miR-545 promoted enterovirus 71 replication via directly targeting phosphatase and tensin homolog and tumor necrosis factor receptor-associated factor 6
Abstract
Enterovirus 71 (EV71) is a small, nonenveloped icosahedral RNA virus and is the predominant causative pathogen of hand-foot-and-mouth disease. Recently, microRNAs (miRNAs) are reported to play important roles in the pathogenesis of EV71 replication. This study investigated the role of miR-545 in the EV71 replication and explored the underlying molecular mechanisms. We showed that miR-545 was upregulated in the EV71-infected human embryonic kidney (HEK) 293 cells and rhabdomyosarcoma (RD) cells. Overexpression of miR-545 promoted the viral replication of EV71 and attenuated the inhibitory effects of EV71 on cell viability in HEK293 and RD cells; while knockdown of miR-545 significantly suppressed the EV71 replication in these two cell lines. Bioinformatics analysis and luciferase reporter assay showed that miR-545 directly targeted the 3′untranslated region of phosphatase and tensin homolog (PTEN) and tumor necrosis factor receptor-associated factor 6 (TRAF6) in HEK293 cells. Furthermore, miR-545 negatively regulated the messenger RNA (mRNA) and protein expression of PTEN and TRAF6. The mRNA and protein expression of PTEN and TRAF6 was also suppressed by EV71 infection, which was attenuated by miR-545 knockdown in HEK293 cells. Overexpression of PTEN and TRAF6 both suppressed the EV71 replication in HKE293 cells, and also attenuated the enhanced effects of miR-545 overexpression on the EV71 replication in HEK293 cells. Collectively, our study for the first time showed that miR-545 had an enhanced effect on the EV71 replication in HEK293 and RD cells. Further mechanistic results indicated that miR-545 promoted EV71 replication at least partly via targeting PTEN and TRAF6.
1 INTRODUCTION
Enterovirus 71 (EV71) that belongs to the Picornaviridae family, is a small, nonenveloped, icosahedral RNA virus (Ayukekbong & Bergstrom, 2014). EV71 mainly causes typical hand-foot-and-mouth disease (HFMD) and often results in severe neurological disorders and high mortality in children (Ventarola, Bordone, & Silverberg, 2015). Up to date, EV71 has caused several outbreaks worldwide and seriously affected the Asia-Pacific region (Mao, Wang, Bian, Xu, & Liang, 2016). In China, more than 10 million children were reported to suffer from HFMD between 2010 and 2016 (Chan, Law, Hamblion, Fung, & Rudge, 2017), which was a serious public health problem. Recently, vaccination has been available in China, but the prevalence of this vaccination is still restricted (W. Wang et al., 2017). Up to date, antiviral drugs or effective therapies against EV71 infection have been developed, due to insufficient understanding of the molecular mechanisms underlying the pathogenesis of EV71 infection. In this regard, further investigations into the pathogenesis of EV71 infection may provide us with a novel strategy for managing the HFMD caused by EV71 infection.
microRNAs (miRNAs) are small, noncoding, single-stranded RNAs with 21–23 nucleotides in length, and miRNAs can repress messenger RNA (mRNA) translation and trigger mRNA degradation via targeting the 3′ untranslated region (3′UTR) of the targeted genes (Ho, Yang, & Yu, 2016; Pathinayake, Hsu, & Wark, 2015; J. Wu, Shen, Chen, Xu, & Mao, 2015). Accumulating evidence has shown that miRNAs functioned as important modulators in cellular, physiological, and pathological processes, including cell proliferation, apoptosis, differentiation, and metabolism (Ni & Leng, 2016). Recent studies also demonstrated that miRNAs may play important roles in EV71 viral entry and replication. For example, miR-548 was found to downregulate the host antiviral response of EV71 via directly targeting interferon-λ1 (Y. Li et al., 2013). Zheng et al. (2017) found that miR-16-5p mediated a positive feedback loop in EV71-induced apoptosis and suppressed virus replication of EV71. Downregulation if miR-526a by EV71 inhibited retinoic acid-inducible gene I-dependent innate immune response (Xu, He, et al., 2014). Clinically, serum miRNA expression was demonstrated to distinguish EV71 and Coxsackievirus 16 infections in patients (Cui, Qi, et al., 2011); elevated expression of circulating miR-876-5p was a specific response to severe EV71 infections (R. Y. Wang, Weng, Huang, & Chen, 2016). Recently, by using a deep sequencing approach, miR-545 was found to be upregulated in EV71-infected human epidermoid carcinoma cells (Cui, Guo, et al., 2010). Although miR-545 has been shown to involve in the development and progression of cancers (Changjun, Feizhou, Dezhen, Zhao, & Xianhai, 2018; Hu et al., 2018; Huang & Lu, 2017; Ma et al., 2018), the molecular mechanisms of miR-545-mediated the pathogenesis of EV71 infection has not been elucidated yet.
