The emerging roles of the interaction between m6A modification and c-Myc in driving tumorigenesis and development
Abstract
N6-methyladenosine (m6A) is an extremely common and conservative posttranscriptional modification, that can specifically target and regulate the expression or stability of a series of tumor-related genes, thus playing critical roles in the occurrence and development of tumors. c-Myc is an important tumorigenic transcription factor that promotes tumorigenesis and development by mainly regulating the expression of downstream target genes. Increasing evidence shows that m6A modification, as well as abnormal expression and regulation of c-Myc, is critical molecular mechanisms driving tumorigenesis and development. Although more evidence has been uncovered about the individual roles of m6A modification or c-Myc in tumors, the interaction between m6A modification and c-Myc in tumorigenesis and development has not been systematically summarized. Therefore, this review is focused on the mutual regulation between m6A modification and c-Myc expression and stability as well as its roles in tumorigenesis and development. We also summarized the potential value of the interaction between m6A modification and m6A expression and stability in tumor diagnosis and treatment, which provides a specific reference for revealing the mechanism of tumor occurrence and development as well as clinical diagnosis and treatment.
Abbreviations
-
- ALKBH5
-
- AlkB homolog 5
-
- AML
-
- acute myelocytic leukemia
-
- BCa
-
- bladder cancer
-
- BCSCs
-
- breast cancer stem cells
-
- CDS
-
- coding sequence
-
- CRC
-
- colorectal cancer
-
- CRD
-
- coding region instability determinant
-
- FTO
-
- obesity-associated protein
-
- GC
-
- gastric cancer
-
- GSCs
-
- glioblastoma stem cells
-
- IGF2BP1/2/3
-
- insulin-like growth factor 2 mRNA-binding protein 1/2/3
-
- LCAT3
-
- lung cancer-associated transcript 3
-
- m6A
-
- N6-methyladenosine
-
- METTL3/METTL14
-
- methyltransferase-like 3/14
-
- MZF1
-
- myeloid zinc finger 1
-
- OSCC
-
- oral squamous cell carcinoma
-
- TC
-
- thyroid cancer
-
- WTAP
-
- Wilms' tumor 1-associated protein
-
- YTHDC1/2
-
- YTH domain-containing protein 1/2
-
- YTHDF1/2
-
- YTH domain-containing family protein 1/2
-
- ZC3H13
-
- zinc finger CCCH-type containing 13
1 INTRODUCTION
In recent years, posttranscriptional modifications, as important regulators of tumor occurrence and development, have received increasing attention in cancer research (Roundtree et al., 2017; Wiener & Schwartz, 2021; B. S. Zhao et al., 2017). N6-methyladenosine (m6A) modification is an extremely common type of posttranscriptional modification that is closely related to tumors (Lan et al., 2019; Livneh et al., 2020). The modification process of m6A mainly involves three kinds of key molecules: m6A writers, m6A erasers, and m6A readers (Z. X. Liu et al., 2018; Ma et al., 2019; Tian et al., 2020). Among them, the m6A writer is a methyltransferase that catalyzes RNA methylation (S. Wang et al., 2018; Zaccara et al., 2019). The m6A eraser is a demethylase that catalyzes the demethylation of RNA (K. Chen et al., 2016; Fu et al., 2014), while the m6A reader is a protein that binds to m6A, which regulates the translation and stability of target RNA by recognizing RNA methylation information (Chang et al., 2020; Yang et al., 2018). Recent studies have found that m6A modification plays a crucial role in the regulation of the stability and expression of c-Myc, and is a crucial molecular mechanism in tumors such as diffuse large B-cell lymphoma, ocular melanoma, head and neck squamous cell carcinoma, esophageal squamous cell carcinoma, hepatocellular carcinoma (HCC), bladder cancer (BCa), and glioma (Chen, Jiang, et al., 2021; Cui et al., 2021; Du et al., 2021; H. Han et al., 2021; Mu et al., 2021; Yang et al., 2021; Yu et al., 2021). In addition, c-Myc has been proven to be an important tumorigenic transcription factor that can regulate key proteins related to m6A modification (Baluapuri et al., 2020; M. Li et al., 2019), such as EIF4A3 and YTH domain-containing family protein 1 (YTHDF1) (Wang et al., 2021; G. Wu et al., 2021), to promote the occurrence and development of tumors. Although much is known about the independent role of m6A modification and c-Myc in tumors (Ganguly et al., 2021; H. Li et al., 2021; Y. Liu et al., 2018; R. Zhao et al., 2021; Zhong et al., 2021), there is still a lack of a systematic review and summary of their mutual regulation and their role in tumorigenesis and development. To address this point, this review focuses on the regulation of m6A modification, c-Myc stability and expression, and their role in tumorigenesis and development, and provides important ideas and potential molecular targets for revealing the principle of tumor carcinogenesis, clinical diagnosis, and treatment.
