Low expression of CRISP3 predicts a favorable prognosis in patients with mammary carcinoma
Abstract
The discovery of cysteine-rich secretory protein 3 (CRISP3) has been made in human neutrophils for the first time. Cloning of the complementary DNA (cDNA) for CRISP3 was performed from a cDNA library of human bone marrow. In patients with mammary carcinoma, we found that lower expression of CRISP3 was connected to a significantly improved DFS (disease-free survival) and OS (overall survival). Furthermore, the CRISP3 protein level was significantly associated with negative ANXA1 protein level. In addition, the heterogeneous expression of CRISP3 had been exhibited in diverse mammary carcinoma cells. A significant higher mRNA and the protein level of CRISP3 were seen in T-47D as well as SK-BR-3 cells compared with those in other types of mammary carcinoma cells. Knockdown of CRISP3 in T-47D or SK-BR-3 cells resulted in the weakened migration or invasion abilities. Furthermore, CRISP3 knockdown significantly inhibited the ERK1/2 MAPK signaling pathway in T-47D or SK-BR-3 cells. Research results indicated that the lowering in the expression of CRISP3 is likely to serve as an unprecedented approach for the treatment of mammary carcinoma.
1 INTRODUCTION
In women, the three most common types of cancer are mammary carcinoma, pulmonary carcinoma, and colorectal carcinoma, which together account for 50% of all cancer cases. Mammary carcinoma alone accounts for 30% of new cancer cases in women. Among women from ages 20 to 59 years, the leading cause of cancer death is mammary carcinoma (Oeffinger et al., 2015; Siegel, Miller, & Jemal, 2018). Most patients with mammary carcinoma die from the constant spread of the metastatic tumor cells and the gradual formation of distant secondary tumors (Fan et al., 2018bb). The invasion of mammary carcinoma cell is mainly due to its regulation by many kinds of signaling pathways, like mitogen-activated protein kinase (MAPK) pathways (Meng et al., 2013). Routine MAPKs comprise extracellular signal-regulated kinase 1 and 2 (ERK1/2), c-Jun N-terminal kinases/stress-activated protein kinases (JNK/SAPK), p38 MAPK, and ERK5 (Fan et al., 2015aa; Wang et al., 2017). The activation of ERK1/2 MAPK pathways plays an essential part in invasion and metastasis of mammary carcinoma cells (Qin et al., 2017; Zanotto-Filho et al., 2018; Zuo, Wu, & Chakraborty, 2012).
Cysteine-rich secretory protein 3 (CRISP3) contains a hinge region, antigen 5, and pathogenesis-related 1 proteins domain, an N-terminal CRISP as well as a C-terminal ion channel regulator domain (Dhaulakhandi, Kumawat, Thakran, & Parshad, 2017). According to research, we can detect high levels of CRISP3 messenger RNA (mRNA) in human pancreas, salivary glands, and prostate, and we can also detect CRISP3 protein in human body fluids, like blood and saliva (Bjartell et al., 2007). CRISP3 possesses an inhibitory effect upon hepatitis C virus during the early infection (Lee et al., 2014). Furthermore, CRISP3 is likely to participate in the carcinogenesis of oral squamous cell carcinoma (Ko et al., 2012) or myeloma (Leng, Miao, Huang, & Wang, 2018). A relatively high-level expression of CRISP3 is in relation with undesirable prognosis and therefore it is considered a prognostic marker for prostate cancer (Al Bashir et al., 2014; Bjartell et al., 2006; Dahlman et al., 2010; Grupp et al., 2013; Pathak, Breed, Apte, Acharya, & Mahale, 2016; Ribeiro et al., 2011). Low-level PTEN expression together with high-level CRISP3 expression is apparently featured with the subgroup of prostate carcinomas possessing undesirable prognosis for biochemical recurrence (Noh, Sung, Kim, Chang, & Park, 2016). In prostate cancer cells, CRISP3 expression is related to the level of androgen, whereas, in androgen receptor positive cells, CRISP3 promoter is subject to the epigenetic regulation of androgen receptor (Pathak, Breed, Deshmukh, & Mahale, 2018). CRISP3 was determined to be a prognostic factor for ovarian cancer, with decreased expression of CRISP3 predicting poor prognosis (Gao et al., 2017).
