Concerted regulation of skeletal muscle metabolism and contractile properties by the orphan nuclear receptor Nr2f6
Abstract
Background
The maintenance of skeletal muscle plasticity upon changes in the environment, nutrient supply, and exercise depends on regulatory mechanisms that couple structural and metabolic adaptations. The mechanisms that interconnect both processes at the transcriptional level remain underexplored. Nr2f6, a nuclear receptor, regulates metabolism and cell differentiation in peripheral tissues. However, its role in the skeletal muscle is still elusive. Here, we aimed to investigate the effects of Nr2f6 modulation on muscle biology in vivo and in vitro.
Methods
Global RNA-seq was performed in Nr2f6 knockdown C2C12 myocytes (N = 4–5). Molecular and metabolic assays and proliferation experiments were performed using stable Nr2f6 knockdown and Nr2f6 overexpression C2C12 cell lines (N = 3–6). Nr2f6 content was evaluated in lipid overload models in vitro and in vivo (N = 3–6). In vivo experiments included Nr2f6 overexpression in mouse tibialis anterior muscle, followed by gene array transcriptomics and molecular assays (N = 4), ex vivo contractility experiments (N = 5), and histological analysis (N = 7). The conservation of Nr2f6 depletion effects was confirmed in primary skeletal muscle cells of humans and mice.
Results
Nr2f6 knockdown upregulated genes associated with muscle differentiation, metabolism, and contraction, while cell cycle-related genes were downregulated. In human skeletal muscle cells, Nr2f6 knockdown significantly increased the expression of myosin heavy chain genes (two-fold to three-fold) and siRNA-mediated depletion of Nr2f6 increased maximal C2C12 myocyte's lipid oxidative capacity by 75% and protected against lipid-induced cell death. Nr2f6 content decreased by 40% in lipid-overloaded myotubes and by 50% in the skeletal muscle of mice fed a high-fat diet. Nr2f6 overexpression in mice resulted in an atrophic and hypoplastic state, characterized by a significant reduction in muscle mass (15%) and myofibre content (18%), followed by an impairment (50%) in force production. These functional phenotypes were accompanied by the establishment of an inflammation-like molecular signature and a decrease in the expression of genes involved in muscle contractility and oxidative metabolism, which was associated with the repression of the uncoupling protein 3 (20%) and PGC-1α (30%) promoters activity following Nr2f6 overexpression in vitro. Additionally, Nr2f6 regulated core components of the cell division machinery, effectively decoupling muscle cell proliferation from differentiation.
Conclusions
Our findings reveal a novel role for Nr2f6 as a molecular transducer that plays a crucial role in maintaining the balance between skeletal muscle contractile function and oxidative capacity. These results have significant implications for the development of potential therapeutic strategies for metabolic diseases and myopathies.
Introduction
Muscle contraction is a highly coordinated process initiated by the transmission of the action potential from efferent neurons to the muscle. Subsequent depolarization of the muscle fibre propagates along the sarcolemma, stimulating the release of calcium ions from the sarcoplasmic reticulum, and promoting myosin-actin cross-bridge cycling and force production. An accessory metabolic machinery that generates energy is necessary to support these processes, and disruption in muscle structure or metabolism leads to pathologies, such as Duchenne's syndrome,1 sarcopenia,2 and cachexia.3 Dynamic crosstalk between energetic status, muscle development, mechanical stress, and transcriptional changes is crucial to maintaining muscle function. In this context, the nuclear receptor family of transcription factors (NRs) is of particular interest as they are potentially regulated by small molecules, such as metabolites and hormones.4
Although the transcriptional landscape for metabolic-functional signalling has been extensively studied in pathological and physiological conditions, a broader role of some NRs has only recently been recognized and the role of many members remains vague.5 NRs have a modular architecture displaying a ligand-binding domain (LBD) and a zinc-finger DNA binding domain (DBD) and can be further grouped in endogenous, orphan, or adopted NRs according to the presence of an endogenous ligand, the absence of a known ligand or if a new ligand was recently identified, respectively.6 The classical mechanistic model proposes that a small molecule binds to the LBD, changing its conformation to one of higher affinity for a transcriptional co-regulator, such as the PPARγ coactivator-1 alpha (PGC-1α), which can mediate transcriptional modulation through the recruitment of chromatin modifiers and basal transcriptional apparatus.7 The orphan nuclear receptor Nr2f6, also named Ear2, has been characterized in several tissues and organs, such as adipose, thyroid, liver, brain, and the immune system, where it plays different and even antagonistic roles.8 Nr2f6 can impair adipocyte differentiation, increase cancer cell proliferation, and promote the development of fatty liver disease by upregulating CD36.9 So far, the most extensively defined role of Nr2f6 is in the immune system, in which it directly and strongly suppresses interleukins 17, 21, 2, and interferon γ transcription by interacting with the NFAT/AP-1 complex at the promoters of these genes.10, 11
Nr2f6 has been reported both as a transcriptional repressor and activator, but the context that defines its activity state is unknown. Recently, Nr2f6 was classified as a stripe transcription factor,12 that is, it can bind to low-complexity motifs together with other transcription factors in a broad range of promoters, regulating chromatin accessibility. This suggests that Nr2f6's function in different environments depends on its DNA occupancy and interaction with other transcription factors. Therefore, it is uncertain if the current understanding of Nr2f6's role can be extended to other tissues like skeletal muscle. Hence, we sought to characterize, for the first time, the molecular mechanisms, and functional roles of Nr2f6 in skeletal muscle biology both in vitro and in vivo. In vitro approaches identified Nr2f6 as a repressor of PGC-1α and uncoupling protein 3 (UCP3) gene expression, consistent with the improvement in cells' resilience to lipid toxicity and enhanced oxidative capacity in Nr2f6 loss-of-function models. Nr2f6 gain-of-function in vivo promoted an inflammatory transcriptional signature, and upregulation of core genes of cell cycle progression, culminating in loss of muscle mass, changes in fibre type, and impairment of force production.
