RETRACTED: Long noncoding RNA LEF1-AS1 silencing suppresses the initiation and development of prostate cancer by acting as a molecular sponge of miR-330-5p via LEF1 repression
Da-Chuang Liu and Lin-Lin Song have contributed equally to this work.
Abstract
Prostate cancer (PCa) is one of the major cancers affecting males with high mortality around the world. Recent studies have found that some long noncoding RNAs play a critical part in the cellular processes of PCa. In our study, aberrant expressed lymphoid enhancer-binding factor-1 antisense RNA 1 (LEF1-AS1), microRNA-330-5p (miR-330-5p), and lymphoid enhancer-binding factor-1 (LEF1) were screened out from a microarray database, the role of the novel noncoding RNA regulatory circuitry in the initiation and development of PCa was investigated. LEF1-AS1 and LEF1 were highly expressed while miR-330-5p was poorly expressed in PCa. Following that, the PCa PC-3 cell line was adopted for subsequently experiments, in which the expression of LEF1-AS1 and miR-330-5p was subsequently altered by means of exogenous transfection. After that, the effects of up- or downregulation of LEF1-AS1 and miR-330-5p on epithelial–mesenchymal transition (EMT) and the cell ability for proliferation, invasion, migration in vitro, and tumorigenesis and lymph node metastasis (LNM) in vivo were evaluated. RNA crosstalk revealed that LEF1-AS1 bound to miR-330-5p and LEF1 was the target gene of miR-330-5p. Silenced LEF1-AS1 or elevated miR-330-5p exhibited inhibited EMT processes, reduced ability of proliferation, invasion and migration, coupling with decreased tumorigenesis and LNM in nude mice. The key findings of this study collectively propose downregulation of LEF1-AS1 competing with miR-330-5p to inhibit EMT, invasion and migration of PCa by LEF1 repression.
1 INTRODUCTION
Prostate cancer (PCa) is a common fatal cancer diagnosed in male patients worldwide and there is about 15% of male cancers diagnosed as PCa globally (Davidsson et al., 2018). PCa is featured by an abnormal digital rectal examination outcome or an elevated prostate specific antigen level (Huang, Reilly, Zhang, & Wang, 2015). There are various factors involved in the pathology of PCa, including age, hereditary factors, being androgen-dependent, sex hormones, environmental factors, diet, as well as immune and inflammatory responses (Facina et al., 2018). Chronic inflammation is also found to be related to the development and progression of PCa (Zhang et al., 2017). Recently, there have been increasing novel approaches to treat advanced metastatic PCa and the conventional androgen deprivation treatment has been improved (Gomella, Petrylak, & Shayegan, 2014). It has been found that statins play antitumor roles in PCa with a better effect before diagnosis (Yu et al., 2014). Besides that, increasing evidence shows that long noncoding RNAs (lncRNAs) have the potential to be new targets in the treatment of cancers (Li & Chen, 2013).
LncRNAs play pivotal parts in numerous biological processes and their dysregulation can result in many human diseases such as cancer (Khandelwal, Bacolla, Vasquez, & Jain, 2015). Furthermore, lncRNAs exert great influence on the proliferation, apoptosis, invasion, and metastasis of cancer cells (Cai, Song, Cai, & Zhang, 2014). At present, some lncRNAs functioning on tumorigenesis have been identified such as metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), PCa-associated noncoding RNA 1 (PRNCR1), and PCa gene expression marker 1 (PCGEM1; Park et al., 2014). Increasing evidence has found that lncRNAs as competing endogenous RNAs (ceRNAs) of microRNA (miR) are implicated in the cell cycle, metabolism, and progression of PCa (D. Liu et al., 2016). MiR-330-5p is also reported to be involved in the development of prostate, neuronal, and pancreatic cancers through the regulation of cell proliferation, migration, invasion, and epithelial–mesenchymal transition (EMT; Yoo, Kim, & Yoon, 2016). Lymphoid enhancer-binding factor-1 (LEF1) is reported to be important for the increase of the mesenchymal marker vimentin and plays a critical upstream role in cancer development and EMT (Liang, Li, Li et al., 2015). Besides that, LEF1 functions as an important transcription factor involved in the Wnt signaling pathway and is associated with regulating cell proliferation and invasion (Liang, Li, Daniels et al., 2015). A recent study found that LEF1 knockdown resulted in inhibition of EMT, migration and invasion in PCa cells (Liang, Li, Li et al., 2015). Moreover, a new type of lncRNA lymphoid enhancer-binding factor-1 antisense RNA 1 (LEF1-AS1) is located in chromosome 4q25 and found to be upregulated in glioblastoma (GBM) and silencing LEF1-AS1 suppresses proliferation and invasion of GBM cells (J. Wang et al., 2017). Therefore, we identified and characterized the mechanism by which LEF1-AS1 and its target miR-330-5p regulate LEF1. The tumor suppressor function of miR-330-5p is represented in vivo and in vitro, the elevated expression of which inhibits EMT and the ability for proliferation, invasion, and migration of PCa cells. In addition, metastasis of PCa is prevented by upregulated miR-330-5p or downregulated LEF1-AS1. Hence, it is implicated that the LEF1-AS1/miR-330-5p/LEF1 axis may involve in PCa.