In this study, we found obvious differences in the expression of CRISP3 between different mammary carcinoma cells, which is related to the prognosis of patients with mammary carcinoma, and further explored the role of CRISP3 involved in migration and invasion.
2 MATERIALS AND METHODS
2.1 Clinical specimens and immunohistochemical staining
Altogether 220 female patients confirmedly diagnosed with mammary carcinoma underwent treatment in Xuzhou No. 1 People's Hospital and Xuzhou No. 2 People's Hospital. With the acquisition of ethical permission from the ethic committees of a local hospital, the informed consents were also attained from all the patients before the collection of specimens. Immunohistochemistry was performed for CRISP3 as previously described (Fan et al., 2013; Fan et al., 2018aa, 2018bb). Upon nuclear counterstaining with hematoxylin, classification of cytoplasmic immunostaining intensity was performed in a semi-quantitative manner (Fan et al., 2013; Fan et al., 2018aa, 2018bb).
2.2 Cell lines
T-47D (Cat# HTB-133, RRID: CVCL_0553), SK-BR-3 (Cat# HTB-30, RRID: CVCL_0033), MCF7 (Cat# HTB-22, RRID: CVCL_0031), MDA-MB-468 (Cat# HTB-132, RRID: CVCL_0419), and MDA-MB-453 (Cat# HTB-131, RRID: CVCL_0418) were bought from the American Type Culture Collection (ATCC, Manassas, VA).
2.3 Interference sequence
The specific sequences are as follows: Small interfering RNA (siRNA)-control, 5′-UUCUCCG AACGUGUCACGUTT-3′ (forward), 5′-ACGUGACACGUUCGGAGAATT-3′ (reverse); siRNA1-CRISP3, 5′-AGUAACCCAAAGGAUCGAAdTdT-3′ (forward), 5′-UUCGAUCCUUUGGGUUACUdTdT-3′ (reverse); siRNA2-CRISP3, 5′-CAGAUAACUGUGACGAUGGdTdT-3′ (forward), 5′-CCAUCGUCACAGUUAU CUGdTdT-3′ (reverse); siRNA3-CRISP3, 5′-GUUGGAUGUGGAAAUGCCUdT dT-3′ (forward), 5′-AGGCAUUUCCACAUCCAACdTdT-3′ (reverse); siRNA4-CRISP3, 5′-AAGCUCACAUUAACCUGUAdTdT-3′ (forward), 5′-UACAGGUUAAUGUGAGCUUdTdT-3′ (reverse). siRNA1-ANXA1, 5′-AUAAGGCCAUAAUGGUUAAdTdT-3′ (forward), 5′-UUAACCAUUAUGGCCUUAUdTdT-3′ (reverse). siRNA2-ANXA1, 5′-CCUUGUAUGAAGCAGGAGAdTdT-3′ (forward), 5′-UCUCCUGCUUCAUACAAGGdTdT-3′ (reverse). siRNA3-ANXA1, 5′-CAUUCUAUCAGAAGAUGUAdTdT-3′ (forward), 5′-UACAUCUUCUGAUAGAAUGdTdT-3′ (reverse). siRNA4-ANXA1, 5′-GUGUUGGAACUCGCCAUAAdTdT-3′ (forward), 5′-UUAUGGCGAGUUCCAACACdTdT-3′ (reverse).
2.4 Quantitative real-time PCR
Based upon the StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA), the SYBR Green I assay (Takara, Dalian, China) was used to make a quantitative and real-time polymerase chain reaction (PCR). The CRISP3 primers were: 5′-GCACCTTCCTTCTGTCA-3′ (Forward), 5′-CAGCCTCTTTGTTCCATT-3′ (Reverse).