Methods
The main reagents, tools, and models necessary for replicating the reported results are listed in Supporting Information S3. Details of all methods are described in the Methods section of the Supporting information S1.
Cell culture
Human primary skeletal muscle cells were isolated from healthy donors,13 55 ± 5 years old, 25.6 ± 1.5 kg m−2 BMI. After confluence, growth media was switched to fusion media, and cells were cultivated for another 9 days. C2C12, MEF, and HEK cells were maintained in DMEM high glucose with supplements. Bovine serum was switched to horse serum to induce myogenesis in C2C12, and experiments were performed 5 days later.
Primary mouse skeletal muscle cells
Myoblasts were isolated as described14 and maintained for 2 days in DMEM high glucose with supplements. Myogenesis was induced for 5 days by removing foetal bovine serum when confluence was reached.
Animals
Male C57Bl6/J mice were maintained at 12/12 h light/dark cycle under controlled temperature and humidity, and ad libitum access to food and water. For high-fat diet experiments, 4-week-old C57Bl6/JUnib mice were fed for 16 weeks.
Reactive oxygen species measurement
Cells were incubated with 5 nM MitoSOX or 5 μM dihydroethidium and fluorescence intensity was measured in a plate reader. Assays were normalized using Crystal Violet.
Reverse transcription-quantitative PCR
Total RNA was extracted with TRIzol and cDNA was synthesized with a high-capacity reverse transcription kit. Primers and probes are listed in Supporting Information S2. Expression is displayed as fold-change over the indicated control.15, 16
RNA sequencing
Library was constructed with TruSeq Illumina Total RNA Stranded and a HiSeq X used for sequencing. Data was processed using Trimmomatic,17 RNA Star, featureCounts, and EdgeR. Pathway enrichment analysis was done using g:Profiler and interaction networks were analysed with CytoScape.
Fatty acid treatment
Palmitate (500 μM) and oleate (500 μM) were conjugated with 1% fatty acid-free bovine serum albumin in cell media and cells were treated for 20 h.
Promoter transactivation assays
MEF cells were transfected with UCP3 7 kbp18 or PGC-1α 2 kb promoters, Renilla luciferase, and either empty vector or Nr2f6 coding plasmid. Luciferase activity was measured with the DualGlo Luciferase Reporter kit. In knockdown assays, cells were transfected with siRNAs 1 day before the transfection with reporter plasmids.
siRNA knockdown
C2C12 were transfected with 200 nM non-target siRNA or siNr2f6 concomitantly with the differentiation media switch and experiments were performed 3 days later. Primary human skeletal muscle cells were transfected twice with 5 nM siScr or siNr2f6 and the experiments were performed on fully differentiated myotubes.
Stable cell lines
Retroviral particles were used to generate stable shNr2f6 (TRCN0000026147) and Nr2f6 overexpressing C2C12 cells.
Electroporation
Mice tibialis anterior were injected with empty or Nr2f6-myc-flag plasmids and voltage was applied. Experiments were performed 9 days after electroporation on 13-week-old mice.
Ex vivo contraction
Mouse flexor digitorum brevis were electroporated and dissected 8 days later. Measurements were conducted at optimal muscle length and maximal force was calculated as the difference between peak force and baseline. Time to fatigue was measured as the time to reach 50% peak intensity.
