miRNA in Parkinson's disease: From pathogenesis to theranostic approaches
Abstract
Parkinson's disease (PD) is an age associated neurological disorder which is specified by cardinal motor symptoms such as tremor, stiffness, bradykinesia, postural instability, and non-motor symptoms. Dopaminergic neurons degradation in substantia nigra region and aggregation of αSyn are the classic signs of molecular defects noticed in PD pathogenesis. The discovery of microRNAs (miRNA) predicted to have a pivotal part in various processes regarding regularizing the cellular functions. Studies on dysregulation of miRNA in PD pathogenesis has recently gained the concern where our review unravels the role of miRNA expression in PD and its necessity in clinical validation for therapeutic development in PD. Here, we discussed how miRNA associated with ageing process in PD through molecular mechanistic approach of miRNAs on sirtuins, tumor necrosis factor-alpha and interleukin-6, dopamine loss, oxidative stress and autophagic dysregulation. Further we have also conferred the expression of miRNAs affected by SNCA gene expression, neuronal differentiation and its therapeutic potential with PD. In conclusion, we suggest more rigorous studies should be conducted on understanding the mechanisms and functions of miRNA in PD which will eventually lead to discovery of novel and promising therapeutics for PD.
1 INTRODUCTION
Parkinson's disease (PD) is a neurodegenerative disorder consisting of multifaceted origin commonly seen in the aged population, affecting over 6 million people globally, the numbers are likely to increase over 8.3 million by 2030 (Pringsheim et al., 2014; Venkatesan, Iyer, Krishnan, et al., 2021; Venkatesan, Iyer, Wilson, et al., 2021). PD is accompanied with the degeneration of nigrostriatal dopaminergic (DArgic) pathway, decrease in dopamine (DA) neurotransmitter signaling and production loss of 50%−70% of DA neurons and this can result in motor dysfunction such as rigidity, postural instability, resting tremor and bradykinesia (Iyer et al., 2021; Venkatesan et al., 2022; Venkatesan, Iyer, Krishnan, et al., 2021; Venkatesan, Iyer, Wilson, et al., 2021). Intracellular inclusions enriched with aggregated α-synuclein (αSyn) named as Lewy bodies are often detected in PD patients and is one of the main biomarker for PD which involves in neuroinflammation or vesicle trafficking pathways (Jayaramayya et al., 2020; Mohana Devi et al., 2020; Vellingiri et al., 2022). At present, the diagnosis is based on PD clinical signs therefore there is no definitive biomarker predicted for PD till now. Currently, studies have suggested that microRNA (miRNA) regulates genes and molecular pathways associated with disease pathogenesis. miRNAs are non-coded small RNAs and endogenous managers of gene expression. The alignment of the RNA-induced silencing complex (RISC) to target the messenger RNA (mRNA) is the function of mature miRNA which binds at the complementary seed sequence in the 3'UTR region (Heman-Ackah et al., 2013; O'Brien et al., 2018). Many studies have explained that miRNA biogenesis and its mechanism of action into canonical and noncanonical pathway, where the Drosha/Dicer -dependent means canonical pathway while Drosha/Dicer independent pathway refers noncanonical pathway for miRNA biogenesis (Heman-Ackah et al., 2013; O'Brien et al., 2018). In PD, Kim et al. (2007) conducted one of the initial studies that demonstrated miRNAs roles in the maintenance of midbrain DA neurons. Contribution of Dicer and miRNA has been examined in PD and demonstrated as a hallmark in neurodegeneration (Schaefer et al., 2007). miRNA-7 and miRNA-153 are abundant in brain especially in midbrain, cortex and hippocampus where their expression downregulates αSyn miRNA levels and in contrast the miRNA depletion leads to increase in αSyn levels in PD (L. Ma et al., 2013). Therefore these growing pieces of evidence prove that dysregulation of miRNA has a pivotal role in PD pathogenesis (Goh et al., 2019). Here, in the current review, we have highlighted the importance of miRNA in PD through mechanistic approaches, ageing and therapeutic aspect in PD. In specific, we explained the miRNAs in ageing process, DArgic loss, αSyn expression, neuronal differentiation, oxidative stress, inflammatory mechanism and autophagy inhibition in PD, also, we have deliberated the in-vitro and in-vivo works in PD on miRNA expression. From the studies, we had given an overview on therapeutic ability of miRNA in PD research and suggested miRNAs as a beneficial biomarker in PD diagnosis and the need for advance research in miRNA.
2 ROLE OF MIRNA IN PD PATHOGENESIS
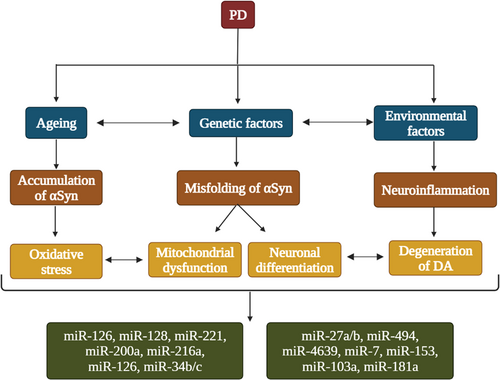
miRNAs were discovered in 1993 since then there were mounting evidence stating that the miRNAs are vital biological molecules that involves in various processes to uphold standard cellular activities (Gaudet et al., 2018; Leung & Sharp, 2010; Shivdasani, 2006). miRNAs can be observed in a collection of biological fluids such as plasma, serum, bodily waste excretions such as tears, urine, and cerebrospinal fluid (CSF) also in the breast milk (Batistela et al., 2017). Dysfunction in miRNAs are found to be involved in various neurological disorders as well as in cancer diseases (Kamal et al., 2015; Palanichamy & Rao, 2014). In vitro experiments suggests that miR-7 and miR-153 was found to be the regulator of αSyn expression in the nervous system (Doxakis, 2010; Junn et al., 2009). One of the specific expressed miRNA in the midbrain DArgic neurons is the miR-133b which indicates the essentiality of miR-133b in conservation of the DArgic neurons, hence evidently takes part in PD pathogenesis. (de Mena et al., 2010; Margis et al., 2011). In PD, there was significant upregulation of miR-24, miR-148b, miR-223, miR-324-3p, and downregulation of miR-339-5p. These discoveries advocate that all these circulating miRNAs were correlated with alterations in PD (Vallelunga et al., 2014). Paul et al. (2020) stated that various genes, including SNCA, LRRK2, DJ-1, PARKIN, NURR1, PINK1, PTEN, and PITX3, plays role in increasing the risk of developing PD and were targeted by several miRNAs (Paul et al., 2020). miR-30, miR-29, let-7, miR-485, and miR-26 are the specific miRNAs that are correlated with PD pathogenesis which are of particular interest and are likely to be interacted with PD risk gene PTEN and LRRK2, which may result in mitochondrial dysfunction and dysregulated immune pathways in PD. (Goh et al., 2019). miR-27a and miR-27b suppress the PINK1 expression, miR-133b with PITX3, let-7a-5p, miR-184-5p, miR-205 with LRRK2, miR-132 with NURR1 and miR-34b/c indirectly with PARKIN and DJ1, which will probably result in oxidative stress (Briggs et al., 2015). Nrf2 expression can be downregulated by miR-153, miR-27a, miR-142-5p, and miR-144 in SH-SY5Y cells, doubtlessly contributing to compromised oxidative stress reaction (Konovalova et al., 2019). In PD brain, miR-34b and miR-34c are downregulated along with DJ-1 and PARKIN levels (Kabaria et al., 2015). Inhibition of DJ-1 due to the upregulation of miR-4639-5p in PD leads to cell death. Mitochondrial miRNA has been recently detected and establishes an association amongst miRNA regulation and mitochondrial function, in an in vitro study it's clear that in the absence of adenosine triphosphate (ATP) RISC loading with small duplex RNA was ineffective (Yoda et al., 2010). And in other study there was a decrease in RISC function triggered by failing of RISC assembly due to the interference of ATP production by mitochondria in human cell lines (Huang et al., 2011). The above-mentioned miRNAs their target genes and their pathways have been represented as Table 1. So, pathways of miRNA gets weakened by the dysfunction of mitochondria in PD (Huang et al., 2011; Yoda et al., 2010). The deregulation of these miRNAs advocates how these molecules can influence PD pathogenesis in many forms such as Dicer formation, oxidative stress, apoptosis, expression of αSyn, and cytokines (Santos-Lobato et al., 2021). The role of miRNAs in DArgic loss, αSyn expression, neuronal differentiation and oxidative stress has been portrayed in Figure 1.
| S. No | miRNA | Target gene | In vitro/in vivo | Mechanism of action | Conclusion | Reference |
|---|---|---|---|---|---|---|
| 1. | miR-30 | LRRK2 | Mice | ↓ BDNF expression | miR-30 exerts a neurotoxic role in PD by associating with the BDNF level. | Mellios et al. (2008) |
| 2. | miR-29 | Mice | Dysregulated immune system | PD progression is mediated by miR-29 by targeting immune response such as differentiation of T helper cells and transcription factor. | Steiner et al. (2011) | |
| 3. | let-7 | LRRK2 | Mice | ↑ Oxidative stress ↑ Lipid storage |
Downregulation of let-7 leads to synuclein accumulation and increased autophagy ultimately leading to neuronal damage in PD. | Goh et al. (2019) |
| 4. | miR- 485 | Mice & SH-SY5Y cells | Apoptosis regulation | miR-485 targets TRADD to prevent TNF-α induced neuronal cell apoptosis. | Z. Chen, Zhang, et al. (2016); C. Y. A. Chen, Chang, et al. (2016) | |
| 5. | miR-26 | PTEN & PINK1 | Mice | ↓ TGF-β/JNK signaling pathway | miR-26 also modulates NF-κB signaling ultimately modulating immune system and neuroinflammation in PD | Z. Chen, Zhang, et al. (2016); C. Y. A. Chen, Chang, et al. (2016) |
| 6. | miR-27a, miR-27b, miR-133b, miR-132 | PINK1, PITX3, NURR1 | Mice & SH-SY5Y cells | ↓ PINK1, PITX3, NURR1expression | By targeting these genes miRs indirectly contributes to Oxidative stress in PD. | Briggs et al. (2015) |
| 7. | miR-153, miR-27a, miR-142-5p, miR-144 | NrF2 | SH-SY5Y cells | ↓ NrF2 | By the downregulation of NrF2 expression the oxidative stress is compromised. | Konovalova et al. (2019) |
- Abbreviations: BDNF, Brain derived Neurotrophic Factor; PD, Parkinson's Disease; TRADD, TNF receptor (TNFR)-associated death domain.