2.5 RTCA migration and invasion assays
Cellular migration, as well as invasion, was monitored by utilizing an xCELLigence Real-Time Cell Analysis (RTCA) Instrument (Roche Applied Science, Indianapolis, IN; Fan et al., 2015aa, 2015b). All the cells were seeded to the upper chamber of the CIM-plate16 (Roche Applied Science, Indianapolis, IN). Electrical impedance refers to a dimensionless parameter, which is also known as cell index. Changes can be measured on the bottom of a membrane separating the upper and lower chambers to assess cellular migration. The RTCA invasion assay was performed in a similar manner, excepting for covering the upper chamber with Matrigel (Catalog Number: 356234, BD Biosciences, MA). The detection of cell index values was performed every 15 min for 24 hr or 48 hr in cell migration or invasion assays, respectively.
2.6 Western blot analysis
Assays were performed as described previously by us (Fan et al., 2018aa). The specific antibodies are as follows: CRISP3 (Sigma-Aldrich, St. Louis, MO, Cat# HPA054392), ANXA1 (Sigma-Aldrich, Cat# HPA011271, RRID: AB_1844883), p-ERK1/2 (Cell Signaling Technology, Inc., Danvers, MA, Cat# 4370, RRID: AB_2315112), p-p90RSK (Ser380; Cell Signaling Technology, Inc., Cat# 11989, RRID: AB_2687613), p-Elk-1 (Cell Signaling Technology, Inc., Cat# 9181, RRID: AB_2099016), ERK1/2 (Cell Signaling Technology, Inc., Cat# 4695, RRID: AB_390779), p90RSK (Cell Signaling Technology, Inc., Cat# 8408, RRID: AB_10828594), Elk-1 (Cell Signaling Technology, Inc., Cat# 9182, RRID: AB_10695670), and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, Cat# sc-517582).
2.7 Statistical analysis
With one-way analysis of variance (ANOVA) and Bonferroni post test (upon necessity), an examination of discrepancies between groups was performed for statistical significance. All the p-values under 0.05 were deemed to possess statistical significance. The data are presented as mean ± standard error of mean (SEM).
3 RESULTS
3.1 Expression analysis of CRISP3 in mammary carcinoma tissues
An examination was performed on the relevance of CRISP3 expression status with tumor progression in patients with mammary carcinoma, which is intended to give an elucidation of the effect of CRISP3 upon pathogenesis and progression for mammary carcinoma. Table 1 expounds both the clinical and pathological features among the entire patients with cancer. Observation of staining for CRISP3 was made for all the tumor specimens following immunohistochemistry (Figure 1a–h provide representative immunohistochemical staining images observed in tissues). The CRISP3 staining patterns observed in tumor tissues were cytoplasmic and plasma membranous. It was nearly impossible to make a detection of CRISP3 protein levels in normal breast tissues (Figure 1i,j). Through comparison of the CRISP3 expression level with certain tumor biological factors, it was demonstrated that CRISP3 expression was positively related with lymph node stage and tumor grade (p = 0.027 and p = 0.030, respectively; Table 1). Pathak et al. made an observation of expression level of CRISP3 and ANXA1 in seven microarray datasets in Oncomine database. In six out of seven research on prostate cancer, the upregulation was observed in CRISP3, with observation of the downregulation in ANXA1. Pathak et al. also found that ANXA1 was upregulated upon CRISP3 knockdown in LNCaP cells (Pathak et al., 2016). In this study, we found that the CRISP3 protein level was inversely related to the ANXA1 protein level (p = 0.022; Table 1). Furthermore, CRISP3-negative tumors had obvious connection to prolonged disease-free survival (DFS) as well as overall survival (OS) (p = 0.003 and p = 0.004, respectively; Figure 1k,l).