Myosing heavy chain staining
Tibialis anterior were dissected and immediately frozen in nitrogen-cooled isopentane. Muscle slices were blocked and probed with primary antibodies. Whole sections were imaged in a fluorescent scanning microscope at 20× magnification.
Oxygen consumption assays
Oxygen consumption rates (OCR) were measured in a Seahorse XF24 extracellular flux analyser.
Lactate measurement
Lactate was quantified by the reverse reaction of l-lactate dehydrogenase and normalized using Crystal Violet.
Western blot
Protein was extracted with RIPA buffer and loaded into gradient SDS-PAGE gels. Separated proteins were transferred to PVDF membranes and detected with ECL. Data are shown as fold-change over control.
Microarray
Gene expression in tibialis anterior was assessed with an Affymetrix Whole Transcript (WT) Assay kit probed in a CGAS cartridge for Clariom S (mouse).
Cell death assays
Cell-death assays were performed as described,19 and fluorescence intensity was measured in a plate reader.
Cell doubling time
Cells were plated and counted in a Neubauer chamber every 24 h.
ATP measurement
CellTiter-Glo Luminescent Cell Viability Assay kit was used for measuring absolute ATP concentrations.
Bioinformatic analysis of public datasets
Nr2f6 ChIP-seq data from HepG2 and K562 cells (GSM2797593 and GSM253434320, 21) and C2C12 RNA-seq (GSE4694)22 were used. Pathway enrichment was performed using g.profiler, and Nr2f6 response elements were identified using RSAT.
Statistical analysis and quantification
GraphPad Prism v7.0 was used for plotting and statistical analysis. Data normality was confirmed by the Shapiro–Wilk test.
Results
Nr2f6 regulates the transcriptional landscape of myogenesis and metabolism in skeletal muscle cells
Whole-body and in vitro genetic manipulations of Nr2f6 have been conducted,9, 23-25 but its role in the skeletal muscle is underexplored. We depleted Nr2f6 (75%) in C2C12 myocytes using siRNA and verified the effects on the transcriptome landscape through RNA-seq (Figure S1A). The 1849 differentially expressed genes, 920 upregulated and 939 downregulated, could be grouped into five main classes, with increased expression of genes related to muscle differentiation, contraction, and metabolism and decreased expression of genes with roles in mitosis and DNA packaging (Figure 1A,B,E). Among the 20 most significant genes, 11 are linked to muscle contraction (RYR1, TTN, MYH3, MYH2, ACNT2, ATP2A1, MYL1, TNNC2, MYOM3, CACNA1S, and LRP4) and other three compose the cytoskeleton (NEB, MACF1, and XIRP1), all upregulated by Nr2f6 knockdown (Figure S1B). Accordingly, canonical markers of myogenesis including muscle regulatory factors (MRFs) and myosins (Figure 1C) were broadly upregulated. Ontologies of the upregulated genes were enriched with sarcomere, contractile fibre, and cytoplasm location terms. Our transcriptome directly correlated with a public C2C12 differentiation dataset (Figure S1C), indicating an association between Nr2f6 levels and myogenic potential. Myogenesis demands withdrawal of the cell cycle and both processes are concertedly coordinated26, 27 and downregulated genes were related to cell division, with enrichment DNA replication, packaging, and chromosome segregation genes, including essential components of the mitosis progression, such as CDC25B/C phosphatases and CDK1/4 kinases, that induce quiescence and halt cycle in muscle progenitors when down-regulated.28-30 Considering the differences between mouse and human myogenic transcriptional landscape31 we verified whether Nr2f6 depletion effects are conserved in primary human skeletal muscle cells. Consistently, the expression of MYH1/2/7, muscle creatine kinase, and myosin light chain kinase 1 were upregulated by Nr2f6 knockdown in human myotubes (Figure 1D). Because increased cellular oxidative capacity and the activation of the PI3K pathway are required for the completion of myogenesis,32, 33 we investigated whether the genes of major pathways of glycolysis and β-oxidation were also affected by Nr2f6 depletion and found that several energy sensors such as AKT2, PRKAG3 subunit of AMP-activated protein kinase (AMPK) and the mTOR complex were upregulated (Figure S1D). Altogether, the changes in the myocyte's global transcriptome indicate that Nr2f6 represses key gene networks associated with myoblast differentiation and metabolism.