2 EFFECT OF m6A MODIFICATION ON THE STABILITY AND EXPRESSION OF c-Myc MESSENGER RNA
2.1 m6A modification site on c-Myc messenger RNA
The m6A modification site plays a vital role in the stability, splicing, and translation control of messenger RNA (mRNA) (Xiang et al., 2020; H. Zhang et al., 2020). Numerous studies have found that m6A modification is enriched around the 3′-untranslated region (3′-UTR), stop codon region, and the coding sequence (CDS) of RNAs (L. He et al., 2019; D. D. Yang et al., 2020; W. Zhao et al., 2020). In particular, the region near the stop codon has the highest enrichment, while the m6A modification at the 5′-UTR is relatively low, approximately 5% (Su et al., 2018; Weng et al., 2018). It has been confirmed that the m6A writer complex preferentially deposits m6A on the consistent sequence DRA*CH (D = G/A/U, R = G/A, A* = m6A, H = U/A/C) (Y. Cheng et al., 2019; Dixit et al., 2021), and the consensus motif of most m6A loci is GGACU (T. Chen et al., 2015; Dominissini et al., 2012; W. Zhao et al., 2020).
m6A modification of c-Myc mRNA also has a crucial effect on the stability and translation of c-Myc mRNA. There is a significant m6A abundance near the stop codon and downstream 3′-UTR of c-Myc mRNA (M. Cheng et al., 2019). At the same time, the motif of the m6A modification site on c-Myc mRNA is also GGACU, and the motif is located at the c-Myc mRNA 3′-UTR and the region adjacent to the stop codon (W. Zhao et al., 2020). However, different m6A modification sites on c-Myc mRNA show different functions (Vu et al., 2017), of which the m6A modification of the c-Myc 3′-UTR enhances the stability of mRNA, while the m6A modification of the c-Myc 5′-UTR accelerates the degradation of mRNA (Wang et al., 2014; Xia et al., 2021).
2.2 Effect of m6A writers on the expression of c-Myc
The m6A writers play a role in the m6A modification of target genes, usually in the form of a methyltransferase complex, which mainly consists of three members: methyltransferase-like 3/14 (METTL3/METTL14) and Wilms' tumor 1-associated protein (WTAP) (Bokar et al., 1997). In addition, it has been reported that METTL5, zinc finger CCCH-type containing 13, and RNA-binding motif protein 15 are important in the regulation of m6A modification of target gene mRNA (J. Liu et al., 2014; Ping et al., 2014; Y. Wang et al., 2014). Recent studies have shown that m6A modification mediated by the writer complex plays a vital role in the regulation of c-Myc expression (Figure 1).
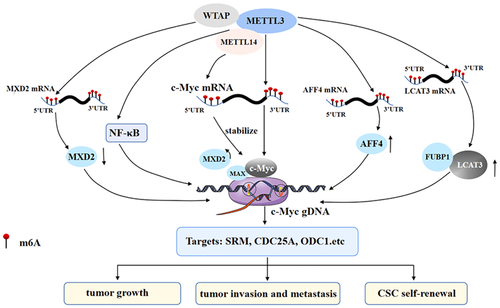
2.2.1 Methyltransferase-like 3
As a core component of the methyltransferase complex, METTL3 has two domains named the methyltransferase domain and the zinc finger domain, which are necessary for the activity of the enzyme. As a catalytic subunit of the complex, it can catalyze the methylation of mRNA (Frye et al., 2018; Zeng et al., 2020). METTL3 is crucial for the direct regulation of c-Myc by m6A modification. For example, the peak of m6A on mRNA is most enriched near the stop codon, and it is also distributed in introns and exons. The consensus motif of the m6A site is GGACU in gastric cancer (GC). In addition, METTL3 is highly expressed in GC. When METTL3 was stably knocked out, it was found that m6A modification, and mRNA and protein levels of c-Myc were significantly decreased. Therefore, the expression of the key target genes MCM5 and MCM6 downstream of c-Myc was decreased, leading to the significant inhibition of the proliferation, clone formation, migration, and invasion of GC cells (D. D. Yang et al., 2020). Similarly, studies on gastric, prostate, and lung carcinoma confirmed this conclusion (H. Wu et al., 2021; Z. Yang et al., 2020; Yuan et al., 2020). Likewise, METTL3 increased the level of c-Myc mRNA m6A modification in colorectal cancer (CRC) and promoted the recognition of m6A by insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1), that is, METTL3 enhanced the stability of c-Myc in CRC cells by relying on