| Variable | Negative CRISP3 N (%) | Positive CRISP3 N (%) | Total | χ2 | p value |
|---|---|---|---|---|---|
| Age (year) | |||||
| ≤50 | 54 (50.0) | 54 (50.0) | 108 | 0.180 | 0.895 |
| >50 | 57 (50.9) | 55 (49.1) | 112 | ||
| Size (cm) | |||||
| <2 | 30 (50.8) | 29 (49.2) | 59 | 3.326 | 0.190 |
| 2–5 | 76 (52.8) | 68 (47.2) | 144 | ||
| >5 | 5 (29.4) | 12 (70.6) | 17 | ||
| LN stage | |||||
| I (Negative) | 65 (57.0) | 49 (43.0) | 114 | 7.244 | 0.027 |
| II (1–3 LN) | 31 (50.8) | 30 (49.2) | 61 | ||
| III ( ≥4 LN) | 15 (33.3) | 30 (66.7) | 45 | ||
| Grade | |||||
| I | 20 (51.3) | 19 (48.7) | 39 | 7.024 | 0.030 |
| II | 72 (56.7) | 55 (43.3) | 127 | ||
| III | 19 (35.2) | 35 (64.8) | 54 | ||
| ER | |||||
| Negative | 50 (52.1) | 46 (47.9) | 96 | 0.181 | 0.671 |
| Positive | 61 (49.2) | 63 (50.8) | 124 | ||
| PR | |||||
| Negative | 60 (50.0) | 60 (50.0) | 120 | 0.022 | 0.883 |
| Positive | 51 (50.0) | 49 (50.0) | 100 | ||
| HER2 | |||||
| Negative | 62 (54.9) | 51 (45.1) | 113 | 1.810 | 0.179 |
| Positive | 49 (45.8) | 58 (54.2) | 107 | ||
| Ki67 | |||||
| Negative | 41 (44.1) | 52 (55.9) | 93 | 2.614 | 0.106 |
| Positive | 70 (55.1) | 57 (44.9) | 127 | ||
| P53 | |||||
| Negative | 66 (47.5) | 73 (52.5) | 139 | 1.334 | 0.248 |
| Positive | 45 (55.6) | 36 (44.4) | 81 | ||
| ANXA1 | |||||
| Negative | 44 (42.3) | 60 (57.7) | 104 | 5.237 | 0.022 |
| Positive | 67 (57.8) | 49 (42.2) | 116 |
- Note. Significant p values are indicated in bold.
- Abbreviations: CRISP3: cysteine-rich secretory protein 3; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; LN: lymph node; PR: progesterone receptor.
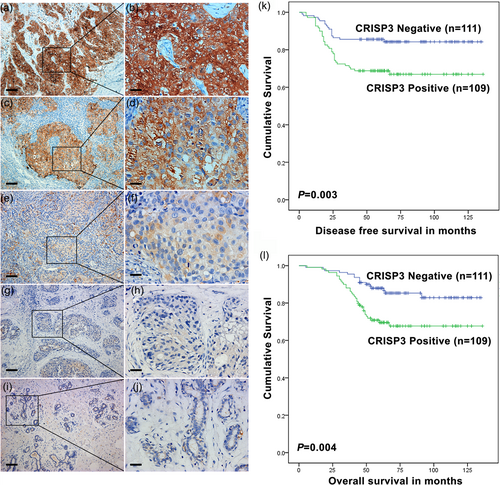
Relevance of CRISP3 expression with its prognostic significance in patients with mammary carcinoma. (a–h) Representative immunohistochemical staining images for CRISP3 in mammary carcinoma tissues. (a,b), Strong staining, (score 3); (c,d), moderate staining (score 2); (e,f), weak staining (score 1); (g,h), negative staining (score 0). The eventual score was represented as either negative or positive as presented hereinafter: negative (score of 0–1), positive (score of 2–3) (i,j) Representative immunohistochemical staining images for CRISP3 in healthy breast tissues. (k) DFS (disease-free survival); (l) OS (overall survival). Scale bars: (a,c,e,g, and i) 30 µm; (b,d,f,h, and j) 120 µm. CRISP3: cysteine-rich secretory protein 3 [Color figure can be viewed at wileyonlinelibrary.com]
3.2 CRISP3 and ANXA1 expressions correlate with prognosis
Through an application of Kaplan–Meier analysis, it was indicated that the ANXA1-negative group possessed shorter DFS and OS, whereas the ANXA1-positive group had the longer DFS and OS (Figure 2a,b). Furthermore, the CRISP3-positive ANXA1-negative group had the shortest DFS and OS, whereas the CRISP3-negative ANXA1-positive group had the longest DFS and OS (Figure 2c,d).