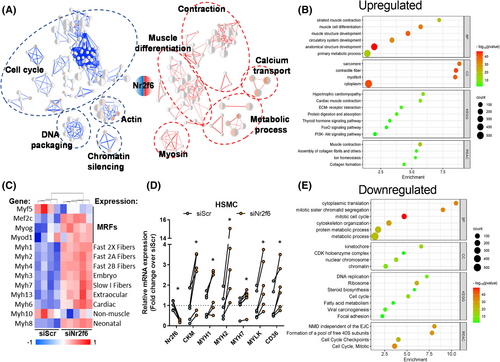
Nr2f6 depletion improves oxidative metabolism and enhances lipid handling capacity
Given the enrichment of genes related to energetic metabolism, we examined the functional effects of Nr2f6 knockdown on oxidative capacity. Maximal respiration and spare capacity were increased in oxygen consumption (Figure 2A,B) using palmitate as the major substrate. In high-glucose media, there was no difference between control and knockdown (Figure S2A, B). However, knockdown cells showed reduced extracellular acidification rates and lower extracellular lactate concentration, without decreasing ATP concentrations (Figure S2C, D, E). We hypothesized that these cells would be protected against lipid overload, and generated stable Nr2f6 knockdown (70%) C2C12 cell line to study the effects of sustained Nr2f6 depletion (Figure 2D). Nr2f6 knockdown attenuated palmitate-induced cell death (Figure 2E) and prevented the palmitate-induced increase in mitochondrial and cytosolic superoxide (Figure 2C). Accordingly, Nr2f6 knockdown increased the expression of the glucose transporter GLUT4, the anaplerotic enzyme pyruvate carboxylase (PC), and fatty acid transporters (Figure 2D). Next, we verified whether Nr2f6 is modulated by palmitate treatment in vitro and by high-fat diet in mice, as models of increased lipid oxidation and supply.34 These conditions reduced Nr2f6 expression, indicating a role as an energy stress response gene that facilitates metabolic adaptations to improve lipid oxidation (Figures 2F and S2F-I). To characterize the Nr2f6 cistrome, we explored ENCODE Nr2f6 chromatin immunoprecipitation-sequencing (ChIP-seq) data from two contrasting cell lines, human hepatocarcinoma (HepG2) and human acute myeloma (K562). There were 5610 common Nr2f6 binding sites, corresponding to 1868 unique genes with peaks within the promoter. Gene ontology (Figure S2J) indicated enrichment in metabolic processes, respiration, stress response, organelle organization, and lipid metabolism terms. Consistently, there was a significant enrichment of kinases of the insulin signalling pathway in the Nr2f6 knockdown transcriptome (Figure S2K). Collectively, the data provide evidence that Nr2f6 inhibition protects against lipid overload by increasing lipid handling capacity, possibly by a concerted upregulation of mitochondrial and cytosolic lipid transporters, mitochondrial proteins, and TCA cycle anaplerotic genes.
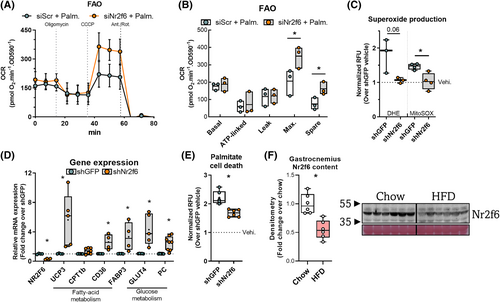
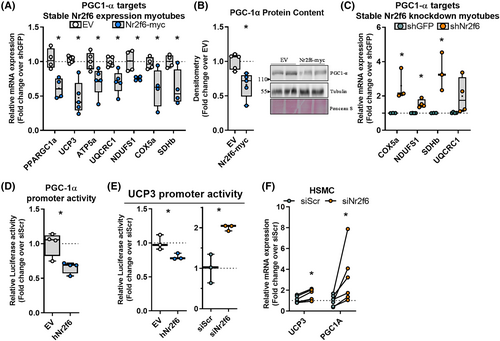
Transrepression of PGC-1α and UCP3 transcription by Nr2f6
We recently provided evidence that PGC-1α regulation of UCP3 transcription is essential for maintaining myotube viability during lipid overload.35 Considering a similar phenotype in Nr2f6 knockdown, we investigated whether Nr2f6 regulates the same pathway. Nr2f6 overexpression (Figure S3C, D) downregulated both UCP3 and PGC-1α in myotubes, also decreasing PGC-1α protein content and its mitochondrial electron transfer chain (ETC) target genes (Figure 3A,B). Conversely, Nr2f6 knockdown increased PGC-1α ETC targets (Figure 3C). PPARGC1A and UCP3 promoters were inhibited by Nr2f6 overexpression, and the latter was increased by Nr2f6 knockdown (Figure 3D,E). Nr2f6 binding motifs were found within both UCP3 and PPARGC1A promoters (Figure S3E, F) and the match in the UCP3 promoter overlapped an open chromatin region and anchoring sites of Myod1 and Myogenin known regulators of UCP3 transcription. Importantly, the inhibitory effects of Nr2f6 on UCP3 and PGC-1α expression were conserved in primary myotubes of both humans (Figure 3F) and mice (Figure S3A). UCP3 transcription is regulated by peroxisome proliferator-activated receptors (PPARs) and oestrogen-related receptors (ERRs) in skeletal muscle36, 37; however, transactivation of their response elements remained unchanged in Nr2f6 knockdown (Figure S3B). Given that UCP3 is a PGC-1α target, these results suggest that Nr2f6 can regulate UCP3 expression indirectly, via PGC-1α transcriptional regulation, or by direct modulation of UCP3 promoter activity.