2 MATERIALS AND METHODS
2.1 Ethics statement
In this experiment, all patients with PCa signed the informed consent, agreed to leave the tissue samples for research and followed the Declaration of Helsinki. Animal experiments were carried out in accordance with the license agreement of the animal experiment committee of Xuzhou Central Hospital, and there was no animal abuse in the experiment.
2.2 Profiling data analysis
The PCa-related microarray expression data and probe annotation file was downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The “limma” package of R language was used to conduct variance analysis for PCa samples and normal samples, with |logFC| > 2, p < 0.05 as the standard for screening differentially expressed genes. At the same time, the R language “pheatmap” package was used to construct differentially expressed gene imaging. Correlation analysis was used to determine the correlation between lncRNA and messenger RNA (mRNA). The RNA22 website (https://cm.jefferson.edu/rna22) was used to predict miR, which can be combined with lncRNA. The MicroRNA.org website (http://34.236.212.39/microrna/getMirnaForm.do) was used to predict the target gene combined with miR.
2.3 Study subjects
A total of 58 cases of radical resection of PCa were collected from September 2014 to September 2017 in the Department of Urology of Xuzhou Central Hospital and their para-cancerous tissues were taken as controls, that is, a control group (para-cancerous tissues) and a PCa group (specimens of radical resection of PCa). (All specimens were verified by pathological diagnosis and there was a complete collection of clinical data.) The subjects were in the age range of 56–84 years old. Among them, there were 32 patients ≥70 years old, and 26 patients <70 years old. There were 27 cases with the transverse diameter of the prostate more than 35 mm and 31 cases with the transverse diameter of prostate <35 mm. There were 20 cases with a Gleason score ≤7 points and 38 cases with a Gleason score >8 points. According to tumor, nodes, metastasis staging, 25 cases were at Stages I and II, and 33 cases were at Stage III and IV (Pasoglou et al., 2014). All 58 patients with PCa were diagnosed as having primary tumors by professional diagnosis and the patients did not receive PCa-related chemotherapy or radiotherapy. There were no pathological changes in the para-cancerous tissues including tumor cell infiltration or any obvious inflammatory reaction. One part of the specimens was fixed with 10% formaldehyde, followed by routine dehydration, paraffin embedding, and slicing (4 μm/slice). Another part of the specimens was immediately placed in liquid nitrogen and then transferred to −80°C for preservation.