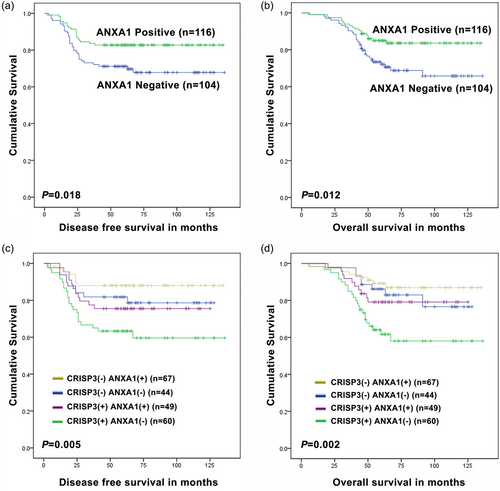
CRISP3 and ANXA1 collectively demonstrated the improvement of prognosis values for patients with mammary carcinoma. The relevance of ANXA1 expression with DFS (a) or OS (b) by using the Kaplan–Meier approach. The relevance of CRISP3/ANXA1 expression with DFS (c) and OS (d) by using the Kaplan–Meier approach. CRISP3: cysteine-rich secretory protein 3 [Color figure can be viewed at wileyonlinelibrary.com]
3.3 Univariate analysis of factors connected to DFS and OS for patients with mammary carcinoma
Through the deployment of a Cox regression model, univariate analysis was performed to make a further evaluation of whether the expression levels of CRISP3 and ANXA1 could be reckoned as an independent prognostic value for patients with mammary carcinoma. As indicated in Supporting Information Table S1, CRISP3 level and ANXA1 level were correlated with DFS of patients with mammary carcinoma (p = 0.005 and p = 0.021, respectively). Apart from the factors mentioned hereinbefore, tumor size (p < 0.001), lymph node (LN) stage (p < 0.001), tumor grade (p < 0.001), estrogen receptor (ER; p = 0.044), progesterone receptor (PR; p = 0.044), and human epidermal growth factor receptor 2 (HER2; p = 0.005) were also correlated with DFS (Supporting Information Table S1). As shown in Supporting Information Table S2, CRISP3 level and ANXA1 level were correlated with OS of patients with mammary carcinoma (p = 0.004 and p = 0.014, respectively). Apart from the factors mentioned hereinbefore, tumor size (p < 0.001), LN stage (p < 0.001), tumor grade (p < 0.001), ER (p = 0.033), PR (p = 0.041), and HER2 (p = 0.002) were also correlated with OS (Supporting Information Table S2).
3.4 Establishment of CRISP3 knockdown cell lines
Then, an examination of the mRNA and protein level of CRISP3 was performed in mammary carcinoma cell lines. CRISP3 mRNA levels in T-47D and SK-BR-3 cells were comparatively higher in comparison with other cells (Figure 3a). Furthermore, T-47D and SK-BR-3 cells displayed comparatively higher protein levels of CRISP3 than other cells (Figure 3b,c). The comparatively CRISP3-highly-expressed cell lines SK-BR-3 and T-47D were selected to perform the functional investigation. We established a siRNA knockdown cell line targeting CRISP3 in the SK-BR-3 cell line, which was named siRNA–CRISP3. Based on western blot analysis data (Figure 3d,e), CRISP3 siRNA1 and CRISP3 siRNA3 demonstrated greater efficiency versus CRISP3 siRNA2 and CRISP3 siRNA4. Therefore, application of the siRNA1-CRISP3 and siRNA3-CRISP3 cells were performed for additional experiment.