In the present study, we further explored the role of miR-545 in the pathogenesis of EV71 infection. We found that miR-545 was upregulated upon EV71 infection in both human embryonic kidney (HEK) 293 cells and rhabdomyosarcoma (RD) cells. Manipulation of miR-545 expression levels in HEK293 and RD cells affected the viral replication of EV71, and further mechanistic studies also revealed the downstream targets of miR-545. The present study may provide novel insights into understanding the role of miRNAs in the pathogenesis of EV71 infection.
2 MATERIALS AND METHODS
2.1 Cell culture
HEK293 cells and RD cells were purchased from the American Type Culture Collection company (ATCC, Manassas, VA). Cells were cultured in DMEM medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) at 37°C in a humidified 5% CO2 incubator.
2.2 miRNAs, plasmids, and cell transfections
The miR-545 mimics (5′-UCAGCAAACAUUUAUUGUGUGC-3′), miR-545 inhibitors (5′-GCACACAATAAATGTTTGCTGA-3′), and the respective negative controls (NCs) that is, mimics NC and inhibitors NC, were designed and synthesized by RiboBio (Guangzhou, China). The pcDNA3.1 plasmid, PTEN overexpressing plasmid (pcDNA3.1-PTEN), and TRAF6 overexpressing plasmid (pcDNA3.1-TRAF6) were purchased from GenePharma company (Shanghai, China). The cell transfections for miRNAs and plasmids were performed by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol, and at 24 hr after transfection, cells were collected for in vitro assays.
2.3 Virus infection and titration
EV71 GDV-103 was purchased from the China Center for Type Culture Collection (Wuhan, China) and was grown in HEK293 cells or RD cells for propagation. Briefly, the confluent cells were infected with the EV71 at different multiplicity of infection (MOI) and the viruses were collected from supernatants at indicated infection time points. EV71 virus titers were determined by the 50% tissue culture infectious dose (TCID50) assay using HEK293 cells or RD cells based on the Reed-Munch endpoint calculation method in previous studies (B. Wang, Zhang, Zhu, Luo, & Peng, 2012).
2.4 Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from cells was extracted by using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The extracted RNA was then reversely transcribed into cDNA PrimeScript RT Master Mix Kit (TaKaRa, Dalian, China). The RT-PCR reaction was performed by using SYBR Green Master Mix (TaKaRa) on an ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). The miR-545 expression level was normalized to U6 and the expression levels of the EV71 viral RNA, phosphatase and tensin homolog (PTEN) and tumor necrosis factor receptor-associated factor 6 (TRAF6) mRNA were normalized to glyceraldehyde-3-phosphate dehydrogenase. The data analysis was carried out by using method. The primers for qRT-PCR were shown in Table 1.
| Genes | Forward | Reverse |
|---|---|---|
| miR-545 | 5′-TCAGTAAATGTTTATTAGATGA-3′ | 5′-GTGCAGGGTCCGAGGTATTC-3′ |
| PTEN | 5′-CAAGATGATGTTTGAAACTAT-3′ | 5′-CCTTTAGCTGGCAGACCACAA-3′ |
| TRAF6 | 5′-AGGGACCCAGCTTTCTTTGT-3′ | 5′-GCCAAGTGATTCCTCTGCAT-3′ |
| U6 | 5′-CTCGCTTCGGCAGCACATATACT-3′ | 5′-ACGCTTCACGAATTTGCGTGTC-3′ |
| GAPDH | 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ | 5′-CATGTGGGCCATGAGGTCCACCAC-3′ |
- Note.GAPDH: glyceraldehyde-3-phosphate dehydrogenase; PTEN: phosphatase and tensin homolog; TRAF6: tumor necrosis factor receptor-associated factor 6.
2.5 Cell viability assessment
Cell viability of HEK293 and RD cells was evaluated by 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) assay. Briefly, after different treatments, cells were incubated with 20 µl MTT (5 mg/ml) for 4 hr at 37°C to allow viable cells to produce formazan. Then, after discarding the medium, 150 µl dimethyl sulfoxide was added for incubation with agitation for 10 min at room temperature. The cell viability was determined by measuring optical density at a wavelength of 570 nm.
2.6 Luciferase reporter assay
Luciferase reporters were generated based on the firefly luciferase expressing pGL3 vector (Promega, Madison, WI). To construct pGL3-PTEN 3′UTR and pGL3-TRAF6 3′UTR, a partial 3′UTR of the PTEN and TRAF6 mRNA containing the miR-545 binding sites was amplified and cloned into the vector pGL3-control. We also constructed luciferase reporters pGL3-PTEN 3′UTR-MUT and pGL3-TRAF6 3′UTR-MUT, which contains a putative miR-545 binding site with a mutant region of 3′UTR. For the luciferase reporter assay, HEK293 cells were seeded in 24-well plates at 5 × 104 cells/well, and 400 ng of luciferase reporter, 40 pmol miRNAs (miR-545 mimics, miR-545 inhibitors, mimics NC, or inhibitors NC) and 40 ng of pRL-TK were added in each well. Cells were collected 48 hr after transfection and luciferase activity was analysed by using the Dual-Luciferase Reporter Assay System (Promega).