m6A-IGF2BP1, thus promoting the proliferation of CRC cells (Xiang et al., 2020). In the study of oral squamous cell carcinoma (OSCC), peak m6A was mainly concentrated around the stop codon of mRNA, including the CDS and 3′-UTR. The motif of METTL3 binding to c-Myc was also GGACU. The overexpression of METTL3 increased the m6A modification of c-Myc mRNA and increased the mRNA stability and protein expression of c-Myc under YTHDF1 recognition, thus promoting the invasion and metastasis of OSCC cells (W. Zhao et al., 2020). In addition, the change in c-Myc expression mediated by m6A modification is important for the differentiation of the hematopoietic system, and the increase in m6A may be a critical factor in sustaining the undifferentiated status of myeloid leukemia. In a study of acute myelocytic leukemia (AML), it was found that the knockout of METTL3 significantly reduced the enrichment of m6A in c-Myc. In contrast, overexpression of wild-type METTL3 led to increased expression of c-Myc, Bcl2, and PTEN proteins in cells, thus promoting cell growth and inhibiting differentiation (H. Wu et al., 2021). The above studies indicate that METTL3-mediated c-Myc stability and expression regulation are crucial in tumorigenesis and development.
m6A modification can not only directly affect the stop codon of c-Myc mRNA and the abundance of m6A near the 3′-UTR through METTL3, but also indirectly regulate the transcription and expression of c-Myc through METTL3, which is critical in tumorigenesis and development. For example, the high expression of METTL3 can increase the m6A modification on lung cancer-associated transcript 3 (LCAT3) mRNA and prolong the half-life of LCTA3 transcripts. Moreover, LCAT3 can recruit far upstream element-binding protein 1 to the c-Myc promoter, thus promoting the transcription and expression of c-Myc as well as the proliferation of lung cancer cells (Qian et al., 2021). Similarly, METTL3-mediated m6A modification can also promote the binding of AFF4 to the c-Myc promoter and increase the transcriptional activity and expression of c-Myc by targeting the regulation of two key regulators (IKBKB and RELA) in the AFF4 or nuclear factor (NF)-κB pathway. On the other hand, NF-κB activation mediated by IKBKB and RELA promotes the transcription and expression of c-Myc, thus leading to the tumorigenesis and development of BCa (M. Cheng et al., 2019). At the same time, the expression of METTL3 was upregulated in breast cancer stem cells (BCSCs), and overexpression of METTL3 improved the self-renewal and maintenance of stem cell properties of breast cancer cells through AFF4-mediated regulation of c-Myc transcriptional activity (Q. Gao et al., 2020). These results suggest that METTL3-mediated m6A modification can indirectly regulate the expression of c-Myc in a variety of ways and play a vital role in tumorigenesis, development, and maintenance of stem cell properties.
2.2.2 Methyltransferase-like 14
METTL14 is also an important part of the methyltransferase complex. METTL14 shows high methyltransferase activity and plays a significant role in the recognition of substrate RNA (P. C. He & He, 2021; X. Yang et al., 2020; Zhou et al., 2021). m6A modification can regulate the expression of c-Myc through METTL14 at the translational level, thus affecting cell fate and differentiation. A high level of METTL14 was found in AML, and ectopic METTL14 can enhance the m6A modification of the 3′ termination codon region of c-Myc mRNA and directly combine with c-Myc transcripts to prolong the half-life of c-Myc transcripts and improve the stability of c-Myc transcripts. Meanwhile, METTL14 can promote its translation by recruiting eIF3A to c-Myc 5′-UTR mRNA, thereby inhibiting the differentiation of hematopoietic stem or progenitor cells, and thus improving the self-renewal and growth of HSCs/LSCs and leukemia (Weng et al., 2018). Therefore, METTL14 is also essential for the process of m6A modification-mediated c-Myc expression regulation.