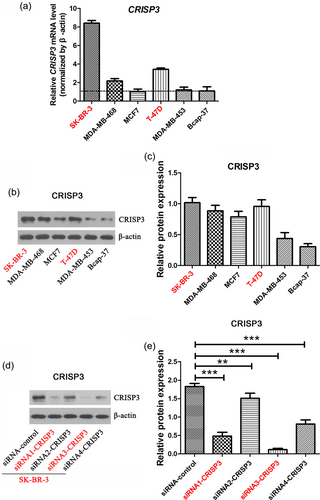
Establishment of CRISP3 knockdown cell line. The expression level of CRISP3 in a variety of mammary carcinoma cell lines were measured with quantitative real-time PCR and western blot (a,b). (c) Analysis of relative density of CRISP3 protein bands. (d) Analysis of CRISP3 level in the CRISP3 knockdown cell lines originated from SK-BR-3 cells by using the western blot approach. (e) Analysis of relative density of CRISP3 protein bands. The data are represented as mean ± SEM of three replicates. **p < 0.01; ***p < 0.001. CRISP3: cysteine-rich secretory protein 3; PCR: polymerase chain reaction; SEM: standard error of mean [Color figure can be viewed at wileyonlinelibrary.com]
3.5 Impacts of CRISP3 upon migration and invasion
The RTCA system was deployed for real-time analysis of cellular migration and invasion. It was revealed from the outcomes that CRISP3 knockdown in SK-BR-3 cells exerted an important inhibitory effect upon cellular migration within 24 hr (Figure 4a,b). CRISP3 knockdown was also found to exhibit a significant inhibitory effect upon cellular migration in T-47D cells (Figure 4c,d). In addition, the results of RTCA assay showed that CRISP3 knockdown in SK-BR-3 cells demonstrated significant inhibition of cellular invasion (Figure 5a,b). Furthermore, it was revealed by RTCA assay that CRISP3 knockdown in T-47D cells had significant inhibitory effects upon cellular invasion (Figure 5c,d).
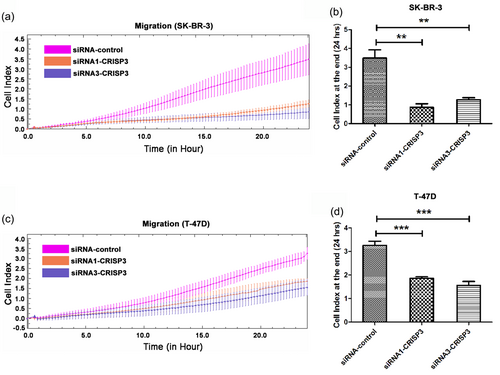
Knockdown of CRISP3 had inhibitory effects upon SK-BR-3 and T-47D cells migration. (a,c) The monitoring of migration was performed for 24 hr by using the xCELLigence RTCA system, with measurement of cellular indexes every 15 min. As a function of time, the alteration rates of cellular indexes were calculated for measurement of migration activity. The cellular indexes at the terminal of 24 hr are indicated in the bar chart (b,d). Data are represented as mean ± SEM of three replicates. **p < 0.01; ***p < 0.001. CRISP3: cysteine-rich secretory protein 3; RTCA: real-time cell analysis; SEM: standard error of mean [Color figure can be viewed at wileyonlinelibrary.com]
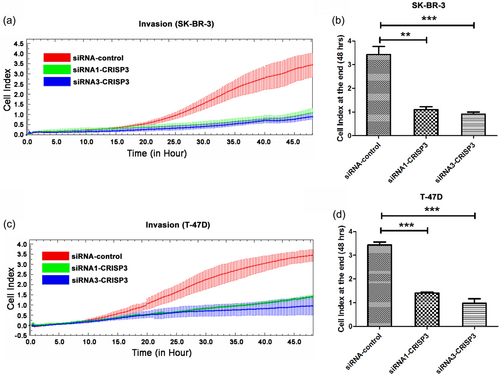
Knockdown of CRISP3 had inhibitory effects upon SK-BR-3 and T-47D cells invasion. (a,c) The monitoring of invasion was performed for 48 hr by using an xCELLigence RTCA system. Measurement of cellular indexes was made every 15 min. As the function of time, the alteration rates of cellular index were calculated for measurement of invasion activity. The cellular indexes at the terminal of 48 hr are indicated in the bar chart (b, d). Data are represented as mean ± SEM of three replicates. **p < 0.01; ***p < 0.001. CRISP3: cysteine-rich secretory protein 3; RTCA: real-time cell analysis; SEM: standard error of mean [Color figure can be viewed at wileyonlinelibrary.com]
3.6 Knockdown of CRISP3 inhibited ERK1/2 signaling pathway