3 MIRNA TOWARD AGING AND NEURONAL DIFFERENTIATION IN PD
Aging is a multifactorial activity that involves most of the biological roles of the organism and increases sensibility to disease and death (Ugalde et al., 2011). miRNAs had been significantly related to brain aging, function deteriorating in brain, and neurodegenerative diseases (Niccoli & Partridge, 2012). (Szafranski et al., 2015). Various miRNAs have shown age-regulated expression and movement (Boehm & Slack, 2005; de Lencastre et al., 2010). Through recognized aging pathways the miRNAs modulates both negatively and positively (Kinser & Pincus, 2020). The miR-29 family (miR-29a, miR-29b, and miR-29c) found to be downregulated within the serum of PD patients and confirmed a reducing rate of acute Parkinsonism (Bai et al., 2017). The expressions of miRNAs were found to be inflated with age, while CR antagonized this increase, suggesting that those miRNAs may also modulate ageing related biological process (Jung & Suh, 2014). The miR-337, miR-302, miR-181, mir-193, miR-135, and miR-24 are likely to be involved in various cellular processes such as cartilage homeostasis, ageing, and oxidative stress (Balaskas et al., 2017). miR-34 determined to be having a link among ageing and neurodegeneration. Conversely, during aging in Caenorhabditis elegans and Drosophila miR-34 was shown to be one of the most predominantly overregulated miRNA (Kato & Slack, 2008; Liu et al., 2012). Downregulation of miR-34 results in increased brain ageing, brain degeneration, and a decline in longevity and durability, whereas overexpression of miR-34 results in a longer lifetime and less neurodegeneration (Evans et al., 2021). let-7 is affected in regulating the E2 transcription factor 1 (E2F1) which lowers let-7 levels, as a result of E2F1 overexpression there is a loss of DArgic neurons (Kumar et al., 2017). During aging let-7 is one of the vastly downregulated miRNAs (Lima et al., 2018) and in PD it might be critical to health of an animal (Kato & Slack, 2008). During aging and advancement the absence of miR-133b, exhibited no modification in the number of DArgic neurons of mice's intact midbrain hence can be implicated as miR-133b loss had no change in the levels of neurons (Schlaudraff et al., 2014; Singh & Sen, 2017). Life span increases with the increased expression of miR-246, but it had no effect on aging when there was upregulation of miR-238 (Evans et al., 2021). miR-151a-5p, miR-181a-5p, and miR-1248 are downregulated during ageing, the reduced levels of miR-181a were discovered in aged people's brains when compared with younger people (Hooten et al., 2013). Therefore, more studies need to be performed on miRNA association with ageing so that, mechanistic approach of miRNA can be determined in ageing process.
Differential miRNAs were expressed in different peripheral fluids in young and aged people, circulating serum miRNAs of old and young patients resulted with a downregulation of miR-181a-5p, miR-1248, and miR-151a-3p (Catanesi et al., 2020). miR-151a-3p, miR-181a-5p, miR-1248 is noticed to be downregulated in human serum because of ageing. miR-1248 and miR-181a are downregulated in interlukin-6 (IL-6) expression (Kou et al., 2020) whereas, miR-181a is highly expressed with downregulation of other miRNAs (Hooten et al., 2013). This shows that miR-181a-5p and miR-1248 may perform a prominent role in signaling pathways of aging process (Hooten et al., 2013) (Table 2). More specifically, it was identified that miR-151a-5p, miR-181a-5p, and miR-1248, were potential biomarkers in human diseases including neurodegenerative diseases (Dimmeler & Nicotera, 2013) and were downregulated in older individuals as the regulators of aging and cellular senescence (Shamsuzzama Kumar et al., 2016). The ectopic expression of miR-181a-5p in medullary thymic epithelial cells (MTEC1) overexpressed the cell cycle-related genes, endorsed cell cycle progression and amplified the cell proliferation (Guo et al., 2016; Ye et al., 2014). In mice, miRNA profiling for expression was executed at tibilais anterior (TA) muscle where the expression of miRNA-181a was downregulated through ageing (Soriano-Arroquia et al., 2016). Compared to ad lib-fed mice most significant downregulation was displayed by miRNA-181a-1 in brains of caloric restriction (CR)-fed mice, in age-dependent manner (Khanna et al., 2011). In low oxygen exposed environment, accumulation of pressure induced stress-based upregulation of miRNA-181 observed throughout ageing (Liang et al., 2009). Therefore, from the studies, miRNA expressions are upregulated rather downregulated during aging, hence, more studies need to be performed on miRNA association with ageing so that, mechanistic approach of miRNA can be determined in ageing process.
| S. No | miRNA | Target | Sample | Techniques | Outcome | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| 1. | miR-181a-5p | IL-1β, IL-1α, IL-6, IL-8, IL-10, TNF α, TGFβ, FGF2, HMGB1, LIF |
|
Targets the mRNA that encodes several inflammatory cytokines and the anti-inflammatory levels positively correlates with the expression of miR-181a-5p. | miRNAs were found to have downregulated with regarding to age advocating its obvious role in mechanism of aging. | Hooten et al. (2013) | |
| 2. | miR-1248 | IL-6 and IL-8 | miR-1248 gets involved in various pathways of cytokine and modulates cytokines of proinflammatory response. | ||||
| 3. | miR-151a-3p | Significant downregulation was observed in aged patients. |
- Abbreviations: miR & miRNA, microRNA; IL, Interlukin; FGF2, Fibroblast growth factor; HMGB1, High mobility group box 1; LIF, Leukemia inhibitory factor; TGFβ, Transforming growth factor beta.
The brain's midbrain, cortex, and hippocampus are particularly high with miR-7 and miR-153 expression, which is restricted to neurons rather than astrocytes. The regulation of αSyn was done by miR-7 and miR-153 (Doxakis, 2010; Junn et al., 2009). miR-7 and miR-153 has a synergic effect in downregulating the protein expressions and αSyn mRNA levels; in contrast the depletion of miRNA levels leads to an increase in αSyn in PD brains (L. Ma et al., 2013). Neuronal differentiation is a process where different types of neurons are formed from the embryonic stem cells, they differentiate into DArgic neurons and it can be regulated and altered by many factors (Chuang et al., 2015). Some of the neurogenesis related genes (e.g., NSE) were regulated by miR-153 which results in the regulation of certain proteins such as SNAPs (N-ethylmaleimide-sensitive fusion attachment proteins). The mechanism for the membranal fusion is encompassed by SNAPs and SNAREs (receptors of SNAPs). SNAPs that are involved in the differentiation and regeneration of neural cells are SNAP25 (Zylbersztejn & Galli, 2011) hence the expression of miR-153 is directly proportional to the neuronal differentiation (Tsuyama et al., 2015). The cell proliferation affected by miR-153 plays in cell differentiation and cell cycle (C. Xu et al., 2019). The study on zebrafish showed that the downregulation of SNAP25 was caused by the overexpression of miR-153 which resulted in the paralysis of zebrafish embryos (Wei et al., 2013) indicating the role of miR-153 as a regulator of neuronal differentiation. SNCA is an exclusive target for miRNA regulation and the fact that SNCA has a long 3J-UTR which is longer than the coding region; the single nucleotide polymorphisms (SNPs) of SNCA in the 3J-UTR aid to have solid connotation with sporadic PD (Mizuta et al., 2006; Simón-Sánchez et al., 2009; Winkler et al., 2007). In a PD mouse model miR-7 and miR-153 binds to the 3'UTR of SNCA mRNA and ultimately downregulates the expression of αSyn (Doxakis, 2010; Junn et al., 2009). The abundance of nucleotides with Adenine and Uracil residues in the neighboring regions of miR-153 and miR-7 facilitates the interaction with SNCA (Grimson et al., 2007). By evaluating the findings from various studies, αSyn expression in neurons were negatively regulated by miR-7 and an upregulation of αSyn was observed in PD models where the levels of miR-7 was downregulated (L. Ma et al., 2013). While treating SH-SY5Y cells with MPP+, there was an rise in the level of αSyn mRNA and a significant decrease in miR-7 levels indicating that the reduction is due to the upregulation of αSyn in MPP+ cells (Kalivendi et al., 2004). There was a reduction in the levels of both mRNA and αSyn protein expression which resulted in transfection of αSyn-expressed cells with miR-7 suggesting that the degradation of the target mRNA can be promoted by miR-7. Hence, miR-7 downregulating αSyn expression indicates the main mechanism of mRNA stability rather than alteration of translational rate. The αSyn protein expression was actively repressed by miR-7 and in contrast by using anti-miRNA inhibitors against miR-7, the levels of αSyn protein is significantly increased (Sekine et al., 2010). Post transcriptional downregulation of αSyn and the role of miR-7 has been proved in many cellular systems. In MPTP treated PD mouse model, the expression level of miR-7 identified to be decreased and leads to an increased expression in the model (Doxakis, 2010).