2.2.3 Wilms' tumor 1-associated protein
WTAP is a nuclear protein that participates in a variety of pathophysiological processes. WTAP is mainly responsible for the localization of nuclear spots, which is related to the alternative splicing of genes. WTAP-mediated splicing regulation is necessary for this unique nuclear localization (Ping et al., 2014; Sorci et al., 2018; T. Wang et al., 2020). WTAP also plays a critical role in the process by which m6A modification affects c-Myc expression (S. Chen et al., 2020). WTAP was upregulated in AML and increased m6A modification of c-Myc mRNA. The increase in m6A modification affected the half-life of c-Myc mRNA and the level of c-Myc mRNA, which promoted the proliferation and colony formation of AML cells as well as drug resistance to daunorubicin (Naren et al., 2021). In addition, WTAP was discovered to increase m6A modification of c-Myc and thus lead to the development and progression of ovarian cancer, which provides new insight into the role of epigenetic m6A modification of c-Myc in ovarian cancer (X. Han et al., 2020).
WTAP can also indirectly affect the m6A modification of other molecules, thus increasing c-Myc transcriptional activity and promoting tumorigenesis and progression. For example, WTAP can decrease the expression of MXD2 by increasing the m6A modification on MXD2 mRNA and destroying its stability, thereby improving the binding of MAX with c-Myc and promoting the transcription of downstream target genes of c-Myc, such as SRM, RPL23, CDC25A, CDC25, and ODC1, leading to cell proliferation and tumor growth (Cho et al., 2021).
2.3 Effect of m6A erasers on the expression of c-Myc
The m6A eraser is a demethylase that is also crucial to the process of m6A modification-mediated c-Myc expression regulation (Yue et al., 2020; Zou et al., 2019). Erasers can directly affect m6A modification of mRNA, thus affecting the expression of c-Myc. Among them, obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) are the most studied erasers (Figure 2).
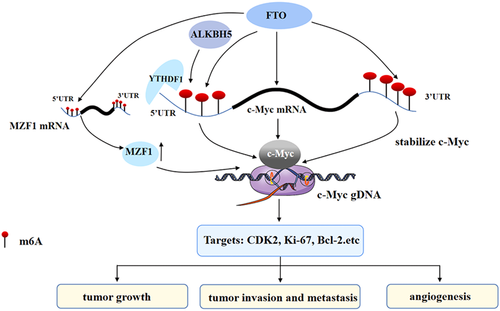
2.3.1 Obesity-associated protein
FTO is located on chromosome 16q12.2, and it is not only received attention in obesity-related diseases but is also the first identified demethylase that catalyzes the demethylation of m6A methylation (J. Chen & Du, 2019; J. Y. Wang et al., 2020). FTO can regulate the stability of c-Myc at different sites, while different m6A modification sites on c-Myc mRNA have different effects on the expression of c-Myc. For example, Yue et al. (2020) found that FTO can upregulate the expression of c-Myc by inhibiting the m6A modification of the c-Myc gene in CRC, thus inhibiting apoptosis and improving cell proliferation, migration, and invasion. It has also been suggested that in the case of FTO overexpression in AML, the reduction in c-Myc mRNA m6A modification can improve the mRNA stability of c-Myc, thus promoting the occurrence of AML (Su et al., 2018). However, the two studies did not explore how FTO upregulated the expression of c-Myc after FTO suppressed the m6A modification of c-Myc mRNA in CRC. Yang et al. found that the upregulation of FTO removed the m6A modification on c-Myc 5′-UTR mRNA in GC cells, while the m6A modification on c-Myc 5′-UTR mRNA could accelerate the degradation of mRNA (Deng et al., 2018). Therefore, the upregulation of FTO made c-Myc mRNA more stable and promoted the expression of c-Myc, thus leading to the occurrence of GC (Z. Yang et al., 2021). However, the expression of FTO decreased in lung adenocarcinoma, which made no difference to c-Myc mRNA, but increased the level of m6A modification on c-Myc mRNA and the protein expression level of c-Myc (X. Yang et al., 2021). This may act on the modification of m6A on c-Myc 3′-UTR mRNA, thus stabilizing c-Myc mRNA and increasing the level of c-Myc expression (Wang et al., 2014; Xia et al., 2021). It has been suggested that different m6A modification sites on c-Myc mRNA have different effects on c-Myc. In addition, the elevated level of m6A on c-Myc mRNA recruits YTHDF1, thus promoting the translation of c-Myc mRNA, and in turn, promoting glycolysis, proliferation, and tumor growth.