2.7 Western blot assay
Total proteins were extracted from treated cells by using total protein extraction kit (KeyGen, Rockville, MD) according to the manufacturer's instruction. The protein concentrations of the lysates were measured by the Protein BCA Assay Kit (Bio-Rad, Hercules, CA). For the western blot assay, proteins (30 µg) were separated with 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by transferring to a nitrocellulose membrane. The membrane was blocked with 5% skimmed milk in Tris-buffered saline with Tween 20 for 1 hr at room temperature. The membrane was then incubated at 4°C overnight with primary antibodies against EV71 VP1 (1:1,000; Abcam, Cambridge, UK), PTEN (1:1,000; Abcam), TRAF6 (1:1,000; Abcam), and β-actin (1:1,500; Abcam) followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 hr at room temperature. Enhanced chemiluminescence western blot analysis detection reagents (New England Biolabs, Ipswich, MA) were used to visualize the target proteins, which were quantified with a BioImage Intelligent Quantifier 1-D (Version 2.2.1; Nihon-BioImage Ltd., Tokyo, Japan).
2.8 Statistical analysis
The statistical analysis was performed by using the GraphPad Prism Software Version 5.0 (GraphPad Software, La Jolla, CA). All data in the study were represented as mean ± standard deviation from at least three independent experiments. Statistical comparisons were made by unpaired Student's t test (for two-group comparisons) or one-way analysis of variance (for multiple group comparisons). p < 0.05 was considered statistically significant.
3 RESULTS
3.1 Effects of EV71 infection on miR-545 expression in HEK293 cells and RD cells
First, HEK293 cells and RD cells were infected with EV71 at a MOI of 1 for 6, 12, and 24 hr, respectively, and qRT-PCR was performed to determine the miR-545 expression in the EV71-infected cells. As shown in Figure 1a,b, EV71 infection significantly increased the expression level of miR-545 in both HEK293 and RD cells, and the effects were in a time-dependent manner (Figure 1a,b). Furthermore, the effects of different MOIs of EV71 infection on the miR-545 expression were also determined by qRT-PCR, and EV71 at an MOI of 0.5 and 1 significantly upregulated the expression of miR-545 in HEK293 cells and RD cells, while EV71 infection at an MOI of 0.1 had no significant effects on the miR-545 expression in HEK293 cells and RD cells (Figure 1c,d). Based on the above results, EV71 at an MOI of 1 infected cells for 24 hr were selected for further mechanistic in vitro studies.
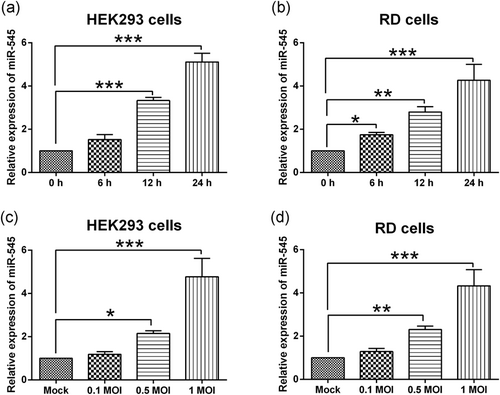
Effects of EV71 infection on miR-545 expression in HEK293 and RD cells. The relative expression of miR-545 was determined by qRT-PCR in (a) HEK293 and (b) RD cells after being infected with EV71 at an MOI of 1 for 6, 12, and 24 hr, respectively. The relative expression of miR-545 was determined by qRT-PCR in (c) HEK293 and (d) RD cells after being infected with EV71 at an MOI of 0.1, 0.5, and 1, respectively, for 24 hr, respectively. N = 3. Significant differences between groups were shown as *p < 0.05, **p < 0.01, and ***p < 0.001. EV71: enterovirus 71; HEK293: human embryonic kidney 293; MOI: multiplicity of infection; qRT-PCR: quantitative real-time polymerase chain reaction; RD: rhabdomyosarcoma
3.2 Effects of miR-545 overexpression on the EV71 replication
To determine the effects of miR-545 overexpression on the EV71 replication, HEK293, and RD cells were transiently transfected with miR-545 mimics or mimics NC, and at 24 hr after transfection, qRT-PCR results showed that miR-545 mimics transfection significantly increased the expression of miR-545 in HEK293 and RD cells by more than 10-folds (Figure 2a,b). Furthermore, at 24 hr after being transfected with mimics NC or miR-545 mimics, HEK293 and RD cells were infected with EV71 at an MOI of 1 for 2, 6, 12, and 24 hr, respectively, and the virus titration results showed that overexpression of miR-545 significantly increased the EV71 replication when compared with mimics NC group (Figure 2c,d). The qRT-PCR results showed that the EV71 copy number as revealed by the viral mRNA expression was significantly increased in the infected HEK293 and RD cells (Figure 2e,f). Furthermore, we examined the viral protein expression of VP1 that is the major components of EV71 capsid (Foo et al., 2008). Consistently, overexpression of miR-545 also increased the VP1 protein expression in the infected HEK293 and RD cells when compared with mimics NC group (Figure 2g,h). These results suggested that miR-545 overexpression promoted EV71 replication in both HEK293 and RD cells. In addition, EV71 infection also suppressed the cell viability as determined by MTT assay in both HEK293 and RD cells and the inhibitory effects of EV71 on cell viability were partially reversed by miR-545 overexpression (Figure S1).