Nr2f6 promotes cell proliferation and represses genes implicated in muscle contraction and oxidative metabolism
We explored the effects of Nr2f6 overexpression in vivo by electroporation of mice tibialis anterior muscle. Microarray transcriptomics revealed 3796 (FDR < 0.05, |fold change| > 2) differentially expressed genes, 1915 downregulated and 1781 upregulated, with Nr2f6 overexpression having a major effect on the hierarchical clustering (Figure S4A). Consistent with reports that highlight Nr2f6 as a gatekeeper of the immune response,24 there was an enrichment of ontologies of processes and pathways associated with the immune system (Figure 4A). RT-qPCR validation indicated that the markers of lymphocyte activation, CD44, macrophage/monocyte activation, CD68, and macrophages, F4–80, were upregulated by Nr2f6 overexpression (Figure 4D). TGFb, a potent inhibitor of haematopoietic cell activation, and the marker for endothelial and non-differentiated haematopoietic cells, VEGFa, were downregulated. These results suggest that Nr2f6 might modulate immune resident cells and/or promote the invasion of circulating cells. Downregulated genes were enriched in energetic metabolism, mitochondria, and muscle contraction terms (Figure 4B), consistent with the functional phenotypes described in vitro. Nr2f6 overexpression also increased the expression of Myogenin and MYOD, however, their downstream targets myosin heavy chains 1 and 2 were decreased (Figure 4C).
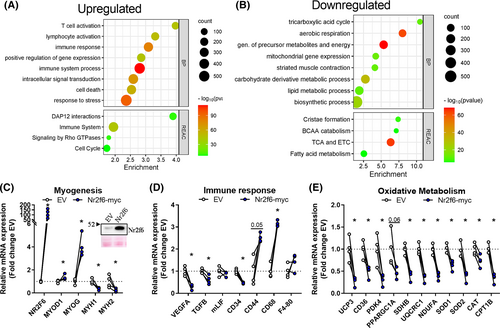
UCP3 and PGC-1α were also repressed by Nr2f6 in vivo (Figure 4E). The lipid transporters CD36 and CPT1B, and subunits of the ETC, upregulated by Nr2f6 knockdown in vitro, were downregulated by Nr2f6 overexpression. Additionally, the expression of reactive oxygen species scavengers SOD1, SOD2, and catalase genes was decreased (Figure 4E). Collectively, these findings further confirm our functional results in vitro and indicate that mitochondrial activity is impaired by Nr2f6 overexpression.
Overexpression of Nr2f6 exacerbates muscle mass wasting and impairs force production
As Nr2f6 negatively affects muscle contraction and development genes, we considered whether Nr2f6 gain-of-function would impair muscle morphology and function. Nr2f6 overexpressing tibialis anterior (TA) muscles weighted less and were visually paler (Figure 5A). Nr2f6 overexpression reduced (20%) the number of myofibres, mainly due to the decrease (25%) in type IIB fibres (Figure 5B–D).