FTO can not only directly affect the level of m6A modification on c-Myc mRNA but also regulate the m6A modification of other molecules, thus indirectly regulating c-Myc mRNA and the expression of c-Myc as well as driving tumorigenesis. For instance, the tumor suppressor gene GSK3β can regulate FTO and inhibit myeloid zinc finger 1 (MZF1) in CRC. FTO can remove the m6A modification on MZF1 mRNA, thereby increasing the expression of MZF1 and promoting the expression of c-Myc downstream of MZF1, thus promoting the proliferation of CRC cells (Z. Zhang et al., 2021). It is suggested that FTO can act on the expression of c-Myc in a direct and indirect way.
2.3.2 AlkB homolog 5
ALKBH5 is a member of the AlkB family. ALKBH5 is essential for biological processes such as tumorigenesis, migration, invasion, and metastasis. In addition, ALKBH5 is also a well-known m6A demethylase (Chao et al., 2020; Malacrida et al., 2020; J. Wang et al., 2020). Some researchers have used different strategies to study the role of ALKBH5 in m6A modification-mediated c-Myc expression. For example, Li et al. fused ALKBH5 with inactivated Cas13b (dCas13b) to create a dCas13b-ALKBH5 fusion protein. As a result, dm6ACRISPR targeting c-Myc could induce c-Myc mRNA demethylation, reduce c-Myc mRNA levels and inhibit its protein expression. At the same time, dm6ACRISPR also reduced the binding of c-Myc mRNA with YTHDF1, thus inhibiting cell proliferation (J. Li et al., 2020). Furthermore, ALKBH5 and the m6A recognition protein YTHDF3 may lead to the degradation of circ3823 by recognizing the m6A modification on circ3823 in CRC. Circ3823 can inhibit the expression of TCF7 promoted by miR-30c-5p and further activate the downstream functional molecule c-Myc, thus leading to the migration and invasion of CRC cells (Guo et al., 2021). In lung carcinoma, the overexpression of ALKBH5 reduced the m6A modification on c-Myc mRNA, thus reducing the recognition of YTYTHDF2 on m6A modification of c-Myc mRNA, thereby promoting the stability and expression of c-Myc mRNA (D. Zhang et al., 2021). These results suggest that ALKBH5 can regulate cell activity by targeting the demethylation of functional gene transcripts on c-Myc.
2.4 Effect of m6A readers on the expression of c-Myc
Common m6A readers include the YTH family (such as YTH domain-containing protein 1 [YTHDC1] and YTHDC2), IGF2BP1/2/3, and heterogeneous nuclear ribonucleoprotein C (Bi et al., 2019; L. C. Wang et al., 2020; B. Zhang et al., 2021). Due to the different sites of m6A modification on mRNA, there are differences in the reaction effect of different readers on m6A modification (Y. Zhao et al., 2020). In the process of modifying and regulating c-Myc by m6A, more studies have covered readers including insulin-like growth factor-binding proteins (IGFBPs) and YTHDF2 (Figure 3).
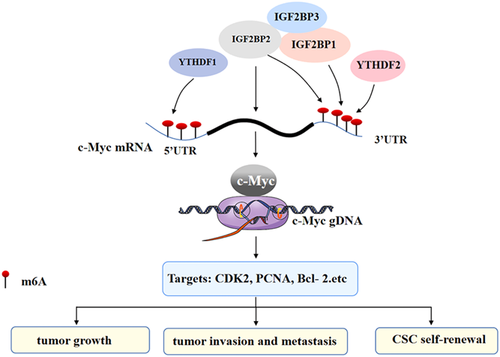
2.4.1 Insulin-like growth factor-binding protein
IGFBPs include IGF2BP1, IGF2BP2, and IGF2BP3, which can recognize m6A modification on RNA (Hu et al., 2020; Huang et al., 2018; Muller et al., 2019). IGF2BP1 and IGF2BP2, members of IGFBPs, can act alone in the effect of m6A modification on the stability of c-Myc mRNA. For example, there is a high abundance of m6A modification in the coding region instability determinant (CRD) region of ∼250 nt at the 3ʹ-end of the c-Myc mRNA coding region in CRC (P. Zhu et al., 2021; S. Zhu et al., 2020). In this study, RBRP promoted the binding of IGF2BP1 and c-Myc CRD mRNA. On the one hand, this binding stabilizes the c-Myc mRNA and promotes the expression of the c-Myc protein, on the other hand, this binding can also recruit RNA stabilizers such as Hur, MATK3, and PABPC1, thus promoting IGF2BP1's recognition of m6A on c-Myc mRNA, thereby improving the stability and level of c-Myc mRNA as well as improving cell proliferation, migration and invasion. Likewise, the c-Myc CRD region is rich in m6A modification in BCSCs. In addition, IGF2BP1 binding to c-Myc CRD mRNA can enhance the spheroid formation and clone formation ability of BCSCs, and maintain the stem cell properties of BCSCs under hypoxia (Huang et al., 2018). Similarly, the upregulated expression of IGF2BP2 enhances the recognition of c-Myc mRNA m6A modification and upregulates the protein level of c-Myc in thyroid cancer (TC), thus promoting the migration and antiapoptosis of TC cells (Ye et al., 2021). These results suggest that m6A can influence c-Myc mRNA alone through the m6A reader and play a role in different types of tumors.