miR-153 is another miRNA which downregulates αSyn and targets 3'UTR with predominancy in expression (Doxakis, 2010; Farh et al., 2005). Upregulation of miR-153 leads to the reduction of αSyn levels and vice versa. miR-7, miR-153 and miR-153/7 produces 33%, 12%, and 47% decrease in luciferase activity compared to controls, thus showing the synergy between these miRNAs in the action of repressing αSyn expression and its mRNA levels. The expression levels of miR-7 and miR-153 deregulation may be essential in the pathogenesis of PD. miR-7 and miR-153 regulate SNCA protein levels by using variable kinetics, with miR-153 for the transient mRNA degradation, while miR-7 for sustained translation inhibition. The interaction effect is said to be cumulative and the inhibition of translation was from the chimeric transcript (Doxakis, 2010). Overall, the seed sequence of miRNA and SNCA is complementary and the downregulation of miR-7 and miR-153 affects the differentiation of neurons by αSyn regulation and mRNA protein levels.
4 ROLE OF MIRNA IN PD PROGRESSION
Sirtuins (SIRT) are proteins that seem to be class 3 histone deacetylases (HDACs) that are reliant on nicotinamide adenine dinucleotide (NAD+), which has a significant impact on neurodegenerative illnesses (Shoba et al., 2009). The family consists of seven members in mammals from SIRT1—SIRT7 (Ivy et al., 1986). It possesses two different activities: an ADP ribosyl transferase activity and/or a deacetylase activity (Landry et al., 2000; Liszt et al., 2005) Sirtuins induces neurodegenerative diseases either by mitochondrial function and energy metabolism regulation. Growing evidence have suggested the role of sirtuins in regulating ageing process and neurodegenerative condition (Jęśko et al., 2017). Sirtuins reported to hold an important role in maintaining the cell proteome where its function is controlled by miRNAs (Hebert et al., 2013; Rardin et al., 2013). In considering the relation between sirtuins and miRNA, various miRNAs are expressed either upstream or downstream by sirtuins-regulated pathways. In PD, miR- 34a induces apoptosis by suppressing SIRT1 in PD induced PC12 cells (Lin et al., 2015; Lou et al., 2015; Nemoto et al., 2004; Raver-Shapira et al., 2007; M. Xu et al., 2013). Similarly, in rotenone induced PD miR-384-5p promoted PD progression by targeting SIRT1 thereby inducing p53 function (Lee & Gu, 2013). Hori et al. (2013) did a study and found miRNA was downregulated, expressing the function of FOXOs and p53 with upregulation of PD. In a Chinese case-control study with 222 PD patients the SIRT2 expression was affected by miR-8061 binding to the 3'UTR which eventually contributed to PD progression (X. Chen et al., 2019). Without the involvement of miRNA, it was stated that either activation of SIRT1 or inhibition of SIRT2 exerts protective effect in PD (Donmez & Outeiro, 2013). In SH-SY5Y PD model miR-212-5p was downregulated as a result the SIRT2 translation was upregulated specifically. Moreover, there was a deacetylation of p53 by decreasing cytoplasmic p53 expression and SIRT2 inhibition promoted autophagy (P. Sun, Liu, et al., 2018). In a PC12 cell model induced with MPP+ (1-methyl-4-phenylpyridinium) significantly upregulated miR-141-3p, which caused reactive oxygen species (ROS) production upliftment, and neurotoxicity with decreased mitochondrial membrane potential, while PC12 cells were protected from MPP+ induced toxicity by miR-141-3p inhibitor, and reduced miR-141-3p regulation with increased SIRT1 expression, this concludes that miR-141-3p promotes PD progression (Zheng et al., 2020). Another study with miR-200a and miR-204 have been studied for their involvement in PD pathogenesis, in which upregulation of miR-200a and miR-204 induced by MPP+ leads to the SIRT1 downregulation which is an antioxidant gene and oxidative stress intensification. (Talepoor Ardakani et al., 2019). The link between chlorpyrifos and PD has been identified in a case study were it was found that the downregulation of miR-181 could upregulate the expression of SIRT1 with increased oxidative stress and pyroptosis (Zhao et al., 2019). In differentiated PC12 cells with MPP+ induction, significant reduction of SIRT1 with dramatic upregulation of miR-34a, miR-141, and miR-9 was observed with oxidative stress (Rostamian Delavar et al., 2018). The elevated expression of miR-9-5p in MPP+ treated SH-SY5Y cells showed that apoptosis, inflammation, cell viability and suppression and oxidative stress resulted with overexpression of SIRT1, which indicates the neuroprotective effect of SIRT1 in PD (Z. Wang et al., 2019). In a study on miR-132, the overexpression of long noncoding RNA (LncRNA) and myocardial infraction associated transcript enhanced the MPP+ induced cell viability and reduced cell apoptosis, alleviated the oxidative stress in PC12 cells treated with MPP+ as evidenced by the decrease of ROS accumulation with an increase in superoxide dismutase (SOD) and plasma glutathione peroxidase (GSH-PX) activities and these activities are carried out by promoting SIRT1 activities (X. Xu et al., 2021). miR-486-3p targets SIRT2 and their binding efficiency was influenced by SNP rs2241703. Moreover, by disrupting miR-486-3p binding sites in 3'UTR of SIRT2 this SNP mechanically influences SIRT2 level expression and hence miR-486-3p influences SIRT2 levels, aggregation and α-syn neurotoxicity (alpha synuclein), thereby impacting the risk and progression of PD (Y. Wang et al., 2018). In another study SIRT3 expression was decreased with increased miR-494-3p expression in SH-SY5Y MPP+ induced PD cells (Bause & Haigis, 2013). From the above discussed studies, it was clear that miRNA asserts a major dominant role in either upregulation or downregulation of PD by targeting the sirtuin proteins. Research has yet to be focused on exploring the efficiency of miRNA and sirtuins in PD which aid in recommending miRNAs as a potential biomarker for therapeutic approaches. Table 3 depicts the studies related to miRNA and sirtuins in PD.
