Intracellular siRNA and precursor miRNA trafficking using bioresponsive copolypeptides
Abstract
Background
Small interfering RNAs (siRNAs) can induce specific gene silencing through cytoplasmic mRNA cleavage and nuclear transcriptional silencing, necessitating delivery to different cellular compartments. This study presents a reducible copolypeptide (rCPP) carrier containing different molar ratios of a histidine-rich peptide (HRP) and nuclear localization sequence (NLS) peptide to modulate intracellular trafficking of transfected siRNA and primary RNA transcripts (pri-miRNA).
Methods
Polyplex formation using siRNA and rCPP was demonstrated using photon correlation spectroscopy and atomic force microscopy. Confocal and fluorescence microscopy were used to investigate cellular uptake and nuclear trafficking whilst endogenous enhanced green fluorescent protein (EGFP) knockdown in H1299 cells was evaluated using flow cytometry. Transcriptional gene silencing of endogenous EF1A was verified using real-time reverse-transcription polymerase chain reaction (RT-PCR) and pri-miRNA nuclear processing was demonstrated using Northern analysis.
Results
rCPP-based polyplexes showed rapid cellular uptake and low cytotoxicity. Labelled components revealed intact polyplexes after 2 h that exhibited directed movements consistent with endosomal trafficking. Polyplex-mediated knockdown of EGFP increased with greater HRP content. The inclusion of NLS promoted nuclear localization of transfected siRNAs and pri-miRNAs to the nuclear compartment allowing for transcriptional silencing of EF1A and Drosha and Dicer dependent expression of mature miRNA, respectively.
Conclusion
Our results demonstrate that reducible copolypeptides can be used as carriers for the non-toxic cellular delivery of siRNA and pri-miRNA. The nuclear targeting of rCPPs can be utilized for delivery of siRNAs and pri-miRNAs to the nuclear compartment for transcriptional gene silencing or endogenous processing into mature miRNA, respectively, which could potentially lead to improved therapeutic approaches. Copyright © 2007 John Wiley & Sons, Ltd.
Introduction
Small interfering RNAs (siRNAs) are 21–25 nucleotide duplexes that have been found to induce silencing of specific genes through the mechanism of RNA interference (RNAi) 1, 2. In RNAi one strand of the duplex base pairs with a complementary mRNA strand and through the action of the RNA-induced silencing complex (RISC) induces cleavage of the mRNA. This process is highly specific allowing targeting and silencing of single genes upon introduction of siRNAs into cells and has been used extensively as a tool for investigating cellular processes 3, 4. Originally thought to have been evolved as defence against viral infections, the discovery of a closely related class of endogenously expressed small RNAs, the microRNAs (miRNAs) of ∼22 nucleotides, has proven that the mechanism of RNAi has more widespread functions in the cells where it play key roles in diverse regulatory pathways such as developmental timing, cell proliferation and tumorigenesis 5, 6. The miRNAs mediate their effects on post-transcriptional gene silencing in a manner very similar to the siRNAs, through the use of the RISC complex. In mammalian cells miRNAs are transcribed as long primary RNA transcripts (pri-miRNAs) in the nucleus which are sequentially processed into mature miRNAs 7. The endogenous sequential processing of miRNA has been shown to improve the potency of gene silencing compared to externally introduced synthetic siRNAs 8 and avoids activation of the interferon system, which has complicated the use of siRNA 9.
In addition as a tool to investigate cellular processes and target validation in drug discovery, siRNA and miRNA can be potentially used as therapeutic drugs to silence genes implicated in viral pathogenesis, inflammatory conditions and cancer 3. In order to maximize the therapeutic efficacy, however, several drug delivery barriers dependent on the route of administration have to be overcome. Extracellular barriers, including the mononuclear phagocyte system 10 and the endothelial lining of blood vessels, have evolved to protect the human organism from pathogens and foreign material. In addition, intracellular factors, including compartmentalization and the availability of RNA to interact with intracellular targets, must be considered. Poor release from the endosomal compartments and subsequent degradation by the lysosomes restrict translocation into the cytoplasm where the siRNA and miRNA direct target mRNA degradation or translational repression 11, 12. Recent findings have shown that siRNAs are capable of inducing transcriptional gene silencing (TGS) through involvement of DNA methylation 13, 14 and that siRNAs mediate knockdown of nucleus restricted transcripts 15-17. In order to engage the nuclear RNAi pathways, the siRNA must enter the nucleus, a translocation process restricted by the nuclear envelope. Synthetic polycation-based vectors (polyplexes) such as polyethylenimine (PEI) or poly-L-lysine (PLL) have been widely used for in vitro and in vivo delivery of nucleic acids as a safer alternative to viral delivery systems 18-20. The effectiveness of these polyplexes has been shown to increase with increased polymer molecular weight; however, increased cytotoxicity is encountered 21. ‘Bioresponsive’ polycations containing reducible disulfide bridges that respond to intracellular redox conditions have proven advantageous in delivering nucleic acids into cells 22-25. Upon cellular internalization, the intracellular reducing environment facilitates polymer breakdown into low molecular weight components that exhibit reduced cytotoxicity. Furthermore, polymer breakdown activates polyplex decomplexation and consequent release of free nucleic acids allowing interaction with their intracellular targets 26.
Development of a non-toxic polyplex system capable of modulating RNA delivery and facilitating release within cytoplasmic and nuclear compartments will clearly improve the therapeutic potential of RNA-therapeutics. In this study we use a reducible copolypeptide (rCPP) composed of different molar ratios of a histidine-rich peptide (HRP) and a nuclear localization sequence (NLS) peptide. The HRP contains three lysine residues which due to their protonation at neutral and acidic pH are able to bind the negatively charged phosphate backbone of nucleic acids through electrostatic interactions. Six histidyl residues were included to promote endosomal escape due to the buffering capacity of histidyl residues in the endosomal and lysosomal pH range 27. In order to allow targeting of the nuclear compartment a NLS peptide derived from the importin α binding SV40 large T antigen was included in various amounts 28. This work describes formulation and physicochemical characterization of siRNA and miRNA polyplexes using the rCPP carrier containing different molar ratios of the HRP and NLS peptide and the ability of the complexes to modulate intracellular siRNA and pri-miRNA trafficking, transcriptional gene silencing and enhanced green fluorescent protein (EGFP) knockdown.