A positive correlation was observed between CRISP3 level and LN staging of mammary carcinoma; whereas, CRISP3 protein level also had a negative correlation with the ANXA1 protein level (Table 1). We observed that CRISP3 knockdown in SK-BR-3 and T-47D cells remarkably increased the ANXA1 protein levels (Figure 6a). Maschler et al. found that ANXA1 knockdown could active the ERK1/2 MAPK pathway (Maschler et al., 2010). So, we next investigated whether CRISP3 level could influence the activation of ERK1/2 MAPK pathway. The ERK1/ERK2 serve as primary protein constituents for ERK1/ERK2 MAPK pathway; the extent of phosphorylation of those proteins plays an important role for activation of ERK1/ERK2 pathway (D'Espessailles, Mora, Fuentes, & Cifuentes, 2018). An obviously lower p-ERK1/2 level was found in siRNA1-CRISP3 and siRNA3-CRISP3 cells in contrast with siRNA–control cells (Figure 6a). The p90RSK and Elk-1 are key protein components of ERK1/2 MAPK pathway (Kajiya et al., 2009; Khan, Ahmad, & Haqqi, 2018; Ma et al., 2017). The extent of phosphorylation of those proteins plays an important role in ERK1/2 MAPK pathway activation. The p-p90RSK and p-Elk-1 levels were found to be significantly lower in siRNA1-CRISP3 and siRNA3-CRISP3 cells in contrast with siRNA–control cells (Figure 6a). According to the outcomes mentioned hereinabove, CRISP3 knockdown in SK-BR-3 and T-47D cells could increase the levels of ANXA1, and inhibit ERK1/2 MAPK signaling pathway (Figure 6b).
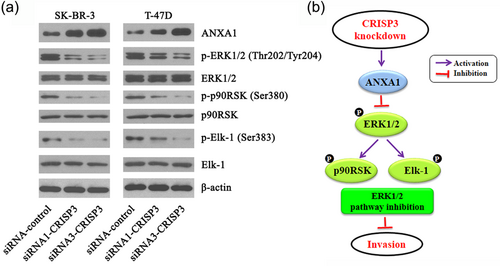
Knockdown of CRISP3 inhibited ERK1/2 MAPK signaling pathway in SK-BR-3 and T-47D cells. (a) Analysis of the essential proteins of ERK1/2 MAPK signaling pathway by using western blot. (b) Schematic diagram. Knockdown of CRISP3 in SK-BR-3 and T-47D cells could increase the ANXA1 level and inhibit ERK1/2 MAPK signaling pathway. CRISP3: cysteine-rich secretory protein 3; MAPK: mitogen-activated protein kinase [Color figure can be viewed at wileyonlinelibrary.com]
3.7 Influence of CRISP3 and ANXA1 knockdown on the migration and invasion abilities of SK-BR-3 cells
We established a siRNA knockdown cell line targeting ANXA1 in the SK-BR-3 cell line, which was named siRNA–ANXA1. Based on western blot analysis data (Supporting Information Figure S1A, B), ANXA1 siRNA3 demonstrated the greater efficiency versus ANXA1 siRNA1, ANXA1 siRNA2, and ANXA1 siRNA4. Intriguingly, the RTCA migration assay showed that the reduced expression of ANXA1 in siRNA3-CRISP3 (SK-BR-3) cells could significantly increase cell migration compared with the siRNA3-CRISP3 (SK-BR-3) cells (p < 0.01; Supporting Information Figure S1C, D). RTCA invasion assay also showed that reduced expression of ANXA1 in siRNA3-CRISP3 (SK-BR-3) cells could significantly increase cell invasion compared with the siRNA3-CRISP3 (SK-BR-3) cells (p < 0.05; Supporting Information Figure S1E, F). In addition, western blot analysis assay showed that reduced expression of ANXA1 in siRNA3-CRISP3 (SK-BR-3) cells could significantly increase the ratios of p-ERK1/ERK1 (p < 0.01; Supporting Information Figure S1G, H), and p-ERK2/ERK2 (p < 0.05; Supporting Information Figure S1G, I) compared with the siRNA3-CRISP3 (SK-BR-3) cells.