2.4 Immunohistochemistry
The tissue sections were placed in a 60°C oven and heated for l hr, followed by conventional xylene dewaxing and gradient alcohol dehydration. Then the sections were added with 0.5% Triton phosphate-buffered saline (PBS) and incubated at room temperature for 20 min. The sections were rinsed with PBS two to three times, 5–10 min per time. High-pressure antigen retrieval was conducted for 2 min. After that, the sections were placed in citric acid buffer solution (0.01 M, pH 6.0) boiling at 95℃ for 20 min, and were soaked in 3% H2O2 reacting for 15 min to block the activity of exogenous peroxidase. The sections were then blocked with 3% bovine serum albumin (BSA) sealing solution at 37℃ for 20–30 min. An appropriate amount of primary antibodies rabbit anti-human E-cadherin (ab15148, 1:30; Abcam Inc., Cambridge, MA), N-cadherin (ab18203, 1:100; Abcam Inc., Cambridge, MA), and vimentin (ab45939, 1:80; Abcam Inc., Cambridge, MA) was added for incubation at 37℃ for 2 hr, followed by a PBS rinse. After drying, an appropriate amount of horseradish peroxidase (HRP)-labeled goat antirabbit immunoglobulin G (IgG) secondary antibody working solution (ab6721, 1:1,000; Abcam Inc., Cambridge, MA) was added for incubation in a wet box at 37℃ for 30 min. After that, hematoxylin (Shanghai Fu Sheng Industrial Co. Ltd., Shanghai, China) was used for redying at room temperature for 4 min, followed by running water rinsing excess dye liquor. Then the sections were sealed with 10% glycerol/PBS. The primary antibody was replaced by PBS as the negative control (NC) and the known positive sections were considered as the positive control. Photographs of the prepared tissue specimens were obtained using a general optical microscope (XSP-36; Bosda Optical Instrument Co. Ltd., Shenzhen, China). Five high-power fields (×400) were selected from each section with 100 PCa cells being counted in each field. The number of positive cells <5% was regarded as negative and ≥5% as positive (Gupta et al., 2010). E-cadherin is expressed in both the cell membrane and cytoplasm, and N-cadherin is expressed in the nucleus and cell membrane, and vimentin is mainly expressed in the glands and stroma. The sections with brown particles were considered to be positive. The results of immunohistochemistry were conducted by two people independently by the double blind method. The experiment was repeated three times.
2.5 Cell culture and screening
Dulbecco's modified Eagle medium (DMEM) containing 10% calf serum was used to cultivate the PCa cell lines PC-3, 22RV1, and the human normal prostate epithelial cell line RWPE-1 (all were purchased from the cell bank of the typical culture preservation Committee of the Chinese Academy of Sciences). Then, penicillin–streptomycin mixed with a percentage of 1:1 was added to the culture solution and 100 U/ml was considered as the final concentration followed by culturing in a 37℃, 5% CO2 incubator with the cells. The cells were treated with 0.25% trypsin and underwent subculture based on the ratio of 1:3. The cells were inoculated into a six-well plate (3 × 105 cells/well) and when the cell confluence reached 70–80%, detection was conducted. The expression level of LEF1-AS1 in each cell was detected by real-time fluorescence quantitative polymerase chain reaction (PCR), and the cell line with the highest expression was selected for the follow-up experiment. The experiment was repeated three times.
2.6 Cell treatment
The third generation of subculture cells were taken, treated with trypsin and inoculated into a 24-well plate. Then the cells were cultured into a monolayer. After discarding the culture solution, the cells were assigned into 12 groups according to the design requirements, including the vector group (PC-3 cells transfected with blank plasmids), LEF1-AS1 group (PC-3 cells transfected with LEF1-AS1 plasmids), si-NC group (PC-3 cells transfected with LEF1-AS1 negative insignificant sequences), si-LEF1-AS1 group (PC-3 cells transfected with si-LEF1-AS1 plasmids), blank group (PC-3 cells transfected with no sequence), NC group (PC-3 cells transfected with blank plasmids), miR-330-5p mimic group (PC-3 cells transfected with miR-330-5p mimic plasmids), miR-330-5p inhibitor group (PC-3 cells transfected with miR-330-5p inhibitor plasmids), LEF1-AS1 + miR-330-5p mimic group (PC-3 cells transfected with LEF1-AS1 and miR-330-5p mimic plasmids), si-LEF1-AS1 + miR-330-5p inhibitor group (PC-3 cells transfected with si-LEF1-AS1 and miR-330-5p inhibitor plasmids), si-LEF1 group (PC-3 cells transfected with si-LEF1 plasmids), and LEF1-AS1 + si-LEF1 group (PC-3 cells transfected with LEF1-AS1 and si-LEF1 plasmids). The transfected sequences were constructed by Shanghai Sangon Biological Engineering Technology & Services Co. Ltd. (Shanghai, China). The cells were seeded into a six-well plate at 24 hr before transfection and transfected with Lipofectamine 2000 (11668-019; Invitrogen, Carlsbad, CA) when the cell density reached 90%–95%. Next, a total of 100 pmol of plasmids were diluted with 250 μl of serum-free Opti-MEM (51985042; Gibco Company, Gaitherburg, MD, final concentration of 50 nM in the cells), mixed, and incubated at room temperature for 5 min. The Lipofectamine 2000 (5 μl) was diluted with 250 μl of serum-free Opti-MEM, mixed and incubated for 5 min at room temperature. The above two were mixed and incubated at room temperature for 20 min, followed by adding into cell culture wells. Following incubation at 37℃ and 5% CO2 culturing for 6–8 hr, the cells were further cultured in complete culture medium for 24–48 hr for the follow-up experiments. After that, photographs of cells in each group were obtained from an inverted microscope (MI13; Guangzhou Mingmei Photoelectric Technology Co. Ltd., Guangzhou, China) and the change in the cell morphology in each group were observed. PC-3 cells with good growth status were selected for the follow-up experiment.