Materials and methods
Materials
Dulbecco's modified Eagle's medium (DMEM) and RPMI 1640 culture medium + GlutaMAX™, DMEM without riboflavins and phenol red, penicillin/streptomycin (Pen/Strep), G418 selection factor, trypsin-EDTA (1×), foetal bovine serum (FBS) and 10 × TBE buffer were purchased from Invitrogen Corporation (Carlsbad, USA). CellTiter 96® AQueous One Solution cell proliferation assay (MTS) was purchased from Promega Corp. (Madison, USA). TransIT-TKO and TransIT-Oligo transfection reagents were obtained from Mirus Corp. (Madison, WI, USA). An EGFP-specific siRNA duplex (Dharmacon, Boulder, CO, USA) containing the sequence: passenger, 5′-GACGUAAACGGCCACAAGUUC-3′, guide, 3′-CGCUGCAUUUGCCGGUGUUCA-5′ and an EGFP-mismatch siRNA duplex containing the sequence: passenger, 5′-GACGUUAGACUGACAAGUUC-3′, guide, 3′-CGCUGAAUCUGACCUGUGGUUCA-5′ was used for nanoparticle characterization, cytotoxicity and EGFP knockdown studies. An EGFP-specific siRNA labelled with a Cy5 fluorophore on the 5′ end of the guide strand was used for cellular uptake and trafficking studies. EF1A promoter specific siRNA were purchased from DNA Technology A/S (Aarhus, Denmark) and contained the sequence: passenger, 5′-AAGGUGGCGCGGGGUAAACUG-3′ and guide, 3′-UUCCACCGCGCCCCAUUUGAC-5'.
Cell lines
The human lung cancer cell line (H1299) stably expressing EGFP (2 h half-life), kindly provided by Dr. Anne Chauchereau (CNRS, Villejuif, France), was grown in RPMI 1640 + GlutaMAX™ supplemented with FBS, Pen/Strep and G418 selection factor. Human cervical cancer cells (HeLa) were grown in DMEM + GlutaMAX™ supplemented with FBS and Pen/Strep. All cell lines were incubated at 37 °C in a 5% CO2 humidified environment.
Preparation of reducible polycations and polyplex formation
The HRP, CKHHHKHHHKC, and the NLS peptide, CGAGPKKKRKVC, were synthesized and purified and reducible copolypeptides (rCPPs) prepared through oxidative copolymerization. Five rCPPs containing different amounts of NLS were prepared by adding HRP and NLS peptide at different molar ratios in the reaction mix (Table 1). The synthesis, composition and molecular weight of the polymers have previously been described 29. rCPP-D was labelled by succinimidyl ester conjugated Alexa488 according to the Invitrogen protocol.
| Polymer Identity | NLS peptide in reaction (mol%) | NLS peptide in polymer (mol%) | HRP peptide in reaction (mol%) | HRP peptide in polymer (mol%) | Weight per Charge | MW |
|---|---|---|---|---|---|---|
| A | 100 | 100 | 0 | 0 | 325 | ∼100.000 |
| B | 75 | 73 | 25 | 27 | 315 | |
| C | 50 | 48 | 50 | 52 | 300 | ∼60.000 |
| D | 25 | 20 | 75 | 80 | 280 | |
| E | 0 | 0 | 100 | 100 | 255 | ∼200.000 |
RNA or siRNA was added to 20 mM sodium acetate buffer adjusted to pH 5.0 using acetic acid (0.2 M). rCPP was added to the nucleic acids at the desired N : P ratio and mixed by gently pipetting. Polyplexes were allowed to form for at least 1 h at room temperature prior to use. To calculate specific N : P ratios (defined as the molar ratio of reducible polycation amino groups/RNA phosphate groups) a mass per phosphate of 325 Da was used for RNA. The mass per charge of the rCPPs is listed in Table 1. The hydrodynamic diameter of the polyplexes was determined by photon correlation spectroscopy (PCS) using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). PCS was performed at 25 °C in sodium acetate buffer in triplicate with sampling time and analysis set to automatic.
Stability of polyplexes under redox conditions using polyacrylamide gel electrophoresis
Polyplexes (N : P ratio 5 and 10) with or without prior incubation with 1,4-dithiothreitol (DTT) for 30 min at 37 °C were analyzed by electrophoresis using a 10% polyacrylamide gel (50 mM Tris-borate, pH 7.9, 1 mM EDTA) at 150–230 V for 2 h; they were then stained with ethidium bromide and visualized using a UV illuminator.
Determination of polyplex morphology using atomic force microscopy (AFM)
Polyplex D (N : P ratio 10) with or without 25 mM DTT incubation for 30 min was deposited onto freshly cleaved micas for 10 min. After the solvent had evaporated, the sample was transferred for AFM. Freshly prepared samples were imaged using a commercial Nanoscope IV MultiMode atomic force microscope (Veeco Instruments, Santa Barbara, CA, USA) under ambient conditions. AFM imaging was performed in tapping mode at scan frequencies of 1–2 Hz with minimal loading forces applied and optimized feedback parameters. Several images were obtained from separate locations across the surfaces to ensure reproducibility. All the images were first flattened using the NanoScope™ software (Digital Instruments), while excluding the particles from the flattened area, and then analyzed automatically using the commercial Scanning Probe Image Processor (SPIP™) software (Image Metrology ApS, Lyngby, Denmark) to yield the particle diameter histograms from the images.