| S. NO | miRNA | in vivo/in vitro | Inducers | Methods | SIRT family | Outcome | Reference |
|---|---|---|---|---|---|---|---|
| 1 | miR-384-5p | Mice SH-SY5Y cells |
Rotenone | RT-qPCR Western blot analysis Cell apoptosis assay Evaluation of α-synuclein aggregation Cell apoptosis assay |
SIRT1 | miRNA-384-5p promotes the progression of PD by targeting SIRT1PD ↑ α-synuclein ↑ Apoptosis ↑ |
Tao et al. (2020) |
| 2 | miR-8061 | 222 PD patients | - | Genotyping Plasmid constructs Luciferase Assay Western blot analysis qRT-PCR |
SIRT1 SIRT2 | mRNA level of SIRT2 and SIRT1 ↑ | X. Chen et al. (2019) |
| 3 | miR-212-5p | Mice SH-SY5Y cells |
MPTP | TEM Oligonucleotide and plasmid transfection Western blot analysis qRT-PCR Cell viability assay SIRT2 activity assay |
SIRT2 | miR-212-5p ↓ SIRT2 ↑ |
S. Sun, Han, et al. (2018) |
| 4 | miR-141-3p | PC12-cell model | MPP+ | MTT assay RT-qPCR Western blot Apoptosis assay ROS production assay ELISA |
SIRT1 | SIRT1 ↓ and miR-141-3p ↑ Apoptosis ↑ ROS ↑ |
Zheng et al. (2020) |
| 5 | miR-200a and miR-204 |
PC12-cell model | MPP+ | Cell survival evaluation Measurement of intracellular ROS production Flow cytometry analysis of cell apoptosis RNA isolation RT-PCR |
SIRT1 | miR-204 and miR-200a ↑. ROS ↑ Apoptosis ↑ |
Talepoor Ardakani et al. (2019) |
| 6 | miR-181 | SH-SY5Y cells | CPF | Transfection CCK-8 assay Measurement of ROS Cell pyroptosis analysis qRT-PCR Western blot analysis |
SIRT1 | SIRT1/PGC-1α/Nrf2 ↓ pathway miR-181 ↑ ROS ↑ Oxidative stress and pyroptosis ↑ |
Zhao et al. (2019) |
| 7 | miR-34a, miR-141, and miR-9 | PC12 | MPP+ | Flow cytometry Measurement of intracellular ROS measurement RNA isolation and RT-PCR |
SIRT1 | SIRT1 ↓ MPP+ toxicity ↑ miR-34a, miR-141, and miR-9 ↑ Apoptosis ↑ ROS ↑ |
Rostamian Delavar et al. (2018) |
| 8 | miR-9-5p | SH-SY5Y cells | MPP+ | RT-qPCR Western blot analysis Cell apoptosis assay ELISA Measurement of ROS, LDH SOD |
SIRT1 | miR-9-5p was ↑ and SIRT1 ↓ ROS, LDH and SOD ↑ | Z. Wang et al. (2019) |
| 9 | miR-200a | SH-SY5Y cells | MPP+ | Flow cytometric analysis of apoptosis RNA extraction cDNA synthesis RT-PCR |
SIRT1 | miR-200a ↑ Apoptosis ↑ |
Salimian et al. (2018) |
| 10 | miR-132 | PC12 cells | MPP+ | MTT assay Apoptosis test Determination of ROS, SOD and GSH-PX qPCR Western blot |
SIRT1 | LncRNA MIAT overexpression ROS, SOD and GSH-PX ↓ miR-132 ↑ |
X. Xu et al. (2021) |
| 11 | miR-486-3p | Clinical case study with 304 PD patients | qRT-PCR western blot analysis Evaluation of α-synuclein aggregation Genotyping |
SIRT2 | miR-486-3p α-synuclein ↓ | Y. Wang et al. (2018) | |
| 12 | miR-494-3p | Mice, SH-SY5Y cells |
MPTP MPP+ |
RNA isolation and qRT-PCR SOD activity LDH release assay ROS levels Western blot analysis ELISA Apoptosis analysis |
SIRT3 | miR-494-3p ↑ SIRT3 expression ↓ LDH ↓ Apoptosis ↓ ROS ↓ |
Geng et al. (2018) |
- Abbreviations: CCk-8, Cell Counting Kit-8; CPF, Chlorpyrifos; ELISA, Enzyme Linked immunosorbent assay; GSH-PX, Plasma Glutathione peroxidase; LDH, Lactate dehydrogenase; LDH assay, Lactose Dehydrogenase assay; miRNAs or miRs, MicroRNAs; MPTP, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; MPP+, 1-Methyl-4-Phenylpyridinium; MTS, [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide] assay; MTT assay, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium assay; Nrf2, Nuclear Factor erythroid 2; PCR, polymerase chain reaction; PC12 cells, Adrenal Phaeochromocytoma; PD, Parkinson's Disease; qRT-PCR, Quantitative Real Time reverse transcription PCR; ROS assay, Reactive oxygen species assay; RT-qPCR, Real Time Quantitative PCR, SIRT1 2 3, Sirtuins 1 2 3; SOD, Superoxide dismutase; TUNEL staining, Terminal deoxynucleotidyl transferase dUTP nick end labeling staining; ↑, upregulation; ↓, downregulation.
5 MIRNA DRIVEN INFLAMMATORY RESPONSE IN PD PATHOGENESIS
One of the important contributions of PD pathogenesis is the neuroinflammation noticed in the nigrostriatal region which are particularly vulnerable to inflammatory attack. Interleukins (IL-1, 6, 10) and tumor necrosis factor-alpha (TNF-α) are the factors of neuro inflammation (Chao et al., 2014; Ferrari & Tarelli, 2011). TNF-α is a primary cytokine where its biological action is related to the effects of IL (Frei et al., 1987). Both IL-1β and IL-6 is secreted by microglial cells and astrocytes, and they have important regulatory effects on differentiation and growth of various brain cells (Fontana et al., 1982; Kasahara et al., 1990). In postmortem brain with PD, the activated microglia along with the activated nuclear factor kappa light chain enhancer of activated B cells also been reported primarily in substantia nigra (Ghosh et al., 2007; Politis et al., 2012). In the brain of PD patients, IL-1β, IL-6, and TNF-α level increase was observed in region of striatal DArgic (Mogi et al., 1994). Studies have shown that a range of miRNA seem to have important roles in the monocytes and macrophages inflammatory responses and regulations (C. Z. Chen et al., 2004; Hou et al., 2009; Lim et al., 2003; O'Connell et al., 2012). miR-29c ameliorates increased proinflammatory cytokines in MPTP treated mice where it inhibits inflammatory cytokine production, suppresses proapoptotic regulator activity and reduces apoptotic rate. miR-29c directly targets the specific protein 1 (SP1) to defend against the apoptotic responses and neuro inflammation in PD (R. Wang, Yang, et al., 2020; R. Wang, Li, et al., 2020). miR-30e downregulation in the substantia nigra pars compact (SNpc) region of the mice prompted with MPTP lead to an increase in TNF-α levels (D. Li, Yang, et al., 2018). miR-494-3p inhibition regulated TNF-α, showing the neuroprotective role of miR-494-3p inhibition in MPP+ treated mice in PD (Geng et al., 2018). The function of miR-146a was restored by IL-6 or TNF-α blockade and also reduced myeloid malignancy. Due to aging, the expression of miR-146a seem to be downregulated by promoting inflammation with the loss of miR-146a (Grants et al., 2020). Although there are no substantial studies covered on the role of IL-6 and TNF-α with miRNAs in PD, further research is necessary to understand the mechanistic approach and emerge with therapeutic aspects. The above discussed studies were explained in detail in Table 4.
| S. No | miRNA | Inducer | PD model | IL-6 and TNF- α | Discussion | Reference |
|---|---|---|---|---|---|---|
| 1. | miR-29c ↓ | MPTP MPP+ |
Mice SH-SY5Y cells |
IL-6 ↑ TNF-α ↑ |
miR-29c directly targets the SP1 to protect against the apoptotic responses and neuro inflammatory in PD and provides a biomarker for therapy and diagnosis. | Z. Wang et al. (2019) |
| 2. | miR-30e ↑ | MPTP | Mice | TNF-α ↑ | Increase of inflammatory cytokines was due to the miR-30e agomir administration. By genetic approach and by targeting miR-30e may provide a novel strategy for treatment of PD. | Li et al. (2013) |
| 3. | miR-494-3p ↓ | MPP+ | SH-SY5Y cells | TNF-α ↑ | miR-494-3p inhibition promotes the Tumor Necrosis Factor alpha–TNF- α | Geng et al. (2018) |
| 4. | miR-146a ↓ | Aged mice | IL-6 ↑ TNF ↑ |
The cell- intrinsic and extrinsic pathway was regulated by the loss of miR-146a which links the HSC inflammation to the myeloid malignancy development. | Grants et al. (2020) |
- Abbreviations: HSC, Hematopoietic stem cell; IL, Interlukin; miRNAs or miRs, MicroRNAs; MPTP, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; MPP+, 1-Methyl-4-Phenylpyridinium, PD, Parkinson's Disease; SP1, Specific protein 1; TNF, α - Tumor Necrosis Factor alpha; ↑, upregulation, ↓, downregulation.
6 PATHOGENIC INFLUENCE OF DICER TOWARD PD
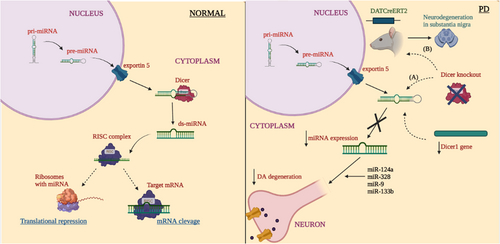
The dicer belongs to the ribonuclease (RNase) III family, it functions as a cleaving factor of long double stranded RNA (dsRNA) into short dsRNA such as miRNA and siRNA and thus it is the key factor of small regulatory RNAs biogenesis (Song & Rossi, 2017). A duplex RNA of ∼22 nucleotide with 3'end having a two-nucleotide overhang is processed by dicer from the pre-miRNA present in cytoplasm (Bernstein et al., 2001; Grishok et al., 2001). A small molecular RNA, ACA45 can also generate a functional miRNA in a DROSHA-independent manner but not in DICER-independent manner (Ender et al., 2008). Binding of miRNA to specific targets of mRNA results in inhibition translationally or mRNA deprivation (Jackson & Standart, 2007). In PD, the disruption of Dicer1 gene severely affects DA development in the embryonic DA neurons (Kim et al., 2007). In the ventral midbrain, degradation of axonal projections to the striatum and progressive loss of DA neurons is observed, which follows the Dicer ablation (Chmielarz et al., 2017). Studies have implemented the prominence of Dicer and miRNA in maintenance and survival of DA neurons. In an earlier study, the authors conducted a study in mice model of PD, where they found that the changes in miRNAs level in aged neurons follows the downregulation of Dicer to about 20% in older wild type mice than the younger one (Chmielarz et al., 2017). In an alternative study selective expression of Cre recombinase in either ventral tegmental area or SNpc of adult Dicerflox/flox mice was examined to observe the Dicer expression in survival and DA neurons maintenance and as a result common neuronal miRNAs namely miR-124a, miR-328, miR-9, miR-133b was significantly downregulated (Pang et al., 2014). miR-133b is a specific and abundant in DAergic neurons and brain of PD patients is Dicer dependent and targets Pitx3 and is necessary for the survival and conservation of DA neurons and its integrity. There is a feedback loop relation between Pitx3 and miR-133b in maintenance, once disturbed it results in downregulation of DA neurons by improper feedback loop (Kim et al., 2007). In a nutshell, the Dicer and miRNA expressions are indeed crucial for the maintenance and existence of DA neurons in the midbrain region, as in the absence, there will be degradation of DAergic neurons, cell apoptosis, disturbance in balance and motor controls, reduction in brain development (H. Zhang et al., 2004). From the studies discussed, the role of Dicer and miRNA in PD pathogenesis has major contribution in DA degeneration hence research studies on various models need to be investigated. Figure 2 represents the impact of Dicer on miRNA expression in PD and normal condition.