4 DISCUSSION
CRISPs (Cysteine-rich secretory proteins) are assumed to play a part in prostate pathophysiology and male reproduction. Three members of the CRISP family, that is, CRISP1, CRISP2, and CRISP3 have been identified. Fourth member of CRISP family, CRISP4 has been discovered only in mice (Dhaulakhandi et al., 2017). Among the mammalian CRISPs, the levels of CRISP3 in prostate cancer in particular have been proved to be obviously upregulated (Anklesaria, Pandya, Pathak, & Mahale, 2016). Many studies have shown that CRISP3 can serve as a prognostic indicator of clinical outcomes for the patients with prostate carcinoma. However, whether CRISP3 exerts an effect upon the development of mammary carcinoma is unknown.
The immunohistochemistry was deployed to perform an analysis of CRISP3 protein levels in mammary carcinoma specimens for validation of the association between CRISP3 protein levels and outcomes in patients with mammary carcinoma (Figure 1a–h). With the utilization of Kaplan–Meier approach for the analysis of patients with mammary carcinoma, higher CRISP3 protein levels were revealed to have a link to shorter DFS (Figure 1k) and OS (Figure 1l). In this study, the number of specimen analyzed (220 examples) may not be sufficient to conclude that CRISP3 can be reckoned as an effective predictor in clinical outcomes for patients with mammary carcinoma. It is of great necessity to perform additional in-depth research for elucidation of an explicit connection of CRISP3 protein levels with the outcomes in the clinical setting. Through comparison of CRISP3 protein level with tumor biological factors, the CRISP3 expression was found to be positively related with LN staging and tumor grade (Table 1). Furthermore, the heterogeneous expression of CRISP3 was observed in a variety of mammary carcinoma cells (Figure 3a). The CRISP3 in T-47D and SK-BR-3 cells displayed significantly higher levels of mRNA and protein than those in the other types of mammary carcinoma cells (Figure 3a–c). CRISP3 knockdown in SK-BR-3 and T-47D cells could exert a significant inhibitory effect upon the migration (Figure 4) and invasion (Figure 5) capabilities in comparison with the control group. ERK1/2 MAPK pathway plays an important role in cancer cell migration and invasion (Schroyer, Stimes, Abi Saab, & Chadee, 2018). According to the research, knockdown of CRISP3 in T-47D or SK-BR-3 cells was found to have an inhibitory effect upon ERK1/2 MAPK signaling pathway (Figure 6).
To summarize, it was proven that knockdown of CRISP3 not only has remarkably inhibitory effects upon migration and invasion of mammary carcinoma cells, but also has a connection to the inhibition of ERK1/2 MAPK signaling pathway. In addition, CRISP3 expression in clinical samples was analyzed, demonstrating the association between highly-expressed CRISP3 and unfavorable prognosis in patients with mammary carcinoma. It is indicated according to our observation that CRISP3 may be reckoned as a marker for clinical outcomes in patients with mammary carcinoma.
ACKNOWLEDGMENTS
This study was sponsored by the National Natural Science Foundation of China [grant numbers 81672731 (S. F), 81571055 (Y. Z)], 2018 “333 Project” Award of Jiangsu Province (Y. W), Xuzhou Municipal Science and Technology Innovation Project [grant number KC15SH029 (Y. W)], Postgraduate Research & Practice Innovation Program of Jiangsu Province [grant number KYCX17_1622 (N. S)], and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) (Y. Z).
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.