Cytotoxicity assay
H1299 or HeLa cells were plated in a 96-well plate at a density of 4 × 104 cells/well and grown overnight. Medium was replaced by serum-free media and polyplexes and TransIT-TKO were added at a concentration of 50 nM siRNA/well for 4 h and then media was replaced by fresh growth medium. After incubation for 24 h, a CellTiter 96 AQueous One Solution cell proliferation assay (Promega, WI, USA) was used to investigate cytotoxicity. CellTiter proliferation assay solution was added to the wells and left for 3 h before absorbance was measured at 490 nm using a fluorescence plate reader.
Fluorescence microscopy
HeLa or primary macrophage cells were grown in glass-bottomed dishes (MatTek Corp., MA, USA) or on glass coverslips in 12-well plates. In some experiments, HeLa cells were transiently transfected using TransIT-Oligo to express cyan fluorescent protein (CFP)-TI-VAMP according to Mirus protocol. The pCFP-TI-VAMP plasmid was a generous gift from Dr Edouard Bertrand (IGMM, Montpellier, France). For live cell experiments transfected cells were incubated in phosphate-buffered saline (PBS) + 1 µg/ml Hoechst 33 342 (Molecular Probes) for 10 min, washed in PBS after which colourless DMEM + 5% FBS was added and the dish was sealed with Parafilm. Fluorescence images were obtained using a Zeiss Axiovert 200M microscope equipped with a heated stage and objective, a 100× 1.4 NA planapochromatic lens and a Coolsnap HQ camera (Ropers Scientific) and analyzed using Meta Morph software (Universal Imaging Corp.). For time lapse studies, a ‘No Neighbours’ deblurring algorithm was used to improve the signal-to-noise ratio (AutoQuant Deblur, Media Cybernetics). For fixed cell experiments, transfected cells were washed in PBS and fixed in 4% paraformaldehyde at room temperature for 10 min. Coverslips were then mounted onto glass slides using Prolong Gold mounting medium with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescence images were obtained using a Zeiss LSM 510 Meta confocal laser scanning microscope equipped with a 100× 1.4 NA planapochromatic lens.
Gene silencing in an EGFP-expressing human cell line
H1299 green cells were plated on 24-well plates (105 cells/well) 24 h prior to transfection. The medium was replaced with serum-free media and the polyplex formulations added at 50 nM siRNA per well. TransIT-TKO transfections were carried out according to Mirus protocol. After 4 h the media was replaced with fresh growth medium. The cells were left for 44 h and then resuspended in PBS containing 1% paraformaldehyde. The EGFP cell fluorescence was measured using a FACSCalibur flow cytometer (Becton Dickenson). A histogram plot with log green fluorescence intensity on the x-axis and cell number on the y-axis was used to define median fluorescence intensity of the main cell population defined by scatter properties (FSC, SSC, not shown).
Transcriptional gene silencing
HeLa cells were plated in a 12-well plate 24 h prior to transfection. The medium was replaced with serum-free media and polyplexes were added at 50 nM EF1A promoter siRNA per well. TransIT-TKO transfections were carried out according to Mirus protocol. After 4 h the media was replaced with fresh growth medium. The cells were left for 48 h after which total RNA was isolated using TriZol Reagent (Invitrogen). RNA (1 µg) was used as template for the oligo-dT primed reverse transcription polymerase chain reaction (RT-PCR) using a M-MLV RT kit (Invitrogen). The resultant cDNA was used in a real-time PCR using the following primers: EF1A, F 5′-CTGAACCATCCAGGCCAAAT-3′, R 5′-ATGTGAGCCGTGTGGCAA T-3′ and GAPDH, F 5′ GAAGGTGAAGGTCGGAGT, R 5′ GAAGATGGTGATGGGATTTC. The real-time RT-PCR data were normalized to GAPDH controls from the respective samples.
Northern blotting
HeLa cells were plated in a 12-well dish 24 h prior to transfection. The media was removed and replaced with serum-free media and TransIT-TKO or rCPP/Pri-miRNA23A polyplexes added at 15 nM pri-miRNA23a per well. Following 4 h the media was replaced with 1 ml fresh media containing 10% FBS. After 44 h total RNA was isolated using TriZol Reagent (Invitrogen). Amounts of 3 µg were loaded in each lane on a 12% denaturing gel and the amount of resolved RNA in the lanes was checked by ethidium bromide staining. The RNA was then transferred to a Hybond-N+ blotting membrane (Bio-Rad) overnight and probed with an oligonucleotide complementary to miRNA 23a (5′-GGAAATCCCTGGCAATGTGAT-3′). The blotting membrane was pre-hybridized for 2 × 1 h in Church buffer and hybridized overnight at 37 °C after which it was washed in 1× SSC + 0.1% sodium dodecyl sulfate (SDS) and exposed on a Molecular Imager FX (Bio-Rad) and analyzed using Quantity One software (Bio-Rad).
Results
Physicochemical characterization
The ability of the rCPPs to form polyplexes with siRNA (21-mers) and disassemble under redox conditions was investigated using five different types of polymers (A–E) containing a variable ratio of HRP and NLS peptides (Table 1). The molecular parameters including hydrodynamic size and surface charge, which can influence cellular interactions, were measured using dynamic light scattering. At an N : P ratio of 10 all rCPPs were able to form discrete particles below 270 nm (Table 2). The particles formed were monodisperse with polydispersity indices below 0.5 (data not shown).
| Polyplex (rCPP/siRNA) | Hydrodynamic radius (nm) | ζ potential (mV) |
|---|---|---|
| A | 240 ± 13 | 22 ± 2 |
| B | 231 ± 20 | 33 ± 1 |
| C | 261 ± 15 | 32 ± 1 |
| D | 213 ± 18 | 30 ± 1 |
| E | 213 ± 34 | 21 ± 1 |
| D (Cy5) | 200 ± 2 | 30 ± 1 |
| D (A488/Cy5) | 267 ± 13 | 27 ± 2 |
The ζ potential measurements showed that all polyplexes had a net positive charge greater than 20 mV, reflecting excess rCPP at this N : P ratio (Table 2). The size of polyplex D containing Cy5-labelled siRNA and Alexa488-labelled rCPP-D was in the same range as the unlabelled polyplex indicating that the fluorescent groups attached did not interfere with polyplex formation and subsequent trafficking experiments (see below).