m6A modification can also promote tumorigenesis through the synergistic action of IGFBP members on c-Myc. In the study of AML, the YBX1 CDS region can interact with IGF2BP1/2/3, and all of them are preferentially combined in the classical m6A motif containing the UGGAC consensus sequence on mRNA. Through m6A-dependent binding, it can not only stabilize c-Myc mRNA, but also improve the protein level of c-Myc, consequently promoting the clone formation ability of AML cells (Feng et al., 2021).
2.4.2 YTH domain-containing family protein 2
Among the m6A reader members, the YTH family is also significant in the influence of m6A modification on c-Myc. The YTH family includes YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3 (Shi et al., 2017; Wang et al., 2015; Xu et al., 2014). Some studies have shown that YTHDF2 binding may not only destabilize some transcripts (Fang et al., 2021; Paris et al., 2019), but also stabilize important oncogenes, such as c-Myc, by m6A modification. For instance, the expression of YTHDF2 is upregulated in glioblastoma stem cells (GSCs), and YTHDF2 can stabilize c-Myc transcripts through m6A-dependent binding, hence promoting the expression of c-Myc mRNA and its proteins. In addition, IGFBP3, the downstream effector of the YTHDF2-MYC axis, is overexpressed in gliomas because it can obtain multiple m6A peaks of c-Myc. IGFBP3 can support the survival of glioblastoma as well as increase GSC frequency and speed up self-renewal by stabilizing c-Myc transcripts, thus promoting the occurrence of GSCs (Dixit et al., 2021). Therefore, the m6A reader is essential for the influence of m6A modification on c-Myc mRNA.
3 EFFECT OF c-Myc ON THE TRANSCRIPTIONAL REGULATION OF GENES RELATED TO m6A MODIFICATION
The oncoprotein c-Myc is an important transcription factor. c-Myc can target and regulate downstream genes such as CDK4, Gadd45, and SRM (Lee et al., 1997; Miliani de Marval et al., 2004). c-Myc can also regulate m6A modification-related molecules such as m6A erasers and readers, therefore playing a significant role in tumorigenesis and development (Kress et al., 2015). The following is a detailed overview of the specific impact of c-Myc on m6A operators (Figure 4).
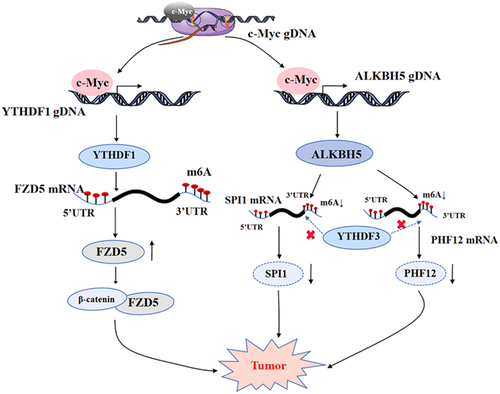
3.1 c-Myc regulates the expression of the m6A eraser ALKBH5
In a small number of studies on the regulation of m6A modification by c-Myc, Wu et al. demonstrated that c-Myc can regulate tumorigenesis by affecting the m6A eraser. Specifically, it was found that c-Myc could downregulate the m6A modification level of total mRNA, and preferentially downregulate the mRNA m6A modification level of c-Myc suppressor genes, such as SPI1 and PHF12. In addition, c-Myc can bind to the promoter region of ALKBH5, making ALKBH5 remove the m6A modification on the m6A-modified SPI1 and PHF12 mRNA, thus reducing the binding of YTHDF3 to the m6A modification on the mRNA of both, and inhibiting the translation of SPI1 and PHF12. While SPI1 and PHF12 are tumor suppressors, the decrease in the protein levels of SPI1 and PHF12 promoted the growth of lymphoid cancer cells (G. Wu et al., 2021). This study showed that c-Myc itself can regulate the expression of the demethylase ALKBH5. This study provides a new perspective on how c-Myc regulates mRNA translation through mRNA m6A modification.