7 EXOSOMAL MIRNA DRIVEN PATHOGENICITY IN PD
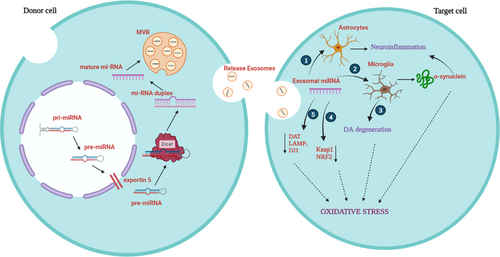
Exosomes are phospholipid bilayer membrane vesicles that ranges from 50 to 500 nm in diameter (Colombo et al., 2014). They are extracellular vesicle variants with molecular transfer and intercellular communication, angiogenesis, cell migration, and antitumor immunity. A wide variety of cells, including blood cells, epithelial cells, cancer cells, and neuronal cells, can move freely within them. The direct extracellular transport of lipids, proteins, mRNA/miRNA/DNA between the cells is aided by exosomes in both in vivo and in vitro studies (Greening et al., 2015). Exosomes takes part in various biological processes and establish intracellular communication among cells, which is essential in diverse diseases such as tumorigenesis, immunological diseases and neurodegenerative diseases hence, exosomes makes up an important cellular component (Kalluri & LeBleu, 2020). The miRNAs can be packed in exosomes and takes place in PD, before the transport of exosomes, the miRNAs should be guided by EXOmotifs to be sorted into exosomes were sorting happens in six potential mechanisms (Dolati et al., 2021; D. Li, Li, et al., 2018). MiRNA binding and sorting in exosomes are regulated by the proteins heterogeneous nuclear ribonucleoprotein (hnRNPA2B1) and synaptotagmin-binding cytoplasmic RNA-interacting protein (SYNCRIP) (Santangelo et al., 2016; Villarroya-Beltri et al., 2013). In neurons the disturbance to the maintenance of the redox is considered as oxidative stress, which takes part with number of biological processes and eventually leads to cell death (Dias et al., 2013). The miRNAs that involve in oxidative stress targets various genes such as NRF2, Keap1, PINK1, VMAT2, Parkin, DAT, and LAMP2.
Since the formation of exosomes are common in disease conditions, following are the studies discussed on exosomal miRNA inducing oxidative stress in PD. In microglia, minimal participation of oligodendrocytes and astrocytes mediates neuroinflammation in PD pathophysiology. Specifically, activated microglia contributes more to oxidative stress as it can produce numerous cytotoxic substances which includes superoxide (Block et al., 2007; Dias et al., 2013). In MPTP PD model, miR-124 enervates stimulation of microglia and increases the survival of DA neurons (Yao et al., 2018). In an in vivo PD model, miR-155 was upregulated mediating αSyn induced inflammation (Thome et al., 2016). Hence, miR-155 acts as proinflammatory and miR-124 as anti-inflammatory (Slota & Booth, 2019). The translocation of nuclear factor–erythroid factor 2-related factor 2 (NRF2) to nucleus is induced by oxidative stress, which encodes SOD1 and GSH proteins; Keap1 regulates NRF2 and facilitates degradation. miR-7 is capable of repressing Keap1 and downregulation of miR-7 which leads to DA loss in in vivo (McMillan et al., 2017). The NRF2 antioxidant response element (ARE) pathway is downregulated in neurodegenerative diseases, and it is an endogenous antioxidant system (Ramsey et al., 2007; Xie & Chen, 2016). In SH-SY5Y cells, the miR-153, miR-142-5p, miR-27a, and miR-144 downregulates NRF2 expression, which ultimately contributes to oxidative stress response (Narasimhan et al., 2012). The Dopamine Transporter (DAT) uptake of DA and downregulation of DAT expression is regulated by miR-491 and miR-137 (Jia et al., 2016) and the inadequate expression of these miRNAs leads to oxidative stress. By the downregulation of Pitx3, miR-133b indirectly stops the expression of VMAT2 (Hwang et al., 2009; Y. Li et al., 2012). Hence the upregulation of miR-133b contributes to PD pathology. In contrast, decreased level of miR-133b may lead to oxidative stress by DAT level increasing. The expression of DAT is controlled by miR-133b and VMAT2 (Hwang et al., 2009).
One of the important sources of ROS is mitochondria dysfunction. The expression of PINK1 is suppressed by miR-27a and miR-27b (Kim et al., 2016). PINK1 exhibits neuroprotective mechanism by obstructing ROS production in DA neurons (H. L. Wang et al., 2011). PINK1 and Parkin initiate the disposal of dysregulated mitochondria, so they are called mitochondrial quality control regulators (Barodia et al., 2017). In a knockout study of PINK1 in humans and mouse, DA neurons revealed the elevation of ROS generation (Wood-Kaczmar et al., 2008). miR-155, miR-27a and mitochondrial complex V subunit ATP5G3, involved in mitochondrial complex I subunit NDSFS4 downregulation (Prajapati et al., 2015). Hence, mutations or dysregulation of PINK1, Parkin and DJ-1 leads to an increase in oxidative stress (Blesa et al., 2015). It is stated that through affecting translation of DJ-1 mRNA miR-hsa-4369-5p represses protein level of DJ-1 (Y. Chen et al., 2017). miR-153, miR-214, miR-7, and miR-34b/c are the miRNAs that were stated to control the expression of αSyn and their downregulation leads to PD (Doxakis, 2010; Junn et al., 2009; Kabaria et al., 2015; Z. H. Wang et al., 2015). The downregulation of LAMP2 expression by miR-373, miR-21, miR-224, and miR-379 were demonstrated and it is stated that the downregulation of Hsc70 is carried by miR-301b, miR-16-1, miR-320a, miR-26b, and miR-106 (Alvarez-Erviti et al., 2013; G. Li et al., 2014; Z. Zhang & Cheng, 2014). Some of these miRNAs seem to be upregulated in PD patients (Xie & Chen, 2016). There are limited studies with assorted findings carried out on exosomal miRNAs in oxidative stress in PD, hence more research is required to focus on molecular mechanistic approach on exosomal miRNA on oxidative stress in PD (Figure 3).
8 ROLE OF MIRNA IN SNCA GENE EXPRESSION
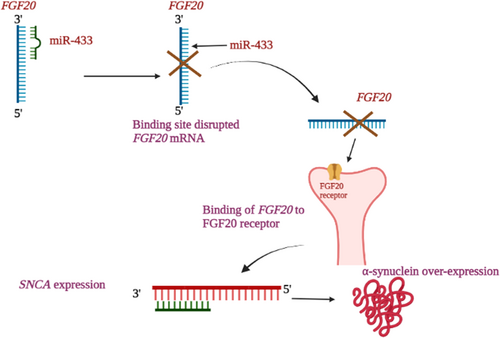
miR-433 indirectly regulate the αSyn without binding to αSyn 3'UTR sequence, by targeting FGF20 (G. Wang et al., 2008). The condensed expression of miR-433 has its contribution in αSyn and FGF20 expression. The expression level of miR-433 was significantly lower in the plasma of PD patients when paralleled to controls (X. Zhang et al., 2017). The high expression of miR-433 is specific in brain and no other body parts (Davis et al., 2005). Both in vivo and in vitro studies on SNP rs12720208 showed that by alternating the miR-433, the FGF20 translation levels can be influenced (G. Wang et al., 2008). miRNA binds to the promotor region of PD associated FGF20. The enriched translation of FGF20 and expression increase in αSyn was a result of disruption of the binding site of miR-433 by the risk allele (G. Wang et al., 2008). The augmentation of DAergic survival and the differentiation of neuronal stem cells to tyrosine-hydroxylase-positive cells were promoted by the binding of FGF20 (Ohmachi et al., 2003). After incubating the SH-SY5Y cells in recombinant human FGF20 for 24 h there was a substantial increase in αSyn protein level when paralleled to controls which proves the involvement of FGF20 in αSyn expression (G. Wang et al., 2008). Thus, it is stated that the increase in FGF20 elevates PD susceptibility via elevation of αSyn. It has been depicted that the FGF20 indirectly regulates the expression of SNCA (G. Wang et al., 2008). The elevated levels of FGF20 caused by the declined expression of miR-433 and ultimately regulates SNCA expression. Decreased miR-433 is closely associated with the elevated FGF20 expression and it was evident that the SNCA and FGF20 are presumed targets for miR-7 and miR-433 (Tarale et al., 2018). Overall, the downregulation of miR-433 leads to the dysregulation of FGF20 binding site and the FGF20 binds with FGFR1 receptor in the neurons and regulates SNCA gene ultimately causing the overexpression of αSyn in the neurons of PD patients. Hence, this mechanistic possibility of FGF20 on miRNA-433 in αSyn expression is represented in Figure 4.