2.7 Dual-luciferase reporter gene assay
The target gene analysis of LEF1-AS1 and miR-330-5p, LEF1 and miR-330-5p was carried out using the biological prediction website https://cm.jefferson.edu/rna22/Interactive/. The luciferase reporter gene assay was used to verify whether there was a targeting relationship between miR-330-5p and LEF1-AS1 and whether LEF1 was a direct target gene of miR-330-5p. The target sequence and the mutation sequence were designed based on the sequence of miR-330-5p combined with the 3′-UTR zone of LEF1-AS1 and LEF1 mRNA. The target sequence was chemically synthesized. When the sequences were synthesized, the XhoI and NotI enzyme sites were added to the both ends of the sequences. The synthesized fragments were cloned into the PUC57 vector (HZ0087; Shanghai Huzhen Industrial Co. Ltd., Shanghai, China). After identification of the positive clones, the recombinant plasmids were identified by DNA sequencing, followed by subcloning into psiCHECK-2 vector (HZ0197; Shanghai Huzhen Industrial Co. Ltd.), and transformed into Escherichia coli DH5α cells. Then the plasmids were amplified. All the above plasmids were extracted according to the Omega plasmid extraction kit (DL100-50T; Beijing Solarbio Science & Technology Co. Ltd., Beijing, China). PC-3 cells were seeded into a six-well plate (2 × 105 cells/well). After cell adhesion, the cells were transfected as per previous methods. Following successful transfection, the cells were cultured for 48 hr and then were collected. The change of the luciferase activity of LEF1-AS1–3′-UTR and LEF1–3′-UTR in cells made by miR-330-5p was detected by using Genecopoeia's dual luciferase assay kit (D0010; Beijing Solarbio Science & Technology Co. Ltd.). The fluorescence intensity was detected by using the Promega's GLomax20/20 Luminometer fluorescence detector (E5311; Shanxi Zhongmei Biotechnology Co. Ltd., Shanxi, China). The experiment was repeated three times.
2.8 Fluorescence in situ hybridization (FISH)
The expression of LEF1-AS1 in PC-3 cells was predicted by the lncRNA cell sublocation website http://lncatlas.crg.eu/. The subcellular localization of LEF1-AS1 was detected by using the FISH kit (Roche Ltd., Basel, Switzerland). PC-3 cells cultured after transfection in each group were selected, washed with cold PBS two times and then fixed with 4% polyformaldehyde. Next, the LEF1-AS1 probe (Sigma-Aldrich Chemical Company, St. Louis, MO) hybrid solution, incubated with digoxin, was added to the cell culture plate. The antagonistic LEF1-AS1 probe was established as a NC. The nucleus was stained by using 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich Chemical Company) at room temperature for 10 min, followed by washing with cold PBS two times. The fluorescence images were recorded by using a confocal laser scanning microscope (FV1000; Olympus, Tokyo, Japan). The experiment was repeated three times.