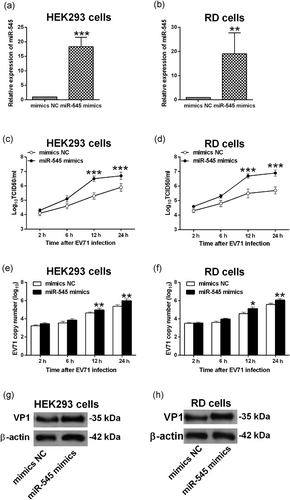
Effects of miR-545 overexpression on the EV71 replication. The relative expression of miR-545 was determined by qRT-PCR in (a) HEK293 and (b) RD cells at 24 hr after being transfected with mimics NC or miR-545 mimics. (c−h) HEK293 cells and RD cells were transfected with mimics NC or miR-545 mimics and at 24 hr after transfection, cells were infected by EV71 at an multiplicity of infection (MOI) of 1 for 2, 6, 12, and 24 hr, respectively. Virus titers in culture supernatants from (c) HEK293 and (d) RD cells were measured at the indicated time postinfection. The EV71 viral RNA levels in (e) HEK293 and (f) RD cells were determined by qRT-PCR. The protein expression of VP1 in (g) HEK293 and (h) RD cells were determined by western blot assay. N = 3. **p < 0.01 and ***p < 0.001. EV71: enterovirus 71; HEK293: human embryonic kidney 293; NC: negative control; qRT-PCR: quantitative real-time polymerase chain reaction; RD: rhabdomyosarcoma
3.3 Effects of miR-545 knockdown on the EV71 replication
The knockdown of miR-545 in HEK293 and RD cells were achieved via transfecting cells with miR-545 inhibitors, and at 24 hr after transfection, the expression of miR-545 was markedly suppressed in HEK293 and RD cells transfected with miR-545 inhibitors when compared with inhibitors NC group (Figure 3a,b). At 24 hr after being transfected with inhibitors NC or miR-545 inhibitors, HEK293 and RD cells were infected with EV71 at an MOI of 1 for 2, 6, 12, and 24 hr, respectively, and the viral replication of EV71 was significantly suppressed in HEK293 and RD cells with miR-545 knockdown (Figure 3c,d). In addition, qRT-PCR results showed that knockdown of miR-545 significantly suppressed the EV71 copy number in the infected HEK293 and RD cells (Figure 3e,f). The western blot assay further showed that the protein expression of VP1 in the infected HEK293 and RD cells with miR-545 knockdown was significantly downregulated when compared with inhibitors NC group (Figure 3g,h). Collectively, the results indicated that knockdown of miR-545 suppressed the viral replication of EV71 in both HEK293 and RD cells.
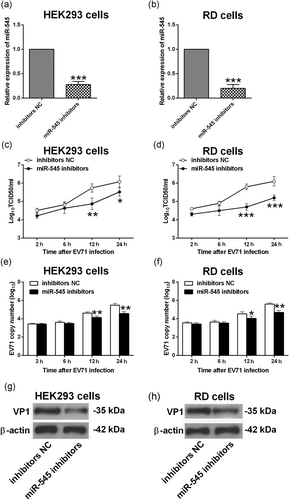
Effects of miR-545 knockdown on the EV71 replication. The relative expression of miR-545 was determined by qRT-PCR in (a) HEK293 and (b) RD cells at 24 hr after being transfected with inhibitors NC or miR-545 inhibitors. (c−h) HEK293 cells and RD cells were transfected with inhibitors NC or miR-545 inhibitors and at 24 hr after transfection, cells were infected by EV71 at an multiplicity of infection (MOI) of 1 for 2, 6, 12, and 24 hr, respectively. Virus titers in culture supernatants from (c) HEK293 and (d) RD cells were measured at the indicated time postinfection. The EV71 viral RNA levels in (e) HEK293 and (f) RD cells were determined by qRT-PCR. The protein expression of VP1 in (g) HEK293 and (h) RD cells were determined by western blot assay. N = 3. *p < 0.05, **p < 0.01, and ***p < 0.001. EV71: enterovirus 71; HEK293: human embryonic kidney 293; NC: negative control; qRT-PCR: quantitative real-time polymerase chain reaction; RD: rhabdomyosarcoma; TCID50: 50% tissue culture infectious dose
3.4 PTEN was a direct target of miR-545