9 ΑSYN AS A FACTOR AFFECTING MIRNA EXPRESSION IN PD PATHOGENESIS
The translational process of miRNA can be consequently changed by the genetic variants in 3'UTR and can also affect the activity of miRNA (Mouradian, 2012; Tan et al., 2015). The 3'UTR has a dominant role in translational efficiency, polyadenylation, mRNA stability and localization and also have an essential role in the post-transcriptional modification (Barrett et al., 2012). In PD patients and controls high expression of target miRNAs in the region of gyri cinguli was observed in a study that aimed at well understanding in the role of epigenetic influences (Tatura et al., 2016). Binding sites are provided by the variants in 3'UTR of PD genes for miRNA and RNA binding proteins thus they have a role in the translational regulation. The contribution was clear in the neurodegeneration and the regulation for PD risk by the variants in 3'UTR of PD-related genes (de Mena et al., 2013; Junn et al., 2009).
SNPs within miRNA target sites in the 3'UTR of mRNA affects the interaction amongst miRNAs and their targets, ultimately impacts the disease risk and alteration of SNCA protein expression. The miR-34b mediated protein repression was depressed by rs10024743 in 3'UTR of SNCA, which suggests that the downregulation of miR-34b and miR-34c in the brain can contribute to PD pathogenesis by increasing the expression of αSyn polymorphic variation (rs10024743) in the αSyn 3'UTR makes resistant to miR-34b (Kabaria et al., 2015). Three PD linked SNPs (rs1045722, rs356165, and rs3857053) in the SNCA 3'UTR were studied using data from the biggest GWAS on miRNA-related genetic variants. The overlapping was limited, and the conserved miRNA-binding site at rs356165 is one of the top allied SNPs in PD which results in disrupting the binding site of miR-6508 with an elevated level of SNCA expression (Ghanbari et al., 2016). The SNP located in the fibroblast growth factor 20 (FGF20) mRNA at the 3'UTR region, a significant association of SNP (rs12720208) with PD has been reported and for miR-433 the binding region genetic variation lies within the projected binding region (G. Wang et al., 2008). The translation of FGF20 protein increase is said to occur by the variant rs12720208 altering the binding of miR-433 (Schmitt et al., 2012). Studies on αSyn affecting miRNA expression is minimally concentrated, more research is needed to investigate on the molecular mechanistic approach since there is no appropriate evidence in PD pathogenesis.
10 MIRNA EXPRESSIONS ON IN VIVO AND IN VITRO MODELS IN PD
miRNA expression was investigated in PD using in vitro and in vivo models. Though many studies have been conducted in PD, there is no known causative factor predicted behind the relation between miRNA influence in PD. Table 5 depicts the list of studies conducted on miRNA expression in different study models of PD.
| S. No | miRNAs | in vivo/in vitro | Inducer | Methods used | Result | Reference | |
|---|---|---|---|---|---|---|---|
| Upregulation | Downregulation | ||||||
| 1. | miR-1, miR-22, miR-29, miR-374, miR-119a, miR-126, miR-151, miR-28, miR-301a, miR-19b-3p, miR-29c, miR-103a, miR-30b, miR-16-2, miR-26a, miR-331-5p, miR-153, miR-132-5p, miR-485-5p, miR-127-3p, miR-409-39, miR-433, miR-370, let-7g-3p, miR-873-3p, miR-136-3p, miR-10a-5p. | 47 PD patient | - |
|
miR-103a, miR-30b, miR-16-2, miR-26a, miR-331-5p, miR-153, miR-132-5p, miR-485-5p, miR-127-3p, miR-409-39, miR-433, miR-370, let-7g-3p, miR-873-3p, miR-136-3p, miR-10a-5p | miR-1, miR-22, miR-29, miR-374, miR-119a, miR-126, miR-151, miR-28, miR-301a, miR-19b-3p, miR-29c | Gui et al. (2015) |
| 2. | miR-26a | PD mice and patients | MPTP |
|
↓miR-26a | Su et al. (2019) | |
| 3. | miR-124 | Mice, MN9D cells, SH-SY5Y cells | MPTP, MPP iodide, MPP+ |
|
↓miR-124 | H. Wang et al. (2016) |
|
| 4. | miR-124-3p | PC12 and SH-SY5Y cells | 6-OHDA |
|
↓miR-124-3p | Dong et al. (2018) | |
| 5. | miR-133b | Human and Dicer flox/floxmice | - |
|
↓miR-133b | Kim et al. (2007) | |
| 6. | miR-7, miR-145, miR-543, miR-144, miR-199b, miR-221, miR-488, miR-544 | Human PD patient | - |
|
↑miR-144, miR-199b, miR-221, miR-488, miR-544 | ↓miR-7, miR-145, miR-543 | Tatura et al. (2016) |
| 7. | miR-16, miR-25 | LRRK2 mouse | - |
|
↓miR-16, miR-25 | Dorval et al. (2014) | |
| 8. | hsa-miR-4508, hsa-miR-4306, has-miR-7704, hsa-miR-151b, hsa-miR-877-5p, hsa-miR-4306, hsa-miR-320d, hsa-miR-320c, hsa-miR-326, hsa-miR-190b, hsa-miR-4791, hsa-miR-6087, hsa-miR-1273g-3p, hsa-miR-192-5p, hsa-miR-1260b, hsa-miR-129-5p, hsa-miR-4454 | SH-SY5Y cells | MnCl2 |
|
↑hsa-miR-4508, hsa-miR-4306, has-miR-7704, hsa-miR-4306, hsa-miR-320d, hsa-miR-320c, hsa-miR-326, hsa-miR-190b, hsa-miR-4791, hsa-miR-6087, hsa-miR-1273g-3p, hsa-miR-192-5p | ↓has-miR-151b, has-miR-877-5p, hsa-miR-1260b, hsa-miR-129-5p, hsa-miR-4454 | He et al. (2017) |
| 9. | miR-30b, miR-30c, miR-26a, miR-335, miR-374a, miR-199a-3p/miR-199b-3p, miR-126, miR-151-3p, miR-199a-5p, miR-151-5p, miR-126*, miR-29b, miR-147, miR-28-5p, miR-30b, miR-374b, miR-19b, miR-30c, miR-29c, miR-301a, miR-26a | PMBCs of 19 patients | - |
|
↑miR-30b, miR-30c, miR-26a | ↓miR-335, miR-374a, miR-199a-3p/miR-199b-3p, miR-126, miR-151-3p, miR-199a-5p, miR-151-5p, miR-126*, miR-29b, miR-147, miR-28-5p, miR-30b, miR-374b, miR-19b, miR-30c, miR-29c, miR-301a, miR-26a | Martins et al. (2011) |
| 10. | miR-222, miR-142-3p, let-7f, let-7a, miR-27a | 25 PD patients | - |
|
↑miR-27a | ↓miR-222, miR-142-3p, let-7f, let-7a, | L. Chen et al. (2018) |
| 11. | miR-30a-5p | 99 PD patients | l-dopa-treated |
|
↑miR-30a-5p | Schwienbacher et al. (2017) | |
| 12. | miR-103a-3p, miR-30b-5p, miR-29a-3p | PMBCs, PD patients | l-dopa-treated |
|
↑miR-103a-3p, miR-30b-5p, miR-29a-3p | Serafin et al. (2015) | |
| 13. | miR-132 | Rat |
|
↑miR-132, | Lungu et al. (2013) | ||
| 14. | miR-29c, miR-146a, miR-214, miR-221 | 138 PD patient |
|
↓miR-29c, miR-146a, miR-214, miR-221 | W. Ma et al. (2016) | ||
| 15. | miR-29c, miR-29a, miR-19b, miR-19a | IPD, LRRK2- associated PD patients |
|
↓miR-29c, miR-29a, miR-19b, miR-19a | Botta-Orfila et al. (2014) | ||
| 16. | miR-64 and miR-65, let-7 | PD-associated Caenorhabditis elegans |
|
↓miR-64, miR-65, let-7 | Asikainen et al. (2010) | ||
| 17. | miR-19b, miR-195, miR-24 | 109 PD patients |
|
↑miR-195, miR-24 | ↓miR-19b | Cao et al. (2017) | |
| 18. | miR-19b, miR-124, miR-126a, miR-133b | Mice | MPTP |
|
↓miR-19b, miR-124, miR-126a, miR-133b | Rosas-Hernandez et al. (2018) | |
| 19. | miR-3200-3p, miR-423-5p, miR-4421, miR-421, miR-155-5p, miR-219-2-3p, miR-382-59, miR-204-5p, miR-485-3p, miR-95, miR-425-5p, miR-3195, miR-221-3p | Postmortem brain samples |
|
↑miR-204-5p, miR-485-3p, miR-95, miR-425-5p, miR-3195, miR-221-3p | ↓miR-3200-3p, miR-423-5p, miR-4421, miR-421, miR-155-5p, miR-219-2-3p, miR-382-59 | Nair and Ge (2016) | |
| 20. | miR-34a, miR-26a, miR-132, let-7a, miR-7 | Parkinsonian | Rotenone |
|
↑miR-34a, miR-26a, miR-132 | ↓let-7a, miR-7 | Horst et al. (2018) |
| 21. | miR-195, miR-185, miR-15b, miR-221, miR-181a | 106 PD patient |
|
↑miR-195 | ↓miR-185, miR-15b, miR-221, miR-181a | Ding et al. (2016) | |
| 22. | miR-144-5p, miR-200a-3p, miR-542-3p | A53T-α-synuclein transgenic mice |
|
↑miR-144-5p, miR-200a-3p, miR-542-3p | Mo et al. (2016) | ||
| 23. | miR-133-3p, miR-137-3p, miR-13b-3p, miR-932-5p, miR-1008-5p | Drosophila |
|
↑miR-133-3p, miR-137-3p, miR-13b-3p, miR-932-5p, miR-1008-5p | Kong et al. (2015) | ||
| 24. | miR-141, miR-146-5p, miR-214, miR-193a-3p | 169 PD patients |
|
↓miR-141, miR-146-5p, miR-214, miR-193a-3p | Dong et al. (2016) | ||
| 25. | hsa-miR-486-5p | 13 PD patients |
|
↑hsa-miR-486-5p | Kurz et al. (2021) | ||
| 26. | miR-103a-3p, miR-485-5p, miR-339-5p | Mouse, SH-SY5Y cells | MPTP, MPP+ | ↑miR-103a-3p, miR-485-5p, | ↓miR-339-5p | J. Zhou et al. (2020) | |