3.2 c-Myc regulates the expression of the m6A reader YTHDF1
c-Myc can also react to m6A readers and affect their expression, thus promoting tumorigenesis. For example, YTHDF1 is regulated by c-Myc transcription in CRC. In addition, c-Myc can bind to the 5ʹ-end of the YTHDF1 gene and promote the expression of the m6A reader YTHDF1, therefore participating in the malignant behavior of CRC and promoting the proliferation of CRC cells as well as developing resistance to anticancer drugs such as fluorouracil and oxaliplatin (Nishizawa et al., 2018). It is suggested that c-Myc induces the carcinogenic effect of transactivation of YTHDF1 in an unknown way, indicating that c-Myc regulates m6A modification and promotes tumorigenesis.
4 THE INTERACTION BETWEEN m6A MODIFICATION AND c-Myc FORMS A DOUBLE POSITIVE FEEDBACK LOOP TO DRIVE THE TUMORIGENESIS AND DEVELOPMENT
m6A modification, as a critical method of gene posttranscriptional regulation, regulates the expression of c-Myc at the mRNA stability and translation levels, which is important for the occurrence, development, and maintenance of stem cell properties of different types of tumors (R. Gao et al., 2021). For example, the m6A writers METTL3, METTL14 and WTAP, the m6A erasers FTO and ALKBH5, and the m6A readers IGFBPs and YTHDF2 can directly affect the stability or translation of c-Myc mRNA, thus promoting the expression of c-Myc. In addition, c-Myc, an important transcription factor, can also regulate the expression of genes related to m6A modification. For example, c-Myc can also directly regulate the transcriptional activity and expression of the m6A eraser ALKBH5 and the m6A reader YTHDF1, hence affecting the m6A modification and expression of downstream genes, thus playing a role in tumorigenesis and development. Therefore, the interaction between m6A modification and c-Myc expression regulation can form a double positive feedback loop.
Meanwhile, m6A modification and c-Myc can regulate each other indirectly. For example, m6A writers, erasers, and readers can regulate the transcription or translation of c-Myc by affecting m6A modification of MXD2, FBXW7, AFF4, LCAT3, MZF1 mRNA, and the NF-κB signaling pathway, while c-Myc can also affect m6A modification through the Wnt/β-catenin signaling pathway. The indirect interaction between them forms a positive feedback loop and jointly drives tumorigenesis and development. Likewise, c-Myc can target the E-box site that binds to the YTHDF1 promoter, thus promoting the transcriptional activity and expression of YTHDF1. Moreover, YTHDF1 can increase its translation efficiency and expression by selectively recognizing the m6A site of FZD5 mRNA, thus activating the FZD5-mediated Wnt/β-catenin signaling pathway. At the same time, the Wnt/β-catenin signaling pathway can also promote the expression of c-Myc, which causes liver cancer by positive feedback (X. Liu et al., 2020). In summary, the direct and indirect interactions between m6A modification and c-Myc form a complex positive feedback regulatory network. c-Myc-mediated transcriptional regulation of downstream target genes and m6A modification-mediated posttranscriptional regulation of mRNA promote the expression of tumor-related effector genes or the activation of signaling pathways to jointly drive tumorigenesis, development, and metastasis (Figure 5).