To further explore the molecular mechanisms underlying miR-545-mediated EV71 replication, we performed the bioinformatics analysis using the online TargetScan Software (www.targetscan.org), and among the predicted downstream targets, PTEN was found to be one of the potential targets of miR-545. As shown in Figure 4a, the 3′UTR of PTEN (position 2747–2753) harbors the complementary sequences with miR-545 (highlighted in light green color). To confirm the interaction between 3′UTR of PTEN and miR-545, we performed the luciferase reporter assay by constructing reporter vector containing the wild-type 3′UTR of PTEN and the mutant 3′UTR of PTEN (the mutant sites were highlighted in red letter in Figure 4a). At 48 hr after cotransfection with pGL3-PTEN 3′UTR-WT or pGL3-PTEN 3′UTR-MUT and different miRNAs, the relative luciferase activity was determined by Dual-Luciferase Reporter system. As shown in Figure 4b, miR-545 mimics transfection significantly suppressed the luciferase activity of pGL3-PTEN 3′UTR-WT when compared with mimics NC group (Figure 4b); while miR-545 inhibitors transfection significantly increased the luciferase activity of pGL3-PTEN 3′UTR-WT when compared with inhibitors NC group (Figure 4b). In contrast, both miR-545 overexpression and miR-545 knockdown failed to affect the luciferase activity of pGL3-PTEN 3′UTR-MUT (Figure 4c). Furthermore, overexpression of miR-545 suppressed the mRNA and protein expression of PTEN when compared with mimics NC group; while knockdown of miR-545 significantly increased the mRNA and protein expression of PTEN when compared with inhibitors NC group (Figure 4d,e). More important, EV71 infection significantly suppressed the mRNA and protein expression of PTEN, and knockdown of miR-545 attenuated the inhibitory effects of EV71 infection on PTEN expression in HEK239 cells (Figure 4f,g).
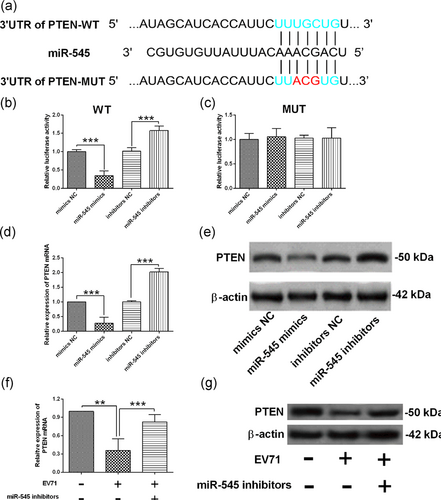
PTEN was a direct target of miR-545 in HEK293 cells. (a) miR-545 binding sites on PTEN 3′UTR predicted by Targetscan. The mutant sites were highlighted in red letters. The relative luciferase activity was determined by luciferase reporter assay in HEK293 cells at 48 hr after being cotransfected with (b) pGL3-PTEN 3′UTR-WT or (c) pGL3-PTEN 3′UTR-MUT and different microRNAs (mimics NC, miR-545 mimics, inhibitors NC or miR-545 inhibitors). The relative expression of (d) PTEN mRNA and (e) PTEN protein was determined by qRT-PCR and western blot assay in HEK293 cells at 24 hr after being transfected with mimics NC, miR-545 mimics, inhibitors NC, or miR-545 inhibitors. (f,g) The HEK293 cells were transfected with inhibitors NC or miR-545 inhibitors and at 24 hr after transfection, cells were infected by EV71 at an MOI of 1 for 24 hr, and the relative expression of (f) PTEN mRNA and (g) PTEN protein was determined by qRT-PCR and western blot assay. N = 3. **p < 0.01 and ***p < 0.001. EV71: enterovirus 71; HEK293: human embryonic kidney 293; MOI: multiplicity of infection; mRNA: messenger RNA; MUT: mutant; NC: negative control; PTEN: phosphatase and tensin homolog; qRT-PCR: quantitative real-time polymerase chain reaction; WT: wild-type [Color figure can be viewed at wileyonlinelibrary.com]
3.5 miR-545 regulated EV71 replication via targeting PTEN in HEK293 cells
To further determine the role of PTEN-mediated EV71 replication, we transiently transfected the HEK293 cells with pcDNA3.1-PTEN, and pcDNA3.1-PTEN transfection significantly increased the mRNA and protein expression levels of PTEN in HEK293 cells when compared with vector NC group (Figure 5a,b). Furthermore, at 24 hr after cotransfection with mimics NC + vector NC, miR-545 mimics + vector NC, mimics NC + PTEN, or miR-545 mimics + PTEN, HEK293 cells were infected with EV71 at an MOI of 1 for 24 hr, the viral titration, qRT-PCR analysis of viral RNA and western blot analysis of VP1 protein results showed that overexpression of PTEN suppressed the EV71 replication in HEK293 cells and also attenuated the enhanced effects of miR-545 overexpression on EV71 replication (Figure 5c−e).