The ability of the polyplexes to decomplex in a reducing environment was examined using the electrophoretic migration of siRNA in a polyacrylamide gel shift assay (Figure 1) and AFM (Figure 2). The polyplexes were incubated with 25 mM DTT for 30 min at 37 °C after which they were loaded onto a gel and the amount of released siRNA assessed by ethidium bromide staining. From Figure 1 it can be seen that small amounts of siRNA were released (Figure 1A, wells 2–11) under non-reducing conditions suggesting that they are in a particulate form and retarded in the gel whereas reduction of the polyplexes by DTT caused a significant increase in the amount of siRNA released (Figure 1B, wells 2–11). The amount of released siRNA was quantified using signal intensity analysis on the gel bands (Figure 1C). Decomplexation was most ineffective for polyplexes containing the A-type polymers suggesting that the highly basic nature of pure NLS polymers may increase complex stability.
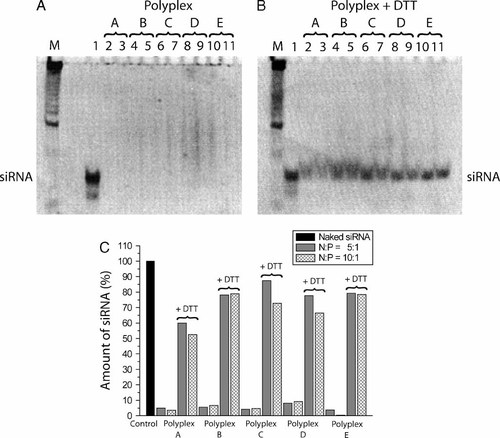
Decomplexation of polyplexes following reduction with DTT. (A) Lane M: DNA marker; lane 1: naked siRNA; lanes 2–3: polyplex A N : P 5 (2), N : P 10 (3); lanes 4–5: polyplex B N : P 5 (4), N : P 10 (5); lanes 6–7: polyplex C N : P 5 (6), N : P 10 (7); lanes 8–9: polyplex D N : P 5 (8), N : P 10 (9); lanes 10–11: polyplex E N : P 5 (10), N : P 10 (11). (B) Polyplexes incubated for 30 min in 25 mM DTT at 37 °C. (C) Signal intensity analysis on the siRNA gel bands in A and B
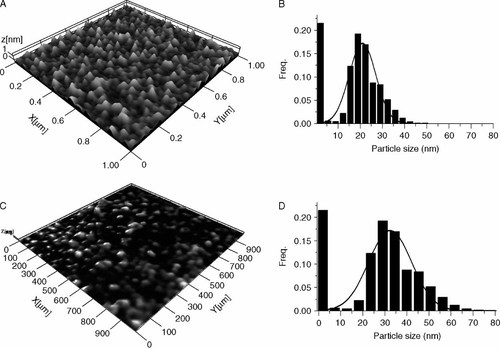
Influence of a reducing environment on polyplex D morphology. Atomic force microscopy images of polyplex D (N : P 10) without (A, B) or with (C, D) incubation for 30 min in 25 mM DTT at 37 °C. (B) and (D) show the particle size distribution with a best fit curve superimposed
AFM analysis of the morphology of polyplex D revealed a monodispersed distribution of nanoparticles (Figures 2A and 2B). Reduction of the copolypeptide by DTT led to significant changes in morphology due to increased polyplex size (Figures 2C and 2D). From Gaussian fitting the average size was found to increase from 19.2 ± 6.4 nm to 32.4 ± 10.1 nm after DTT reduction which suggests decomplexation through a process of particle swelling, most likely due to electrostatic repulsive forces between hydrated polymeric chains during siRNA release. A population of very small complexes (<5 nm) was observed both with and without DTT likely resulting from free polymer and free siRNA.
Cytotoxicity
The cellular cytotoxicity of the polyplex nanoparticles was compared to the commercial TransIT-TKO transfection reagent using a H1299 human lung cancer cell line, HeLa cells and a commercially available tetrazolium-based MTS cytotoxicity assay (Figure 3). At N : P 10, a large excess of free rCPP is likely to be present in the polyplexes; however, no significant cytotoxicity was detected. This was supported by phase contrast microscopy of HeLa cells in which the cells incubated with polyplexes appeared viable, and of similar appearance as untreated cells (data not shown). In comparison, the lipid-based TransIT-TKO transfection reagent caused a significant decrease in cell viability. Decreasing or increasing the N : P ratio of the polyplexes to 5 and 20 had no effect on cell viability (data not shown). These results demonstrate that the rCPPs have negligible cytotoxicity and are significantly less toxic than the lipid-based TransIT-TKO siRNA delivery agent in these cell lines.