| 27. | miR-384-5p | SH-SY5Y cells | Rotenone |
|
↓miR-384-5p | Jiang et al. (2016) | |
| 28. | miR-335 | 40 patients, mice, SH-SY5Y | MPTP |
|
↓miR-335 | Oliveira et al. (2021) | |
| 29. | hsa-miR-626 | 15 PD patients |
|
↓hsa-miR-626 | Qin et al. (2021) | ||
| 30. | miR-7, miR-221 | Rats | Crocin |
|
↑miR-7, miR-221 | Salama et al. (2020) | |
| 31. | miR-374c-5p | Mice SH-SY5Y cells |
MPTP MPP+ |
|
↓miR-374c-5p | Y. Dong, Xiong, et al. (2021); L. I. Dong, Zheng, et al. (2021) | |
| 32. | miR-132-3p, miR-146-5p | 82 PD patients |
|
↓miR-132-3p, miR-146-5p | Shu et al. (2020) | ||
| 33. | miR-421 | SN4741 cells |
|
↑miR-421 | L. I. Dong, Zheng, et al. (2021) | ||
| 34. | miR-216a-3p | Human neuroblastoma cell lines | MPP+ |
|
↓miR-216a-3p | H. Wang, Zhang, Wei, et al. (2021); C. Wang, Zhang, Li, et al. (2021) | |
| 35. | miR-134-5p | SK-N-SH cells | MPP+ |
|
↑miR-134-5p | C. Wang, Zhang, Li, et al. (2021); H. Wang, Zhang, Wei, et al. (2021) | |
| 36. | miR-150 | 80 PD patients |
|
↓miR-150 | H. Li et al. (2020) | ||
| 37. | miR-199a | PC12 cells | MPP+ |
|
↑miR-199a | Ba et al. (2020) | |
| 38. | miR-141-3p | PC12 cells | MPP+ |
|
↑miR-141-3p | Zheng et al. (2020) | |
| 39. | miR-302b-5p | Mice | MPTP |
|
↓miR-302b-5p | Cui et al. (2020) | |
| 40. | miR-214 | Human models |
|
↑miR-214 | L. Li et al. (2021) | ||
| 41. | miR-124 | SH-SY5Y cells | MPP+ |
|
↓miR-124 | H. Wang et al. (2016) | |
| 42. | miR-199a-3p | SH-SY5Y, PC12, mice | MPP+ |
|
↓miR-199a-3p | Q. Zhou et al. (2021) | |
- Abbreviations: BAX, Bcl-2-associated X protein; DA neurons, Dopaminergic neurons; ELISA, enzyme linked immune sorbent assay; FISH, fluorescence in situ hybridization, IPD, idiopathic parkinson's disease; l-dopa, Levodopa; LRRK2, Leucine-rich repeat kinase 2; MEF2D, isoform of myocyte-specific enhancer factor 2; MPTP, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine, PMBCs, peripheral blood mononuclear cells; SG, SYBR green; SNHG- small nucleolar RNA host gene; SN4741 cells, Mouse embryonic substantial nigra-derived SN4741 cells; 6-OHDA, 6-hydroxydopamine.
In a study with 47 PD patients, the CSF with miRNA accumulation in PD was reported with 11 miRNAs downregulated and 16 miRNAs were upregulated (Gui et al., 2015). There was a loss of miR-26a in PD mice which was induced by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and also in the CSF of PD patients (Su et al., 2019). It is considered as a side effect of neuronal death when there is a reduction in miR-124 expression in MPTP induced PD model. Hence there is a downregulation of miR-124 in MPTP induced PD cells of mice and MPP iodide induced MN9D cells of substantia nigra (Kanagaraj et al., 2014). The miR-124-3p showed a neuroprotective function in 6-hydroxydopamine (6-OHDA) treated SH-SY5Y cells of PD by targeting ANXA5 which associates with the ERK pathway stimulation (R. F. Dong et al., 2018). In PD patients the miR-133b found to be downregulated which regulates the function and maturation of the midbrain DArgic neurons (Kim et al., 2007). Five miRNAs found to be upregulated and predicted to control PARK2, SNCA, LRRK2 expressions and downregulated the miRNAs involved in governing DNA damage regulated autophagy modulator 1 (DRAM) (Tatura et al., 2016). In a mutated LRRK2, it was evident that the miR-16 and miR-25 was downregulated among the mis-regulated 24 miRNAs in the absence of LRRK2 (Dorval et al., 2014). In SH-SY5Y cells treated with MnCl2, the miRNAs are dysregulated and targeted several genes such as PARK9, ATP13A2, ATP7B, ZIP8, ZIP14, SLC11A2, and SLC30A10 in PD (He et al., 2017). In an attempt to study the miRNA expression, Martins et al. (2011) used peripheral blood mononuclear cells (PBMCs) where the authors found that 18 miRNAs were differentially expressed in which miR-30b, miR-30c, miR-26a are the chief modulators of gluycospingolipid biosynthesis-ganglioseries and ubiquintination pathway. The upregulation of miR-27a and downregulation of let-7a, let-7f, miR-222, and miR-142-3p was found in a PD study which showed the aberrant expression of circulating miRNAs in PD (L. Chen et al., 2018). The upregulation of miR-30a-5p in levodopa and I-3,4-dihydroxyphenylalanine (l-DOPA) treated PD patients said to have an effectiveness of a plasma biomarker (Schwienbacher et al., 2017). In profiling several candidate miRNAs in PD patients treated with l-DOPA, they found that the miR-103a-3p, 29a-3p, and 30b-5p was overexpressed in PD patients and these miRNAs will be validated for diagnostic potential (Serafin et al., 2015). The upregulation of miR-132 in affected rats was detected with a reduction in both mesencephalon brain tissue and serum of brain-derived neurotropic factor, it was stated that Nurr1 levels were regulated by miR-132 which effected the function and development of midbrain DA neurons (Lungu et al., 2013). In a study where 138 PD patients and 112 controls were recruited for the evaluation and identification of a proper biomarker for the PD disease, in this study the authors observed the downregulation of four miRNAs and stated that the downregulated miR-221 could be a valid biomarker (W. Ma et al., 2016). While analyzing the miRNA serum levels in a set of 10 PD patient's carriers of LRRK2 G2019S mutation and 10 controls by the array techniques, both LRRK2 and idiopathic PD, the miR-29a, miR-29c, miR-19a, and miR-19b were found to be downregulated, and associated with PD (Botta-Orfila et al., 2014).
Asikainen et al. (2010) showed that in animals 12 specific miRNAs were regulated differentially overexpressing αSyn in which three in the pdr-1 mutants, five in cat-1. The under expression of the family miR-64 and miR-65 was found in αSyn transgenic cat-1 strains and the under expression of let-7 family was observed in αSyn and pdr-1 strains indicating the differential expression in C. elegans PD models. In the investigation of 24 candidate miRNAs in PD, the overexpression of miR-195 and miR-24 was observed with a downregulation of miR-19b (Cao et al., 2017). Similarly in another study conducted for identifying a valid PD diagnosis biomarker, Rosas-Hernandez et al. (2018) used MPTP treated mice and found the serum level of miR-19b, miR-124, miR-126a, and miR-133b were downregulated. By the observation of global expression of miRNAs in postmortem putamen tissues of PD patients, six upregulated miRNAs were found and seven downregulated miRNAs were found which were involved in the oxidative stress pathway (Nair & Ge, 2016). Rotenone induced parkinsonian rats were examined for aberrantly expressed miRNA in the striatum, where the downregulation of miR-7 and let-7a, upregulation of miR-34a, miR-26a, and miR-132 was identified and recommended that these miRNAs for neuroprotection and diagnosis of PD (Horst et al., 2018). Ding et al. (2016) stated that the mis-regulation of miRNAs 195, 185, 15b, 221, and 181a can be considered as a serum-based biomarker for PD diagnosis and it is precise as it can be distinguished from normal and PD patients (Ding et al., 2016). In the investigation of A53T- αSyn transgenic mice miRNA profiles in the CSF of PD patients, the miR-144-5p, miR-200a-3p, and miR-542-3p were found upregulated and suggested as a PD biomarker (Mo et al., 2016). In Drosophila expressing A30P PD model, Kong et al. (2015) found that the miR-133-3p, miR-137-3p, miR-13b-3p, miR-932-5p, and miR-1008-5p were upregulated and could provide new insights into PD pathogenesis (Kong et al., 2015). In serum, the levels of miR-214, miR-141, miR-146-5p, and miR-193a-3p were reduced significantly in PD patients when compared with controls could be confirmed as a diagnostic tool (H. Dong et al., 2016). The abundance of hsamiR-486-5p in PD patients have been identified and are stated to have 301 gene targets (Kurz et al., 2021).