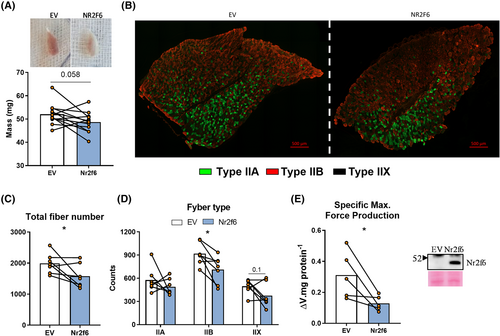
Together with the increase in cell death-related genes (Figure S4B) and the increase in the atrogenes cathepsin and calpain 238 (Figure S4B), this characterizes a state of atrophy and hypoplasia. The functional incurrences of these molecular disturbances were investigated by ex vivo contractions of Nr2f6-overexpressing flexor digitorum brevis (FDB). Considering the length constraints of these experiments, FDB was a suitable model, due to its short size, ease of access for electroporation, and similarity with TA fibre composition.39 Mass-corrected maximal force production was considerably reduced (60%) by Nr2f6 overexpression (Figure 5E). No effects were observed in the time to fatigue between control and treated groups (Figure S4C). As neuronal signals are bypassed by direct electric stimulation, effects on the transmission of the action potential are disregarded and fatigability is mostly induced by detriments in Ca2+ cycling40; therefore, we cannot exclude the possibility that fatigability is affected by Nr2f6 in vivo. Altogether, these findings strongly suggest that Nr2f6 induces muscle loss, worsened by an overactivation of the immune system and an imbalance between satellite cell proliferation and differentiation. Nr2f6 expression showed a strong tendency (P = 0.056) towards an increase in gastrocnemius of elder mice (S5B), which displayed upregulated Myod1 and myogenin.S1,S2 Mining public datasets (Figure S5A), we found that Nr2f6 is increased in biopsies from hereditary spastic paraplegia (HSP)S3 and decreased in dermatomyositis (DM). HSP causes severe muscle atrophy, accompanied by energetic dysfunction.S4 DM muscles are weakerS5,S6 and are characterized by bundles of atrophic and regenerating muscle fibres,S7 suggesting that Nr2f6 may have divergent expression within the same tissue despite overall downregulation. These findings imply a two-way association between Nr2f6 content and muscle function in various physiological and pathophysiological conditions.
Myoblast proliferation rates are governed by Nr2f6
Comparing the transcriptomes of Nr2f6 overexpression in TAs and Nr2f6 depletion in myocytes, we found 706 differentially expressed genes (DEGs) in common, whereby 446 genes were modulated in opposite directions (Figure S6A). Scanning these gene's promoters with Nr2f6 binding motifs available on JASPAR, matched 206 unique genes, whereby 73 were upregulated and 133 downregulated in the Nr2f6 overexpression microarray. Examination of the interaction network of these high-confidence targets had the most connected genes associated with the cell cycle and upregulated by Nr2f6 overexpression (Figure 6A), which emphasizes the role of Nr2f6 as a promoter of cell division and reinforces the dysplastic phenotype observed in the gain-of-function experiments in vivo. Analysis of ENCODE Nr2f6 ChIP-seq data (Figure S7A), provided additional insights into the Nr2f6-driven regulatory networks and addressed possible regulatory partners. De novo motif enrichment analysis was performed in Nr2f6 peaks coincident in both cell types and the most probable transcription factors were attributed to the enriched motifs. As expected, MEME-ChIP shows the highest enrichment of the Nr2f6 binding sequence along with other NRs (Figure S7D,E), although their binding matrices are similar, NRs with key functions in muscle metabolism and differentiation and that interact with Nr2f6, such as RXRA,S24 NR2F2,S24,S25 and NR2C1S25 were also considerably enriched. Outside the NR family, GATA binding protein 5 (GATA5), HNF1 homeobox A (HNF1A), ETS domain-containing protein (ELK1), and FBJ osteosarcoma oncogene (FOS) response elements were also frequent and followed Nr2f6's motif spatial distribution, suggesting a potential interaction at gene promoters to regulate transcription.
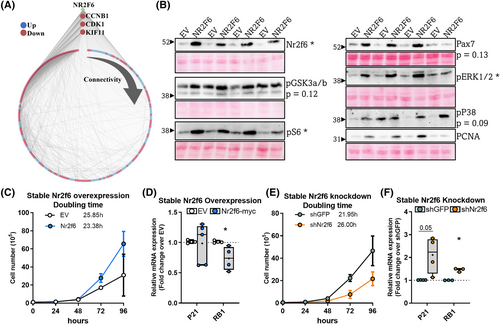
To find potential direct targets, we extracted common peaks in both ENCODE Nr2f6 ChIP-seq experiments and crossed them with our Nr2f6 knockdown transcriptome (Figure S7A), resulting in 231 unique genes significantly modulated by Nr2f6 knockdown, 3 modulated over two-fold change (Figure S7B): Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1), adhesion G protein-coupled receptor F3 (ADGRF3), and augurin precursor (ECRG4). IGF2BP1 is an RNA-binding protein with important functions in organ development and proliferation of cancer cells and myoblasts.S8,S9,S10 ECRG4 is a secreted peptide regarded as a tumour suppressorS11 and is required for proper cardiomyocyte differentiation.S12 These phenotypes are consistent with the presented Nr2f6 knockdown models, and further studies should investigate the functional relationship between Nr2f6 and ECRG4. The scientific literature is scarce regarding ADGFR3, although it has been associated with neuroendocrine tumorsS13. Accordingly, the protein content of several markers of cell proliferation and stemness was increased in Nr2f6 overexpressing TAs (Figures 6B and S6B), including the proliferating cell nuclear antigen (PCNA), a fundamental marker for cell proliferationS14; Pax7, muscle-specific satellite cell marker; the phosphorylation of the stemness marker GSK3a/b,S15,S16 and the proliferation markers ERK1/2, p38,S17,S18 and S6.S19,S20 These modulatory effects were consistent across animals, with a single mouse displaying deviant behaviour; in these cases, comparisons with P-value <0.15 are shown and were considered relevant to explain the observations. In cancer cells, Nr2f6 overexpression and knockdown can promote or inhibit cell proliferation, respectively.S21–S23 Doubling time experiments confirm that this effect is also conserved in C2C12 myoblasts, with an increase (4 h) in the average doubling time by Nr2f6 depletion and a decrease (3.5 h) by Nr2f6 overexpression (Figure 6C,E). RB1 and P21, major markers of cell cycle inhibition, are upregulated by Nr2f6 knockdown and RB1 is downregulated by Nr2f6 overexpression (Figure 6D,F). Collectively, these results demonstrate that Nr2f6 works as a major promoter of cell cycle progression in the skeletal muscle, possibly by direct modulation of transcriptional networks implicated in cell division.