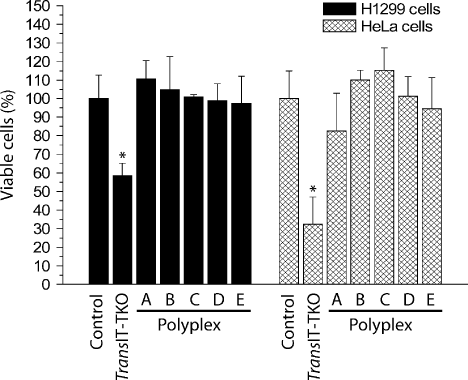
Evaluation of the cytotoxicity of the rCPPs. Polyplexes (N : P 10) or TransIT-TKO/siRNA complexes were added to H1299 and HeLa cells at a concentration of 50 nM siRNA/well and left for 4 h. The medium was replaced and following 48 h of incubation cell viability was determined. The cell viability was normalized with respect to the control (untreated) value and plotted ± standard deviation (SD) (n = 3). *P < 0.01 vs. control
Cellular uptake and intracellular trafficking
Using fluorescence microscopy we investigated the live cellular uptake and intracellular trafficking of the rCPP-D. The use of Cy5-labelled RNA and Alexa488-labelled rCPP-D enabled simultaneous detection of both components of the polyelectrolyte nanoparticle. HeLa cells were transfected for 2 h in serum-free medium with 50 nM RNA and the uptake and intracellular localization of the polyplex was visualized in live cells (Figure 4). Following 2 h of transfection a large amount of intracellular labelled RNA and labelled rCPP was found in the HeLa cells, while some free labelled copolypeptide was observed to be attached to the cell surface. A large proportion of the labelled RNA was found to be co-localized with the bioresponsive rCPP, revealing that polyplex decomplexation was not complete within this time period. It was observed that several of the polyplexes exhibit directed movements in the cell (Figure 5D), suggesting that the polyplexes are trafficking around inside endosomal compartments and, therefore, not exposed to the reducing environment of the cytoplasm. Following 4 h of transfection, polyplexes were localized to compartments containing a TI-VAMP protein known to reside in the late endosomes 30, further supporting endosomal compartmentalization of the polyplexes (Figures 4E and 4F). Directed endosomal trafficking of the polyplexes shown in Fig. 4F was observed (Supplementary movie 1).
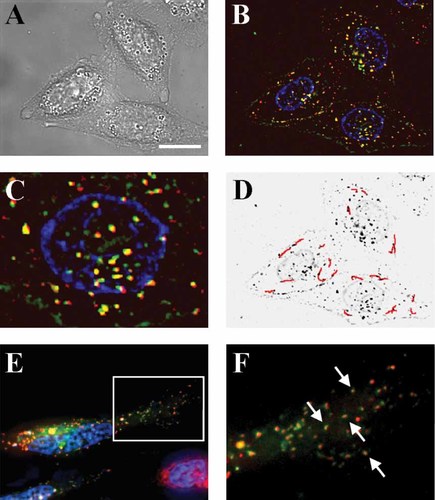
Live cellular uptake and trafficking of RNA in HeLa cells. HeLa cells were transfected with 50 nM Cy5-labelled RNA and Alexa488-labelled rCPP-D (N : P 10) for 2 h. (A) Bright field. (B) Fluorescence overlay of Cy5 RNA (red), Alexa488 rCCP-D (green) and co-localization of Cy5 and Alexa488 (yellow). Nuclei are labelled with Hoechst (blue). (C) Close-up of a nucleus. (D) Directed trafficking of polyplexes over a time period of 90 s shown in red. (E) Co-localization of Cy5 siRNA (red) and TI-Vamp-CFP (green), a marker of late endosomes following 4 h of transfection. (F) Close-up of region shown in E. 100× magnification. Scale bar: 10 µm
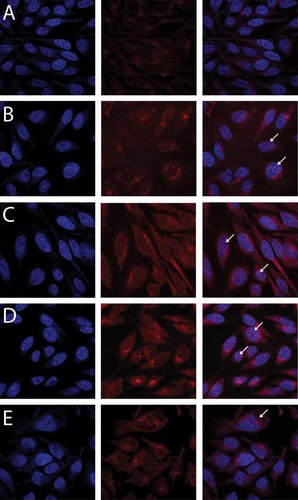
Nuclear localization of siRNA depends on the amount of NLS in the rCPP. HeLa cells were transfected for 1 h with 10 nM Cy5-labelled siRNA and polymers A–E (N : P 10), fixed and visualized using confocal laser scanning microscopy. (A) Polyplex A (100% NLS); (B) polyplex B (75% NLS); (C) polyplex C (50% NLS); (D) polyplex D (25% NLS); and (E) polyplex E (0% NLS). Left panels (DAPI stained nuclei, blue), centre panels (siRNA-Cy5, red) and right panels (overlay). Arrows in B, C and D mark nuclear siRNA. The arrow in E marks perinuclear siRNA
Some polyplexes composed of rCPP-D (25% NLS content) seemed to be found in the nuclear compartment (Figures 4B and 4C). These polyplexes did not exhibit directed movements in comparison to those which did in the cytoplasm in accordance with endosomes being restricted to the latter compartment.
To further verify cellular uptake and nuclear trafficking the effect of various amounts of the NLS peptide in the rCPPs were tested in HeLa cells transfected with 10 nM Cy5-labelled siRNA (Figure 5). Following 1 h transfection, siRNAs complexed with polyplexes B, C and D (with 75% NLS, 50% NLS and 25% NLS, respectively) were all detected in the nucleus using confocal laser scanning microscopy. Polyplex A, composed of 100% NLS, generally showed low cellular transfection efficiency and did not result in detectable amounts of the transfected siRNA in the nuclear compartment. This suggests that the absence of HRP in the polypeptide backbone results in the insufficient release of the polyplex from the endosomal compartments. siRNA transfected into the HeLa cells using polypeptide E, containing no NLS, resulted in a perinuclear distribution of the siRNA which was observed up to 24 h after transfection (data not shown).