Inhibition of miR-103a-3p lead to the improved mitophagy and neuroprotective effects in both in vivo and in vitro PD models and the levels of miR-103a-3p said to be applicable in therapeutic strategies in PD (J. Zhou et al., 2020). In a study, investigation on the part of miR-384-5p in in vitro PD model using rotenone treated DArgic SH-SY5Y cells revealed that miR-384-5p downregulates and governs the expression of GRP78 by targeting its 3'UTR (Jiang et al., 2016). The miR-335 has a novel role on the effects of LRRK2-Wt overexpression and on classical inflammatory stimuli, thus initiates chronic neuro inflammation (Oliveira et al., 2021). In a study to understand the mis-regulated miRNAs in PD patients, Qin et al. (2021) found the downregulation of miR-626 could be used as a CSF based biomarker. In a study after crocin treatment there was a decline in the level of αSyn showing its neuroprotective effects and additionally, the overexpression of miR-7 and miR-221 was reported (Salama et al., 2020). By using a PD mouse model and a cellular model the action and the function of NEAT1 was studied and they observed a downregulated level of miR-374c-5p which indicated NEAT1's effective therapeutic role in treating PD (L. I. Dong, Zheng, et al., 2021; Y. Dong, Xiong, et al., 2021). In a study done with 82 PD patients and 44 healthy controls the miR-132-3p and miR-1465p was dramatically downregulated thereby proposing the idea of PD biomarker (Shu et al., 2020). Restoration of MEF2D or inhibition of miR-421 said to protect neuron toxicity in models of PD where the authors found that miR-421 is decreased and can be used for a putative mechanism for PD pathogenesis (L. I. Dong, Zheng, et al., 2021; Y. Dong, Xiong, et al., 2021). The expression of BAX was controlled by miR-216a-3p by interacting with SNHG1 and this molecular regulatory network is implicated in PD pathogenesis. Yet the molecular regulatory network in animal models is still to be explored (C. Wang, Zhang, Li, et al., 2021; H. Wang, Zhang, Wei, et al., 2021). In the MPP+-induced PD paradigm, miR-134-5p suppression appears to shield neurons from apoptosis, inflammation, and oxidative stress. More data is required for further analysis of the regulatory functions in in vivo studies (C. Wang, Zhang, Li, et al., 2021; H. Wang, Zhang, Wei, et al., 2021). The augmented expression of miR-150 stated to have beneficial effect on PD by targeting AKT3 via the downregulation of neuro inflammation and the downregulated miR-150 used as a potential biomarker for diagnostic purpose (H. Li et al., 2020). The Beclin1 and LC3II decreased protein levels, increased survival rate, reduced autophagy and enhanced cellular viability due to increase in miR-199a expression (Ba et al., 2020). The induction of oxidative stress, apoptosis and mitochondrial dysfunction was performed by increased miR-141-3p in PC12 cells which was treated by MPP+ and stated that this study could provide a strategy for therapeutics for PD (Zheng et al., 2020). miR-302b-5p shown to enhance the neuroprotective effect of insulin-like growth factor (IGF-1) by targeting inducible nitric oxide synthase, and also downregulation of miR-302b-5p said to enhance the neurobehavior of the mice which was treated with MPTP (Cui et al., 2020). In a study, miR-214 was stated to have the ability to differentiate PD patients from the controls (L. Li et al., 2021). Another study showed the DA neurons with the reduced level of miR-124 when exposed to MPTP and MPP+ in mice and SH-SY5Y cells respectively (H. Wang et al., 2016). The expression of miR-199a-3p was decreased while LRRK2 expression was upregulated in accelerating PD (Q. Zhou et al., 2021). Hence, from many studies, miRNAs could act as a promising biomarker in PD. Evidence suggest that miRNAs lie as a potential factor for diagnosis, treatment and evaluation on treatment response.
11 THERAPEUTIC POTENTIAL OF MIRNA IN PD
As discussed above, it is evident that miRNA plays a centered role in the pathogenesis of PD. Thus, Qiu et al. (2015) suggested to be clinically beneficial if the down or upregulated miRNAs were replaced or inhibited in key biological processes such as neuronal development, cell signaling, neural plasticity, apoptosis and maturation (Ambros, 2004). However, improvement and development of synthetic miRNAs with effective delivery systems were needed for the sake of therapeutic approach (Walayat et al., 2018). Artificial miRNAs are synthesized by either chemical processes or by genetic methods and their effects were tested in in vivo and in vitro models. Artificial miRNAs inhibitors that trap miRNAs and inhibit their functions are totally or partially oligonucleotides, and allows the expression of various genes (Tang et al., 2017). The interest on miRNAs role in PD and their therapeutic potential are being increased in the recent years (Nakamori et al., 2019). The studies and the research regarding miRNA as a potential therapeutic approach toward PD is still in the infant stage and discussed below are the research undergone so far with recent ideas.
Therapeutic approaches based on miRNA can be compiled into two cluster; inhibition of miRNA to downregulate the miRNAs disease driven expression and replacement of miRNA to revamp the expression of miRNAs that are disease repressed (Bernardo et al., 2015). By joining the 3'UTR of a gene, a single miRNA has the capability to downregulate several targets and its size of ~22 nucleotides make the miRNA drugs designs effective. Additionally, it has already been approved for human use for the usage of multiple drug delivery system of miRNA (P. Sun, Liu, et al., 2018). Even though there are no ongoing clinical trials using miRNAs and no established therapy, potential therapeutic intervention for PD is suggested to be explored for the miRNAs that targets and represses αSyn for PD therapeutics (Cookson, 2009; Maraganore et al., 2006). Inhibition of neuro inflammation can also be used for impacting PD since the neuro inflammation plays a major role in the pathogenesis of PD (Hirsch et al., 2012) and in this relation miR-155 can be deleted which prevents the activation of microglia and reduce the risk of loss of TH-positive nigral neurons in adeno-associated virus serotype 2-αSyn-injected mice (Thome et al., 2016). One of the method to target the disease-causing miRNAs with designing small molecules and develop new strategies for neurodegenerative studies with the capability of binding miRNAs have been considered and researched recently (Abdelrahman & Gabr, 2021). A recent new method that has advanced as a new class of small molecules that are capable of targeting diverse RNA classes and also miRNAs and mRNAs is RNase targeting chimeras (RIBOTACS) (Dey & Jaffrey, 2019). Ligands that takes part in various pathways that involves in the neurological disorder progression are the multi-targeted directed ligands (MTDLs). Based on the simultaneous suppression of acetylcholinesterase and miRNA synthesis, a novel approach for the detection of MTDLs or neurodegenerative illness was created (Gabr & Brogi, 2020). To re-form the miRNAs physiological level in PD models by miRNA mimics; anti-miRNAs may be considered as a useful tool and be a promising novel therapeutic approach (Nguyen et al., 2022). A study in which miR-124 packed nanoparticles, showed the induced proliferation of neural stem cells, neuronal maturation and neuroblasts in in vitro with an augmentation of olfactory bulb cell layer, as a result the number of neuroblasts which migrates to the cell layer was increased in the damaged striatum of 6-OHDA induced PD mice model with recovering PD symptoms (Saraiva, Ferreira, et al., 2016; Saraiva, Paiva, et al., 2016). Thus, from previous studies, it is evident that miRNA can be recommended as a diagnostic and therapeutic marker in PD and other neurodegenerative disorders.
12 CONCLUSION
The review conveyed the importance of miRNA dysregulation in PD pathogenesis. By understanding the role of miRNA, future studies must focus on investigating the mechanism and therapeutic aspects of miRNA in PD. Thus, shedding light in the field of miRNA regulated mechanisms and explore their functional aspects can be translated into therapeutic aspect in PD and other neurodegenerative disorders.
ACKNOWLEDGMENT
The authors would like to thank the Indian Council of Medical Research (ICMR), Department of Health Research—Grant-In-Aid (DHR-GIA) (grant number: GIA/2019/000276/PRCGIA), Department of Zoology, Central University of Punjab, Bathinda—151401, Punjab, Government of India and Bharathiar University, Coimbatore, to carry out this review work. This work was supported by the Indian Council of Medical Research (ICMR), Department of Health Research—Grant-In-Aid (DHR-GIA) (grant number: GIA/2019/000276/PRCGIA), Department of Zoology, Central University of Punjab, Bathinda—151401, Punjab, Government of India and Bharathiar University, Coimbatore, to carry out this review work.
CONFLICT OF INTEREST
The authors declare no conflict of interest.