Discussion
Numerous nuclear receptors are necessary for the maintenance of muscle mass.S26, S27, S28 For example, whole-body knockout of Nr1d1 (Rev-ERBα and Ear-1) increases atrophic genes and low-diameter fibres and decreases muscle mass.S29 More broadly, nuclear receptor co-repressor 1 (NCoR1) muscle-specific knockout induces hypertrophy and increases oxidative metabolism.S30 Here, using several skeletal muscle models we gathered evidence that Nr2f6 overexpression disrupts myogenesis in vivo and in vitro, and activates myoblast proliferation. Remarkably, Nr2f2 (COUP-TFII), an Nr2f6 interactor, is among the few nuclear receptors known to promote muscle wasting.S31 Nonetheless, Nr2f2 expression in myogenic progenitors impairs muscle differentiation by directly repressing genes associated with myoblast fusion and proliferation,S32 implying that these NRs regulate distinct phases of myogenesis. Future studies should address the redundancy of these NRs in muscle function.
Most of the genetically modified models suggest that NRs are involved in a general activation of oxidative metabolism.S26 For example, Nr4a3 muscle-specific transgenic mice display enhanced mitochondrial density and fast-to-slow fibre switch.S33 Mice lacking NRs coactivators, such as PGC-1α and MED1,S34 or overexpressing the NRs co-repressor RIP140,S35 show decreased mitochondrial density and fewer oxidative fibres. We demonstrate that Nr2f6 is an exception to this model as it reduces myocyte's capacity to oxidize fatty acids, increases reactive oxygen species production, and represses oxidative metabolism genes, such as UCP3 and PGC-1α, leading to a higher susceptibility to lipotoxicity. Muscle-specific UCP3 transgenic mice have improved glucose homeostasis under chow and high-fat diet (HFD) conditions, as well as resistance to obesity-induced diabetes.S36–S38 Moreover, increased levels of circulating lipids increase UCP3 expression.S39, S40 We demonstrate that Nr2f6 is downregulated when C2C12 cells are exposed to fatty acids and when there is an increase in β-oxidation in mice. This reduction may lift UCP3 transrepression, supporting the hypothesis that the downregulation of Nr2f6 is part of an adaptative response to lipid exposure.
Sarcopenia is the age-related loss of muscle mass and function, sustained by reduction of the number and area of myofibresS41 and by mitochondrial dysfunction.S42 Contrasting with most NRs, Nr2f6 overexpression in muscle induces a sarcopenia-like phenotype with muscle atrophy, hypoplasia, inflammation, reduced strength, and altered energy metabolism. Conversely, Nr2f6 knockdown in myotubes improves mitochondrial function and increases MHC expression, indicating that controlling Nr2f6 levels could be effective in treating sarcopenia and other myopathies, which is in line with recent bioinformatic analysis indicating Nr2f6 as a putative regulator of muscle energetic balance and development.S43 Nr2f6 agonists have been proposed for colitis treatmentS44; however, in light of the considerable muscle waste in Nr2f6 gain-of-function muscles reported here, the use of such agonists for patients suffering from myopathies and cachexia should be further evaluated. Although the atrophic phenotype described here could be partially underlaid by the action of Nr2f6 in immune cells, the antagonistic transcriptional and functional changes induced by Nr2f6 overexpression in vivo and its knockdown in vitro in various experimental models strongly indicate a direct action of Nr2f6 in the myofibres as the major driver of the functional changes. A conceivable mechanistic model of Nr2f6-induced force production impairment (Figure S4D), entails the downregulation of contraction-related pathways, including muscle structure, calcium cycling, and action potential transmission. The ryanodine receptor 1 (RYR1) is a major component of the calcium release complex, which mediates calcium efflux from the sarcoplasmic reticulum into the cytosol and in vivo knockdown and mutations of RYR1 lead to severe myopathies.S45 Other putative targets including myosin light chain kinase 4 (MYLK4) and myomesin 1 (MYOM1) are downstream of the androgen receptor (AR) and mediate its effects on force production.S46 The spermine oxidase gene (SMOX), another important target of the AR in muscleS47, S48 is downregulated by Nr2f6 overexpression and upregulated by Nr2f6 silencing. Therefore, possible functional associations between Nr2f6 and AR warrant further studies.