Gene silencing, transcriptional gene silencing and pri-miRNA delivery to the nuclear compartment
The ability of the polyplexes to mediate knockdown of an endogenously expressed enhanced green fluorescent protein (EGFP) was investigated in the H1299 cell line. Cells were transfected for 4 h after which transfection media was replaced by normal growth media. The decrease in EGFP mean fluorescence intensity, detected by flow cytometry at 48 h post-transfection, was used as the measure of knockdown efficiency of the particles. TransIT-TKO was used as a positive control and two different N : P ratios (5 and 10) were used to investigate the effect of copolypeptide content on gene silencing. Figure 6 shows a gradual increase in the EGFP knockdown efficiency of the polyplexes with more HRP content reaching a maximum of 50% reduction in EGFP expression for polyplex E. We tested the specificity of the RNA interference using an EGFP mismatch siRNA (4 bp mismatch) with polyplex E (N : P ratio 10) which produced maximum knockdown with the EGFP siRNA. No reduction in EGFP fluorescence was found using the mismatch siRNA, proving knockdown specificity. Polyplexes with an N : P ratio of 10 showed a slight enhancement in knockdown efficiency compared to N : P 5 polyplexes. This could be due to more stable particles being formed at the higher N : P ratio resulting in improved transfection or due to enhanced endosomal escape efficiency of the polyplexes containing higher amounts of rCPP. However, an increase to N : P 20 did not result in any improvement in EGFP knockdown (data not shown).
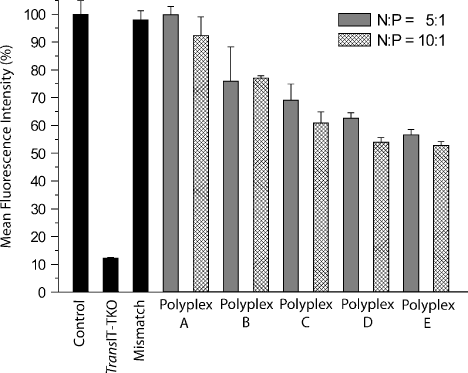
Polyplex-mediated RNAi dependency on histidine-rich peptide content. H1299 cells containing a stably integrated EGFP expression cassette were transfected for 4 h with 50 nM/well EGFP siRNA using TransIT-TKO or polyplex A, B, C, D or E formed at N : P 5 or 10 or EGFP mismatch siRNA using polyplex E at N : P 10 (Mismatch). The decrease in EGFP mean fluorescence intensity, detected by flow cytometry at 48 h post-transfection, was used as the measure of EGFP knockdown (normalized to % control of untreated EGFP cells). Error bars represent ± SD (n = 3)
Experiments were conducted to determine the ability of the polyplexes to facilitate transcriptional gene silencing which occurs within cellular nuclei. siRNA targeting an essential promoter region of the elongation factor 1 alpha (EF1A) gene abundantly expressed in eukaryotic cells 13 was transfected into HeLa cells using polyplexes A–E and TransIT-TKO. After 48 h total RNA was isolated and EF1A mRNA normalized to the GAPDH reporter gene was quantified using real-time RT-PCR. Cells transfected with polyplexes containing NLS showed a reduced EF1A mRNA expression depending on the amount of NLS in the polyplexes (Figure 7). Polyplex E and TransIT-TKO, exhibiting highest transfection efficiencies (Figure 6) but no NLS, showed only a small reduction in EF1A expression (Figure 7), suggesting that the presence of NLS in the polyplexes is necessary for efficient transcriptional gene silencing. The decrease in EF1A expression observed using polyplex E and TransIT-TKO could be due to the division of the HeLa cells during the 48 h incubation period which may allow a small amount of EF1A promoter siRNA access to the nuclear compartment.

Polyplex-mediated transcriptional gene silencing. HeLa cells were transfected for 4 h with 50 nM/well EF1A promoter siRNA using polyplexes A–E (N : P 10) or TransIT-TKO. Forty-eight hours post-transfection total RNA was isolated and EF1A mRNA was quantified by real-time RT-PCR and standardized to GAPDH expression. EF1A expression was normalized to % control of untreated cells. Error bars represent ± SD (n = 3)
Next we investigated the ability of the NLS-containing particles to improve delivery of pri-miRNAs into the nucleus for endogenous processing into mature miRNA effector molecules. HeLa cells were transfected with polyplexes A–E (N : P 10) including a 183 nucleotides (nt) long pri-miRNA. After 48 h total RNA was isolated from the cells and Northern blot experiments were performed to quantify the production of mature miRNA.
A positive NLS-dependent effect was observed on the endogenous processing of pri-miRNA (Figures 8A and 8B), the most efficient production of miRNA being observed using polyplex C that contains equal amounts of NLS and HRP. Polyplexes A and B showed very little miRNA production, a finding consistent with the poor knockdown efficiency observed using the siRNA polyplexes, which probably was due to the low amount of histidine in the polymer backbone. Decreasing the NLS content of the polyplexes (polyplexes D and E) caused a decrease in the produced miRNA confirming the requirement of nuclear targeting of the pri-miRNA for entry into the miRNA biogenesis pathway. Polyplex E and TransIT-TKO containing no NLS resulted in a slight increase in produced miRNA compared to the control value. We conclude that NLS-containing bioresponsive copolypeptides are capable of pri-miRNA delivery to the nuclear compartment allowing for both nuclear and cytoplasmic processing of pri-miRNA into miRNA.