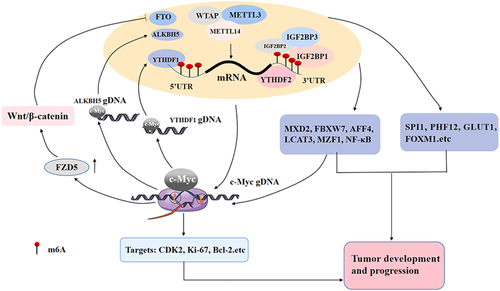
5 APPLICATION PROSPECT OF THE INTERACTION BETWEEN m6A MODIFICATION AND c-Myc REGULATION IN TUMOR DIAGNOSIS AND TREATMENT
Both m6A modification and c-Myc regulation are associated with the occurrence, development, and prognosis of tumors and are potential molecular markers for tumor diagnosis, which can be used for clinical tumor diagnosis and prognosis prediction. For example, the high expression of METTL3 is related to the early diagnosis and poor prognosis of GC, CRC, OSCC, and other tumor types (Xiang et al., 2020; D. D. Yang et al., 2020; W. Zhao et al., 2020). WTAP has a significant correlation with the worse prognosis of AML and ovarian cancer and is expected to be used as a marker in the evaluation of their prognosis (X. Han et al., 2020). At the same time, the m6A eraser FTO has been correlated with the prognosis of AML and other tumors (J. Chen & Du, 2019), and the m6A readers YTHDF2 and YTHDF1 have been significantly correlated with the prognosis of glioma (glioblastoma multiforme) and HCC patients (Fang et al., 2021; X. Liu et al., 2020). As an essential tumorigenic transcription factor, c-Myc has the characteristic of increased copy number and high expression in most tumors, which leads to changes in the expression of downstream-regulated target genes. More importantly, because of the mutual regulation between m6A modification and c-Myc, the combination of the two kinds of molecules will be more conducive to evaluating the early diagnosis and prognosis in tumors and provide important molecular markers and diagnostic strategies for the future diagnosis and prognosis evaluation of tumors.
As m6A modification and c-Myc regulation are important in tumorigenesis and tumor development, targeting key target molecules related to m6A modification and c-Myc regulation is expected to become a critical molecular strategy for tumor therapy. For example, the mTORC1 inhibitors torin1 and rapamycin can decrease the expression of WTAP, thus reducing the m6A modification on MXD2 mRNA, and inhibiting the promotion effect of c-Myc on cell proliferation and tumor growth, suggesting that targeting m6A modification regulators can provide novel treatment strategies for tumor patients (Cho et al., 2021). In GC, both FTO and MYC are highly expressed (Z. Yang et al., 2021). In some clinical related studies, Su et al. (2018) found that R-2-hydroxyglutarate can inhibit the activity of FTO, and then reduce the stability of c-Myc mRNA through m6A modification. This finding suggests that targeting the FTO/m6A/MYC axis can provide a novel and effective treatment strategy for the treatment of leukemia. In addition, c-Myc can bind to the YTHDF1 gene to promote the occurrence of CRC and decrease the sensitivity of YTHDF1 to fluorouracil and oxaliplatin (Nishizawa et al., 2018), suggesting that targeting YTHDF1 or using upstream c-Myc inhibitors of YTHDF1 to treat CRC patients is a good therapeutic strategy. In summary, m6A modification can be associated with c-Myc, and targeting the interaction loop between m6A modification and c-Myc provides a new method for clinical diagnosis and treatment, which has potential clinical application value and prospects.
6 CONCLUSION
Epigenetic modifications have become the focus in the cancer research field. Among them, m6A modification has received great attention in cancer and has gained clinical significance (Chen, Wang, et al., 2021). m6A modification can affect the transcription, stability, and translation efficiency of c-Myc through its regulators, namely writers, erasers, and readers, and thus plays a role in different types of tumors (Meyer & Jaffrey, 2014; Willbanks et al., 2021; Wood et al., 2021). We reviewed the specific effects of m6A regulators on c-Myc from different perspectives, including the direct or indirect mode of action and different modification sites on c-Myc mRNA. In addition, we also found that c-Myc can act on the erasers and readers of m6A and affect the modification of m6A to promote tumorigenesis by regulating downstream target genes and amplifying the action signal. Given the above, it can be concluded that m6A modification and c-Myc can form a double positive feedback loop and jointly drive the malignant process of tumors. To summarize, the interaction of m6A and c-Myc-related genes suggests that targeting m6A-c-Myc-related genes and their downstream regulatory targets will become an important molecular target for tumor diagnosis and treatment as well as provide new ideas for tumor diagnosis, treatment, and drug development in the future.
AUTHOR CONTRIBUTIONS
Ming Zhou, Lemei Zheng, Wei Xiong, and Guiyuan Li designed and conducted this study. Lemei Zheng, Mengna Li, and Jianxia Wei drafted the manuscript. Shipeng Chen, Changning Xue, Yuting Zhan, Yumei Duan, and Hongyu Deng revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
This study was supported by the grants from the National Natural Science Foundation of China (82172592), the Free Exploration Program of Central South University (Grant No. 2021zzts0934), the Program of Introducing Talents of Discipline to Universities (111-2-12), and the Open Sharing Found for the Large-scale Instrument and Equipment of Central South University (Grant No. CSUZC202040).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.