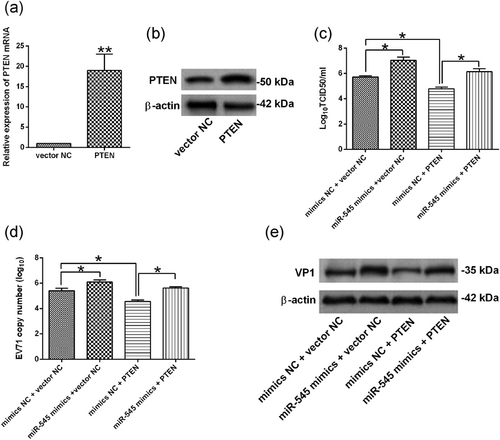
miR-545 regulated EV71 replication via targeting PTEN in HEK293 cells. (a−b) The relative expression of (a) PTEN mRNA and (b) PTEN protein was determined by qRT-PCR and western blot assay in HEK293 cells at 24 hr after being transfection with pcDNA3.1 (vector NC) or pcDNA3.1-PTEN (PTEN). (c−e) HKE293 cells were cotransfected with mimics NC + pcDNA3.1 (vector NC), miR-545 mimics + vector NC, mimics NC + pcDNA3.1-PTEN (PTEN), or miR-545 mimics + pcDNA3.1-PTEN (PTEN), at 24 hr after transfection, cells were infected by EV71 at an MOI of 1 for 24 hr, (c) virus titers in culture supernatants from HEK293 were measured. (d) The EV71 viral RNA levels in HEK293 cells were determined by qRT-PCR. (e) The protein expression of VP1 in HEK293 cells were determined by western blot analysis assay. N = 3. *p < 0.05 and **p < 0.01. EV71: enterovirus 71; HEK293: human embryonic kidney 293; MOI: multiplicity of infection; mRNA: messenger RNA; NC: negative control; PTEN: phosphatase and tensin homolog; qRT-PCR: quantitative real-time polymerase chain reaction; TCID50: 50% tissue culture infectious dose
3.6 TRAF6 was another direct target of miR-545
Apart from the interaction between miR-545 and PTEN, the TRAF6 was also among the predicted targets of miR-545. As shown in Figure 6a, the 3′UTR of TRAF6 (Position 2301–2308) also harbors the complementary sequences with miR-545 (highlighted in light green color). To confirm the interaction between 3′UTR of TRAF6 and miR-545, we also constructed the reporter vector containing the wild-type 3′UTR of TRAF6 and the mutant 3′UTR of TRAF6 (the mutant sites were highlighted in red letter in Figure 6a). At 48 hr after cotransfection with pGL3-TRAF6 3′UTR-WT or pGL3-TRAF6 3′UTR-MUT and different miRNAs, the relative luciferase activity was determined by Dual-Luciferase Reporter system. Overexpression of miR-545 significantly suppressed the luciferase activity of pGL3-TRAF6 3′UTR-WT (Figure 5b); while knockdown of miR-545 significantly increased the luciferase activity of pGL3-TRAF6 3′UTR-WT (Figure 6b). In contrast, the luciferase activity of pGL3-TRAF6 3′UTR-MUT was not affected by either miR-545 overexpression or miR-545 knockdown (Figure 6c). Furthermore, overexpression of miR-545 suppressed the mRNA and protein expression of TRAF6; while knockdown of miR-545 significantly increased the mRNA and protein expression of TRAF6 (Figure 6d,e). Consistently, the expression of TRAF6 was downregulated in EV71-infected HEK293 cells, and the presence of miR-545 inhibitors partially reversed the inhibitory effects of EV71 infection on TRAF6 expression in HEK293 cells (Figure 6f,g).

TRAF6 was another direct target of miR-545 in HEK293 cells. (a) miR-545 binding sites on TRAF6 3′UTR predicted by Targetscan. The mutant sites were highlighted in red letters. The relative luciferase activity was determined by luciferase reporter assay in HEK293 cells at 48 hr after being cotransfected with (b) pGL3-TRAF6 3′UTR-WT or (c) pGL3-TRAF6 3′UTR-MUT and different microRNAs (mimics NC, miR-545 mimics, inhibitors NC or miR-545 inhibitors). The relative expression of (d) TRAF6 mRNA and (e) TRAF6 protein was determined by qRT-PCR western blot assay in HEK293 cells at 24 hr after being transfected with mimics NC, miR-545 mimics, inhibitors NC, or miR-545 inhibitors. (f,g) The HEK293 cells were transfected with inhibitors NC or miR-545 inhibitors and at 24 hr after transfection, cells were infected by EV71 at an MOI of 1 for 24 hr, and the relative expression of (f) TRAF6 mRNA and (g) TRAF6 protein was determined by qRT-PCR and western blot assay. N = 3. **p < 0.01 and ***p < 0.001. EV71: enterovirus 71; HEK293: human embryonic kidney 293; MOI: multiplicity of infection; mRNA: messenger RNA; NC: negative control; qRT-PCR: quantitative real-time polymerase chain reaction; TRAF6: tumor necrosis factor receptor-associated factor 6 [Color figure can be viewed at wileyonlinelibrary.com]
3.7 miR-545 regulated EV71 replication via targeting PTEN in HEK293 cells
To further determine the role of TRAF6-mediated EV71 replication, we also transiently transfected the HEK293 cells with pcDNA3.1-TRAF6, and pcDNA3.1-TRAF6 transfection significantly increased the mRNA and protein expression levels of TRAF6 in HEK293 cells when compared with vector NC group (Figure 7a,b). As expected, at 24 hr after cotransfection with mimics NC + vector NC, miR-545 mimics + vector NC, mimics NC + TRAF6, or miR-545 mimics + TRAF6, HEK293 cells were infected with EV71 at an MOI of 1 for 24 hr, and the in vitro functional assays showed that overexpression of TRAF6 suppressed the EV71 replication in HEK293 cells and also attenuated the enhanced effects of miR-545 overexpression on the EV71 replication (Figure 7c−e).