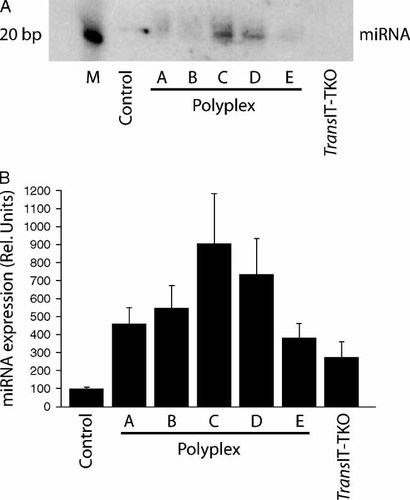
Northern blot analysis of miRNA endogenously processed from polyplex-delivered pri-miRNAs. (A) Polyplexes formed using pri-miRNA-23a and rCPPs (N : P 10) or TransIT-TKO were used to transfect HeLa cells for 4 h. Total cellular RNA was extracted from HeLa cells 48 h post-transfection and separated on a 12% denaturing polyacrylamide gel. Hybridization was performed using a 32P-labelled DNA oligonucleotide probe complementary to the miRNA-23a guide strand. One hour Northern blot exposure. (B) The columns represent the mean miRNA expression normalized to controls from three independent experiments quantified by signal intensity analysis. Error bars represent ± SD
Discussion
Synthetic vectors based on polycations such as polyethylenimine (PEI) or poly-L-lysine (PLL) have been widely used to deliver nucleic acids into cells; however, several drawbacks restrict their use such as high cellular toxicity, sequestration in subcellular compartments and lack of intracellular targeting. In this study we present the use of bioresponsive copolypeptides consisting of lysine-containing histidine-rich peptide (HRP) and nuclear localization sequence (NLS) peptides as carriers for siRNA and primary RNA transcripts.
The HRP and the NLS peptides are capped by cysteine residues facilitating oxidative polycondensation of the low molecular weight (LMW) peptides into a linear high molecular weight (HMW) copolypeptide. This sensitizes the polypeptide to redox potential gradients and confers a bioresponsive property into the polypeptide design 24. Addition of the NLS peptide to the polycondensation reaction in different molar ratios produced five different copolypeptides containing different amounts of HRP relative to NLS (Table 1) 29. The rCPPs were used to form polyelectrolyte complexes with siRNA having a length of 21 nt, a length well above the minimum number of 6–10 salt bonds required for cooperative binding with the positively charged polypeptide 31. The resulting nanoparticles were found to be 200–300 nm in size as measured by dynamic light scattering; within the size range required for eukaryotic cellular uptake 32.
Measurement of ζ potential revealed that the polyplexes have a positive surface charge when formed at an N : P ratio of 10 reflecting the excess copolypeptide in the particles. A positively charged polyplex can interact with negatively charged proteins and sulfated polysaccharides on the cell surface which facilitates cellular entry by endocytosis 33. The rCPP-A consisting entirely of NLS peptide was able to form polyplexes with siRNA likely due to its high lysine content; however, they were found to have a lower surface charge compared to the other polyplexes, a finding that was attributed to the lack of histidine residues, which become protonated at the slightly acidic pH of 5 during polyplex synthesis.
An essential requirement for nanocarrier-mediated delivery of siRNA is the ability of the carrier to release its cargo inside the cells. The use of cleavable disulfide linkers exploiting the redox potential gradient existing between the extracellular and intracellular environment presents a widely used strategy to ensure drug release after cell entry 23. The bioresponsive copolypeptides described in this study were produced by oxidative copolymerization of cysteine-capped HRP and NLS peptide subunits resulting in a copolypeptide backbone connected by covalently bonded disulfides. Using a gel shift assay we demonstrated release of structurally intact siRNA from the polyplexes upon incubation for 30 min at 37 °C with the reducing agent DTT used to mimic the intracellular redox conditions provided by molecules such as glutathione 34. Atomic force microscopy analysis revealed a monodispersed distribution of nanoparticles that increased in size after DTT incubation consistent with changes accompanying decomplexation.
The effectiveness of polycation vectors such as PEI or PLL increases with higher molecular weight; however, a concomitant increase in cytotoxicity has been shown 21. Polycations containing reducible disulfide bridges in the polypeptide backbone have been found to circumvent this problem due to polypeptide breakdown into LMW fractions upon internalization into cells activated by redox conditions 22, 24. In our study, only a minimal cytotoxicity of the bioresponsive copolypeptides was found after polyplex transfection. The observed biocompatibility can be attributed to the intracellular reduction of the disulfide bonds in the polypeptide backbone yielding LMW fragments resulting in decreased binding affinity to membranes, nucleic acids and vital proteins inside cells.
The major cellular uptake pathways including phagocytosis, macropinocytosis, clathrin-mediated endocytosis and caveolae-mediated endocytosis facilitate particle entry into eukaryotic cells. The exact mechanism by which polyplexes are taken up depends on the size of the particles with clathrin-mediated endocytosis the predominant mechanism for particles of ∼200 nm 32, 35. We found that inclusion of fluorescent probes into the polyplexes did not give rise to significant changes in size, promoting the use of such probes for uptake and localization studies. We found a rapid internalization of the polyplexes into both primary macrophages and HeLa cells. Double labelling of rCPP and RNA revealed that a large amount of the internalized polyplexes was still co-localized following 2 h of transfection. A recent study have shown that many of the endosomal compartments are non-reducing 36, which suggests that the histidine-mediated process of endosomal buffering and subsequent escape into the reducing environment in the cytoplasm is the limiting step prior to polyplex dissociation within the cell. Several of the internalized polyplexes in our study exhibited directed movements within cells, a process generally attributed to endosomal transportation along microtubules 37, and co-localized with a marker for the late endosomes further supporting entrapment of polyplexes within endosomal compartments.
Transfected siRNAs mediate degradation of target mRNA within the cytoplasm of the cells 11; however, recent findings show siRNA induced transcriptional gene silencing through involvement of DNA methylation 13, 14 and siRNA dependent knockdown of nucleus restricted transcripts 15-17. The demonstration that nuclear RNAi pathways exist in mammalian cells validates the need for carrier systems capable of delivering siRNA to the nuclear compartment. Using confocal scanning microscopy, we proved that the inclusion of the NLS in the rCPPs resulted in nuclear localized siRNA in the case of rCPP-B, -C and -D containing 75%, 50% and 25% NLS, respectively. Polyplexes formed using rCPP-A, that consists entirely of NLS, had minimal detectable amounts of siRNA in the nuclei, that might be due to inefficient endosomal escape as a consequence of the lack of a histidine component. Free siRNA duplexes, compared to siRNA when in a polyplex, labelled with a fluorophore are below the detection limit of the confocal microscope and could therefore be present in the nuclei. The use of rCPP-E (0% NLS) resulted in cytoplasmic localization of the siRNA exhibiting a perinuclear distribution which has been shown to correlate with RNA interference efficiency 38. The NLS-dependent localization of siRNA to the nuclear compartment suggests the NLS does not fully dissociate from the siRNA within the cytoplasm allowing it to mediate nuclear entry of any bound nucleic acids.