Nr2f6 can activate transcription by tethering to circRHOT1,S49 DDA1,S50 and CD369 promoters. Conversely, it represses the expression of numerous other genes such as IL17, IL21, renin, and oxytocin.10,S22,S51,S52 Our transcriptomics experiments display an equilibrated number of up and downregulated genes, suggesting that Nr2f6 is a dual-function transcription factor. The case of CD36 illustrates this context-dependent regulation as it is activated by Nr2f6 in the liver9 but repressed in skeletal muscle. This suggests that Nr2f6 activity could be modulated by post-translational modifications or interaction partners, including miRNAsS53 and proteins, such as RAR related orphan receptor γ (RORγ), an Nr2f6 interactor that regulates CD36 transcription in muscleS54 and liver.S55
In ovarian cancer, Nr2f6 promotes cell proliferation by tethering the histone acetylase P300 to the Notch3 promoter.25 We found that Nr2f6 induces myoblast proliferation, increases the expression of proliferation markers in vivo, and key regulators of the cell cycle, such as Cdk1 and its activator cyclin B1 (Ccnb1), are placed among high-confidence Nr2f6 targets. Ccnb1 ectopic expression increases cell proliferation rates and it is upregulated in several types of cancer.S56,S57,S58 The simultaneous effect of Nr2f6 modulation on cell cycle and differentiation markers might also be sustained indirectly by RB1, which is responsible for cell cycle arrest through the inhibition of the E2F family of transcription factors. In a feedback loop, E2F transcription factors antagonize MyoD1, which induces RB1 expression, thereby linking the processes of proliferation and differentiation.S59,S60 While Nr2f6 increases the expression of myogenin in the tibialis anterior muscle, myosin heavy chains and other indicators of terminally differentiated myofibres are sharply reduced, sustaining the possibility that Nr2f6 overexpressing myoblasts proliferate but fail to assemble into robust myofibrils due to a dysregulated modulation of the MRFs during myogenesis. Nr2f6 ChIP-seq peaks are associated with important transcriptional regulators of myogenesis, including GATA5, which regulates crucial genes for heart function.S61,S62 Also, ELK1 and FOS, which are downstream targets of ERK1/2, are necessary for proper muscle differentiationS63–S65 and regeneration,S66,S67 respectively. Interestingly, affinity capture-MS experiments identified FOS as an Nr2f6 interactor.S68 Using different approaches, we demonstrate that Nr2f6 is part of a cell cycle regulatory network and partners with other transcription factors to modulate muscle cell differentiation and proliferation.
The evidence gathered here points to a global regulation of pivotal aspects of skeletal muscle biology by Nr2f6, through a concerted control of multiple gene networks. Nr2f6 modulation alone can determine myoblast proliferation rates, consolidating its role as a general regulator of cell cycle progression in different lineages, and, in the skeletal muscle, acting as a fulcrum between myogenesis and cell division. Conversely, our findings amount to the hypothesis that the metabolic outcomes of Nr2f6 modulation are tissue specific. Our discoveries establish Nr2f6 as a novel regulator of muscle contraction and metabolism and allude to a potential therapeutic strategy for muscle wasting and metabolic diseases.
Acknowledgements
We would like to appreciate the technical support of Ann-Marie Petterson. This work was supported by São Paulo Research Foundation (FAPESP) (thematic projects and scholarships: 2016/23008-5, 2017/24795-3, 2021/07411-2, 2018/20581-1, 2017/24851-0), by the Karolinska Institutet Grants with support from Swedish Diabetes Foundation (DIA2021-645), Swedish Research Council for Sport Science (P2019-0140 and P2020-0064), and the Swedish Research Council (2015-00165). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES) – Finance Code 001 and by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Conflict of interest
The authors declare that they have no conflict of interest.