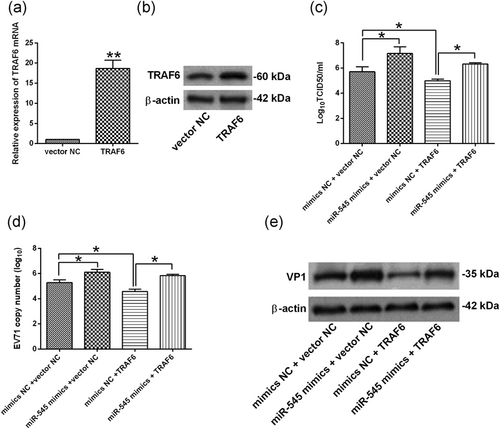
miR-545 regulated EV71 replication via targeting TRAF6 in HEK293 cells. (a,b) The relative expression of (a) TRAF6 mRNA and (b) TRAF6 protein was determined by qRT-PCR and western blot assay in HEK293 cells at 24 hr after being transfection with pcDNA3.1 (vector NC) or pcDNA3.1-TRAF6 (TRAF6). (c−e) HKE293 cells were cotransfected with mimics NC + pcDNA3.1 (vector NC), miR-545 mimics + vector NC, mimics NC + pcDNA3.1-TRAF6 (TRAF6), or miR-545 mimics + pcDNA3.1-TRAF6 (TRAF6), at 24 hr after transfection, cells were infected by EV71 at an MOI of 1 for 24 hr, (c) virus titers in culture supernatants from HEK293 were measured; (d) the EV71 viral RNA levels in HEK293 cells were determined by qRT-PCR; (e) the protein expression of VP1 in HEK293 cells were determined by western blot assay. N = 3. *p < 0.05 and **p < 0.01. EV71: enterovirus 71; HEK293: human embryonic kidney 293; miR: microRNA; MOI: multiplicity of infection; NC: negative control; qRT-PCR: quantitative real-time polymerase chain reaction; TRAF6: tumor necrosis factor receptor-associated factor 6
4 DISCUSSION
In the past few years, the morbidity and mortality caused by EV71 infection have tended to increase (Lee, 2016; Yi, Shin, Kim, Kim, & Chang, 2017). Unfortunately, while the underlying molecular mechanisms underlying the pathogenesis of EV71 infection remain unclear, the effective therapies have not been developed. Recently, miRNAs were found to play important roles in the viral replication of EV71. In the present study, we found that upregulation of miR-545 was induced by EV71 infection in HEK293 and RD cells, where these two lines have been commonly used as an in vitro cell model for EV71 infection (He et al., 2018; B. Wang, Zhang, & Zhao, 2013; Yue et al., 2017). Overexpression of miR-545 enhanced the viral replication of EV71 in HEK293 and RD cells, while knockdown of miR-545 suppressed the viral replication of EV71 in the two cell lines. Further mechanistic investigations showed that miR-545 negatively regulated the expression of PTEN and TRAF6 by targeting the 3′UTR of these genes, and overexpression of both PTEN and TRAF6 attenuated the enhanced effects of miR-545 overexpression on the viral replication of EV71. Collectively, our study for the first time demonstrated the role of miR-545 in the viral replication, and this effect may be mediated via regulating PTEN and TRAF6.
The role of miRNAs in the viral replication of EV71 has been elucidated in various studies. miRNAs such as miR-146a (Fu et al., 2017), miR-16-5p (Zheng et al., 2017), miR-124 (Chang, Wang, Bian, Liu, & Long, 2017), miR-27a (L. Zhang et al., 2014), and miR-21 (Feng et al., 2017) were reported to play important roles in regulating the EV71 replication, suggesting the involvement of miRNAs in the pathogenesis of EV71 infection. Recently, by using a deep sequencing approach, a large number of differentially expressed miRNAs was found in the EV71-infected human epidermoid carcinoma cells (Cui, Guo, et al., 2010). Among these miRNAs, we further investigated the role of miR-545 in the pathogenesis of EV71 infection. Based on the literature, miR-545 exerted inhibitory effects on the lung cancer cell proliferation by targeting cyclin D1 (Du et al., 2014). In addition, miR-545 could upregulate the epidermal growth factor receptor expression to mediate colorectal cancer cell proliferation (Huang & Lu, 2017). miR-545 also contributed to aberrant cell proliferation, invasion, and migration in hepatocellular carcinoma via targeting metallothionein 1M (Changjun et al., 2018). In our study, EV71-induced the upregulation of miR-545 in HEK293 and RD cells, and overexpression of miR-545 promoted EV71 replication attenuated the inhibitory effects of EV71 on cell viability of HEK293 and RD cells, while knockdown of miR-545 had the inhibitory effects, indicating that miR-545 promoted the viral replication of EV71 in these cell lines.
