 (0.72–5.03) | 0.195 | 0 | 0.705 | 0.280 | |||
| Caucasian | 5 | 1497/1639 | 2.57 (0.67–9.80) | 0.167 | 0 | 0.796 | ||||
| Asian | 3 | 381/381 | 3.53 (0.48–26.15) | 0.217 | 0 | 0.438 | ||||
| ε3/ε3 vs. other ApoE genotypes | ||||||||||
| Overall | 9 | 1947/2057 | 0.69 (0.45–1.06) | 0.093 | 74.1 | <0.001 | 0.775 | |||
| Caucasian | 5 | 1497/1639 | 0.90 (0.59–1.39) | 0.636 | 61.2 | 0.036 | ||||
| Asian | 3 | 381/381 | 0.35 (0.09–1.34) | 0.127 | 89.0 | <0.001 | ||||
| ε3/ε4 vs. other ApoE genotypes | ||||||||||
| Overall | 9 | 1947/2057 | 1.52 (0.89–2.59) | 0.126 | 61.0 | 0.008 | 0.165 | |||
| Caucasian | 5 | 1497/1639 | 1.01 (0.71–1.45) | 0.946 | 18.1 | 0.300 | ||||
| Asian | 3 | 381/381 | 5.62 (1.32–23.85) | 0.019 | 55.8 | 0.104 | 0.037 | 0.998 | 0.998 | |
| ε4/ε4 vs. other ApoE genotypes | ||||||||||
| Overall | 9 | 1947/2057 | 1.89 (0.71–5.01) | 0.201 | 0 | 0.784 | 0.928 | |||
| Caucasian | 5 | 1497/1639 | 1.22 (0.36–4.15) | 0.751 | 0 | 0.754 | ||||
| Asian | 3 | 381/381 | 5.37 (0.84–34.19) | 0.075 | 0 | 0.569 | ||||
- * Power to detect a noteworthy finding by false positive report probability (FPRP) when the true OR equals the specified value. RPL, Recurrent pregnancy loss.

3.3 Publication bias
In overall analyses, no evidence of funnel plot asymmetry was detected in the pooled results for the comparison of carriers of the ApoE ε4 allele vs. carriers of other ApoE alleles (Egger’s p-value = 0.143, Figure 2B) or for the comparison of ε4 allele vs. ε3 allele (Egger’s p-value = 0.107, Figure 3B). Similarly, no statistical evidence of publication bias was detected for the other genetic comparisons of ApoE alleles or genotypes, with exception for two genetic comparisons: ε2 allele vs. ε4 allele (Egger’s test p-value = 0.001, Table 2) and ε3 allele carriers vs. carriers other ApoE alleles (Egger’s test p-value = 0.017, Table 2). The non-parametric “trim and fill” method was then applied to adjust for possible publication bias under these two genetic contrasts. The results with and without “trim and fill” did not draw different results, that is the adjusted effect size still remained not significant in both analyses (trim and fill analysis OR for ε2 vs. ε4: 0.84, 95% CI 0.44–1.60, p = 0.595, Supplementary Figure S2; trim and fill analysis OR for ε3 allele carriers vs. other ApoE alleles: 0.76, 95% CI 0.35–1.67, p = 0.499, Supplementary Figure S3), indicating the reliability of these meta-analytic results.
3.4 Subgroup analysis
When subgroup analyses were conducted based on subjects’s ethnic origin, a significant association with RPL was detected in the Asian population under four genetic comparisons: ε4 allele vs. ε3 allele (OR 5.93, 95% CI 1.79–19.61, p = 0.004), ε2 allele vs. ε4 allele (OR 0.14, 95% CI 0.03–0.61, p = 0.009), ε4 allele carriers vs. carriers of other ApoE alleles (OR 8.42, 95% CI 1.47–48.12, p = 0.017), and ε3/ε4 genotype vs. other ApoE genotypes (OR 5.62, 95% CI 1.32–23.85, p = 0.019).
3.5 Bayesian approach analysis by FPRP and BFDP tests
None of the positive findings detected in either overall or subgroup analyses were found to be noteworthy under FPRP or BFDP at a prior probability of 0.001 (Table 2), indicating the possibility of false-positive results.
4 DISCUSSION
The most recent meta-analysis on the association of the ApoE gene polymorphism and RPL risk,19 which was based on six studies published up until 2013, included a total of 975 patients with RPL and 1553 controls. The pooled results suggested that ε4 and ε2 alleles may be risk factors for RPL, whereas the ε3 allele could be a protective factor. Given the publication of a subsequent six additional primary studies, 15, 17, 31-34 we conducted an updated meta-analysis to re-estimate, in a total of 2,231 cases and 2,241 controls, the impact of ApoE genotype on RPL risk susceptibility. In addition, ethnic-based subgroup analyses were performed to provide further insights into the relationship between maternal ApoE genotype and the risk of recurrent pregnancy loss, and FPRP and BFDP tests were also applied to assess the noteworthiness of significant findings.