The use of LMW polycation peptides presents an attractive strategy to avoid the inherent toxicity of HMW polycations. Their effectiveness as nucleic acid carriers, however, depends on efficient release of polymer from the nucleic acid cargo allowing interaction with the intracellular target 22. The inclusion of disulfide bridges in the peptide backbone has been shown previously to improve gene delivery efficiency through polycation degradation resulting in decomplexation of the nucleic acids 22, 24. The rCPPs were used to deliver EGFP siRNA into EGFP-expressing H1299 cells and the specific knockdown of the reporter gene was used to validate the release of biologically intact siRNA. A gradual increase in the EGFP knockdown efficiency with increasing content of HRP in the copolypeptide backbone was found, to a maximum of 50% specific reduction in EGFP fluorescence intensity for rCPP-E. Polyplexes with an N : P ratio of 10 showed a slight enhancement in knockdown efficiency compared to N : P 5 polyplexes. This is likely due to more stable particles being formed at higher N : P ratios enhancing the transfection or it could be caused by the improved pH buffering capacity of the polyplexes containing more excess polypeptide facilitating more escape from the endosomal compartments.
The finding by Morris et al. 13 that siRNAs localized to the nucleus are capable of inducing transcriptional gene silencing led us to test the ability of the rCPPs to deliver siRNAs targeting a promoter region of the EF1A gene into HeLa cells. A gradual increase in the transcriptional gene silencing efficiency with increasing content of NLS peptide in the copolypeptide backbone was observed, to a maximum of 58% reduction in EF1A mRNA for rCPP-A. Polyplex E and TransIT-TKO containing no NLS resulted in a 20% decrease in EF1A mRNA, a reduction which could be due to passive accumulation of siRNA in the nucleus. The efficient transcriptional gene silencing (TGS) observed with polyplex A is in contrast to its poor EGFP reporter gene knockdown. Since the two processes, however, occur in the cytoplasm and in the nucleus respectively, the siRNA delivered by NLS-containing rCPPs could, therefore, translocate to the nucleus possibly limiting the amount available for cytoplasmic targets. In support of this, a study using a HIV-1 gp41 peptide fused with a NLS to deliver siRNA found that a mutation in the NLS enhanced siRNA-mediated knockdown of a reporter gene 39.
In mammalian cells miRNAs are transcribed as long primary RNA transcripts (pri-miRNAs) which are processed in the nucleus by the RNase III protein Drosha/DGCR8 complex into stem-loop miRNA precursors (pre-miRNAs) of ∼70 nt 40, 41. These are then transported to the cytoplasm where they are processed by another RNase III family member, Dicer, into mature miRNAs (∼22 nt) capable of mediating gene regulation 7. The endogenous sequential processing of miRNA has been shown to improve the potency of gene silencing compared to externally introduced synthetic siRNAs 8 whilst avoiding activation of the interferon system 9. Using the rCPPs to deliver pri-miRNA into HeLa cells, we found a NLS-dependent effect in the production of mature miRNA measured by Northern blotting. Polyplexes A and B did not facilitate high levels of production of miRNA, suggesting the need for a high amount of HRP in the polypeptide backbone. The TransIT-TKO reagent which showed high transfection levels of siRNAs did not result in efficient miRNA production suggesting that it is not useful as a nuclear carrier. Pri-miRNA transfected into the cells using polyplex E (100% HRP) also failed to show high amounts of produced mature miRNA, revealing that the correct HRP/NLS balance is required for the successful delivery of pri-miRNA to the nucleus. In contrast, polyplex C containing 50% HRP and 50% NLS peptide was found to yield the highest amount of mature miRNA suggesting that this polymer is the best choice for a strategy involving pri-miRNA delivery.
In conclusion, these results show that the reducible copolypeptides can be used as carriers for the non-toxic delivery of siRNA and pri-miRNA into cells. The nuclear targeting ability of the rCPPs can be used to deliver siRNA into the nucleus enabling transcriptional inactivation by promoter silencing. Furthermore, the rCPPs can be used to deliver pri-miRNAs into the nuclear compartment resulting in the production of mature miRNAs by utilizing the endogenous miRNA biogenesis pathway existing in cells. Currently we are assessing whether the delivery of siRNA to the nucleus can result in a more stable knockdown phenotype or whether endogenously processed miRNA improves the potency of gene silencing. In both cases this could potentially improve the efficacy of RNAi-based therapies.
Acknowledgements
The authors are grateful to Claus Bus and Rita Rosendahl Hansen for excellent technical assistance. We thank Anne Chauchereau (CNRS, Villejuif, France) for providing the EGFP-expressing H1299 cell line used in this work, Edouard Bertrand (IGMM, Montpellier, France) for providing the pCFP-TI-VAMP, Xiudong Liu for gel electrophoresis, Morten Ø. Andersen for cell culture, and Martin Read for helpful discussions. The work was supported by the Danish Technical Research Council, the Danish Strategic Research Council, the Danish Cancer Society, and the EU-FP6 RIGHT Program. David Oupicky and Devika S. Manickam thank NIH for financial support with grant CA109711.
Supplementary Material
The supplementary electronic material for this paper is available in Wiley InterScience at: http://www.inter-science.wiley.com/jpages/1099-498X/suppmat/.