To further explore the molecular mechanisms underlying miR-545-mediated effects on EV71 replication, we then performed the bioinformatics analysis and we found that PTEN and TRAF6 were among the predicted targets of miR-545. These two targets were selected for subsequent studies, as they have been shown to participate in the process of EV71 infection (W. Wang et al., 2017; Zhao et al., 2018). The interaction between miR-545 and PTEN/TRAF6 was further confirmed by the luciferase report assay. PTEN is one of the most frequently altered tumor suppressor genes in cancer, and the tumor suppressive role of PTEN has been reported in various types of cancers (A. Li, Qiu, Zhou, Wang, & Guo, 2017; Ortega-Molina & Serrano, 2013). Recently, the role of PTEN has also been demonstrated in viral replication. Q. Wu et al. (2017) showed that PTEN decreased hepatitis C virus replication and the protein phosphatase activity of PTEN is essential for this function; Kong et al. (2011) showed that upregulated miR-29a by hepatitis B virus X protein enhanced hepatoma cell migration by targeting PTEN in cell culture model; Berndardt et al. (2016) also identified a viral miRNA cluster that regulates the expression of PTEN and p27. More important, miR-494-3p was found to promote EV71 replication by directly targeting PTEN (Zhao et al., 2018). In our study, we showed that miR-545 negatively regulated the expression of PTEN in HEK293 and RD cells, and overexpression of PTEN suppressed EV71 replication and attenuated the effects of miR-545 overexpression on EV71 replication in HEK293 and RD cells, suggesting that miR-545 exerted its enhanced effects on EV71 replication partly via targeting PTEN. Studies also showed that PTEN was closely related to the PI3K/Akt signaling pathways and EV71 infection-induced activation of PI3K/Akt signaling pathway in RD and HEK293 cells (Zhao et al., 2018). Whether miR-545 affects the PI3K/Akt signaling pathways during the EV71 infection may require further investigations.
TRAF6 belongs to the TRAF family and is the key signaling protein in the NF-κB signaling pathways (Liu et al., 2012). TRAF6 could form a homodimer and catalyze K63-linked unbiquitination, which leads to the production of proinflammatory cytokines (Xu, Peng, et al., 2018). In addition, the ubiquitin-specific protease 4 could positively regulate retinoic acid-inducible gene I-like receptor-induced NF-κB activation by targeting TRAF6 for K48-linked deubiquitination and inhibit EV71 replication (Xu, Peng, et al., 2018). Ho et al. showed that inhibition of miR-146a prevented EV71-induced cell death by directly targeting TRAF6 and IRAK1 (Fu et al., 2017). The global quantitative proteomic analysis of human glioma cells in response to EV71 infection identified TRAF2 and TRAF6 as two important differentially expressed proteins in the host cells (L. K. Zhang, Lin, Zhu, Xianyu, & Lu, 2015). TRAF6 was found to activate the NF-κB signaling pathway upon sensing cytosolic viral RNA and DNA (Konno et al., 2009), and NF-κB signaling pathway also played an important role in the EV71 replication (Jin, Zhang, Wu, & Duan, 2018). In the present study, we showed that miR-545 negatively regulated the expression of TRAF6 in HEK293 and RD cells, and overexpression of TRAF6 suppressed EV71 replication and attenuated the effects of miR-545 overexpression on EV71 replication in HEK293 cells, suggesting that miR-545 exerted its enhanced effects on EV71 replication also partly via targeting TRAF6.
Some limitations should be considered in this study. The present study only performed in vitro functional assays, and in vivo future studies may be needed to confirm the role of miR-545 in EV71 infection. The examination of miR-545 in EV71-infected patients may be performed to further elucidate the clinical significance of miR-454 in EV71 infection. As miR-545 targeted multiple genes, other genes targeted by miR-545 should take into consideration, and we should be cautious when interpreting our current findings.
In summary, our study for the first time showed that miR-545 had an enhanced effect on the EV71 replication in HEK293 and RD cells. Further mechanistic results indicated that miR-545 promoted EV71 replication at least partly via targeting PTEN and TRAF6 and may provide a clue to develop preventative and effective therapies for EV71 infections.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (Grant No: 81803863), China Postdoctoral Science Foundation (Grant No: 2017M622352), Henan Province Postdoctoral Science Foundation (Grant No: 00104341), The Key Program of Henan Colleges and Universities (Grant No: 16A310019) and the Doctoral Scientific Fund Project of Henan University of Chinese Medicine (Grant No: BSJJ2015-08).
CONFLICT OF INTERESTS
The authors declare that there are no conflicts of interest.