The overall results of the present meta-analysis showed that the ApoE ε4 allele confers a higher risk of RPL compared with the ε3 allele, and that women carrying the ε4 allele are at higher risk of RPL compared with women carrying the ε2 or the ε3 alleles. While this updated meta-analysis does not support an association of the ε2 allele, our results confirm a possible role of ApoE ε4 as risk factor for RPL. In the subsequent subgroup analysis based on subjects’ ethnicity, no association with ApoE alleles or genotypes was found in women of Caucasian ancestry. Conversely, a significant impact of the ApoE gene polymorphism was detected in Asian women under four genetic contrasts (i.e. ε4 vs. ε3, ε2 vs. ε4, ε4 allele carriers vs. carriers of other alleles, ε3/ε4 vs. other ApoE genotypes). Given the relatively high number of comparisons made, the Bayesian FPRP and BFDP tests were applied to significant pooled ORs in order to assess the credibility of the findings. After application of FPRP and BFDP, none of the significant associations detected in both overall and subgroup analyses were found to be noteworthy at a prior probability of 0.001, suggesting that positive findings of this meta-analysis should be interpreted with caution for possible false positive results.
The Apo E gene polymorphism has been postulated to affect the risk of RPL since it could modulate lipid metabolism in pregnancy35 and contribute to atherosclerosis and thrombogenesis, which are believed to affect fetal intrauterine growth and placental flow. The ApoE alleles and genotypes have been also reported to affect the human implantation process, the ε4 allele and the ε3/ε4 genotype being more frequent in patients with recurrent implantation failure.36 It should be noted that frequency of ApoE alleles strongly varies among ethnic groups, with Asian populations presenting a lower frequency of the ε4 allele (7.1–24%) compared with other populations.37 Worthy of note is that the ApoE genotype modulates female fertility with opposite effects according to patients’ ethnicity. Specifically, Caucasian females carrying the ε3 allele have been reported to display a higher fertile potential compared with female carrying the ε4 and ε2 alleles,38, 39 while in females of Afro-Equadorian origin or Cayapa Indians the highest reproductive efficiency was found among carriers of the ε4 allele.40 Intriguingly, our results also support an ethnic-specific impact of the ApoE gene polymorphism, since an association with RPL was detected in Asians but not in Caucasians. Gene–gene or gene–environment interaction effects might explain the different impact of ethnicity on the relationship between the ApoE polymorphisms and RPL; however, further research should be conducted to support this hypothesis. In spite of the biological plausibility of ApoE genotype involvement in RPL, we failed to find noteworthy genetic associations of ApoE genotypes and alleles, suggesting that large multi-ethnic case–control studies are needed for conclusive evidence of association between the ApoE gene polymorphism and RPL in the Asian population.
This updated meta-analysis has a number of methodological strengths compared with previous meta-analyses19, 41 investigating the association of the ApoE gene polymorphism with RPL susceptibility: (1) study quality was evaluated under the Newcastle–Ottawa scale for case–control studies, allowing determination of the methodological quality of studies included as medium; (2) subgroup analyses were performed according to patients’ ethnicity, allowing detection of ethnic-specific effects of the ApoE gene polymorphism on RPL; and (3) the noteworthiness of statistically significant associations was assessed using the FPRP and BFDP approaches to evaluate the credibility of the findings. A number of limitations should be also taken into consideration when interpreting our results. First, seven out of 12 studies (58%) included in the present systematic review involved fewer than 100 cases and two studies out of 12 (17%) were of low methodological quality. Therefore, given the limited number of patients included in the quantitative analysis and the overall average moderate study quality, conclusions of our meta-analysis should be cautionly interpreted. Second, funnel plot asymmetry was detected in two out of the 12 genetic contrasts assessed in spite of a comprehensive literature search and a rigorous evaluation of primary studies from three different databases. Notwithstanding this, the “trim and fill” method, which was applied to adjust for possible publication bias, has revealed reliability of pooled estimates. Third, significant between-study heterogeneity was still detected in most genetic comparisons within the Asian population. Therefore, we cannot exclude that other factors may partially explain between-study heterogeneity, such as the different definitions used among studies for control group and for RPL diagnosis. Fourth, we conducted a meta-analysis of aggregate data rather than an individual patient data meta-analysis. Therefore, our pooled estimates are not adjusted by variables well known to affect RPL risk, including advanced maternal age, chromosomal and uterine abnormalities, endocrine dysfunctions, thrombophilia, obesity and other lifestyle habits.3 Lastly, given the lack of data in primary studies, we were unable to assess the effect of gene–gene and gene–environment interactions as well as that of maternal–fetal interactions on RPL risk.
5 CONCLUSIONS
The current study provides the most updated pooled OR estimates on the association between the ApoE genotype and RPL. Overall, our findings suggest a possible role of Apo ε4 as susceptibility factor for RPL, especially in Asian population. However, further large and multicentric case–control studies enrolling patients of different ethnicities are nevertheless warranted for conclusive evidence of ApoE ε4 as RPL risk factor and for the investigation of gene–gene, gene–environment and maternal–fetal interactions of ApoE genotype on RPL susceptibility.
CONFLICT OF INTEREST
None of the authors have a conflict of interest to disclose.
ETHICAL STATEMENT
Ethical review and approval is not applicable to this study, due to it being a meta-analysis of existing studies.
AUTHOR CONTRIBUTION STATEMENT
S.C. made substantial contributions to conception, design, data analysis and interpretation, as well as to the writing of the manuscript; F.A. made substantial contributions to data analysis and interpretation. J.I.S. made substantial contributions to data interpretation and to the writing of the manuscript. S.T. made substantial contributions to conception, design, data analysis and interpretation, as well as to the writing of the manuscript. All the authors have read and approved the final version of the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
All the data for the present study are available from the corresponding author upon reasonable request.