2.9 RNA immunoprecipitation (RIP) detection
The PC-3 cells were washed with cold PBS two times, adding l0 ml PBS and the cells were scraped down by curettage and sucked into the centrifuge tube. After centrifugation at 1,500 rpm at 4°C for 5 min and discarding the supernatant, RIP Lysis Buffer (N653—100 ml; Shanghai Hao Ran Biotechnology Co. Ltd., Shanghai, China) was added, fully mixed, lysed on ice for 5 min to prepare cell lysate. Each tube was added 50 μl magnetic beads and turned upside down to fully mix the magnetic beads. Then 0.5 ml RIP Wash Buffer (EHJ-BVIS08102; Xiamen Huijia Biological Technology Co. Ltd., Xiamen, China) was added. After a short vortex, the tube was placed on the magnetic separator and kept for the magnetic beads to aggregate. After that, the supernatant was abandoned, the magnetic beads were retained, and the magnetic beads were washed once again, followed by adding 100 μl RIP Wash Buffer to the suspended beads in each tube. Then 5 μg Ago2 antibody (P10502500; Shenzhen Otwo Biological Technology Co. Ltd., Shenzhen, China) was added and incubated at room temperature for 30 min. Following discarding the supernatant, 0.5 ml RIP Wash Buffer was used to wash the magnetic beads two times for the follow-up experiment. A total of 900 μl RIP Immunoprecipitation Buffer (P10403138; Shenzhen Otwo Biological Technology Co. Ltd.) was added to the mixture of magnetic beads and antibody, followed by centrifugation at 14,000 rpm at 4°C and lysing for 10 min. The supernatant was sucked up to the new EP tube (LBCT015S; Beijing Beifang Tongzheng Biotechnology Co. Ltd., Beijing, China) and 100 μl supernatant was sucked up to the tube of magnetic beads and antibody. Then, 1 ml was regarded as the final volume of the immunoprecipitation reaction, and the mixture was rotated and incubated at 4°C overnight, followed by washing the magnetic beads with 0.5 ml RIP Wash Buffer six times. Finally, 150 μl proteinase K buffer was added and incubated at 55°C for 30 min to purify RNA. The conventional TRIZOL method was used to extract RNA followed by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) detection.
2.10 RNA-pull down assay
PC-3 cells were transfected with WT biotinylated miR-330-5p (50 nM) and MUT biotinylated miR-330-5p (50 nM). After transfection for 48 hr, the cells were collected and washed with PBS, following vortex. Then, the specific cell lysate (Ambion, Austin, TX) was used for incubation for 10 min, followed by split charging the 50 ml sample cell lysate. The remaining lysates were incubated with M-280 streptomycin magnetic beads (Sigma-Aldrich Chemical Company) precoated with RNase-free and yeast tRNA (Sigma-Aldrich Chemical Company) at 4℃ for 3 hr. Then the cold lysate was used to wash the cells two times, the low salt buffer solution was used to wash the cells three times, and then the high salt buffer solution was used to wash the cells once. The antagonistic miR-330-5p probe was established as a NC. The total RNA was extracted with Trizol and then the expression of LEF1-AS1 was detected by RT-qPCR. The experiment was repeated three times.
2.11 Immunofluorescence assay
After routine digestion and transfection, the cells in each group were counted and spread in the immunofluorescence chamber for culture, 2 × 105 cells/well. When the cell fusion rate reached 90%, the cells were washed with PBS three times (this process was operated on ice). The cells were fixed with 4% polyoxymethylene at normal temperature keeping static for 15 min, 1 ml per well. After three times of PBS washing, the cells were perforated with 0.3% Triton. After 10 min, the cells were washed with PBS three times, and sealed with goat serum, keeping static for 30 min. The primary antibody prepared by PBS was added and incubated at 4℃ overnight. After washing with PBS three times, the second antibody was added and incubated at room temperature for 1 hr, avoiding exposure to light, followed by three times of PBS washing. After that, DAPI was added to stain for 15 min, avoiding exposure to light. Following PBS washing three times without exposure to light, the fluorescence quenching agent was added for mounting. Photographs of the cells were obtained using a fluorescence microscope. The experiment was repeated three times.
2.12 RT-qPCR
The transfected cells in each group were placed into l ml Trizol (15596-018; Beijing Solarbio Science & Technology Co. Ltd.), and the cells were subsequently rinsed in an ice bath for 2 min to produce the homogenate, followed by centrifugation at 12,000 rpm for 15 min, and discarding precipitation. Then, 200 μl of chloroform was added, shaking and mixing manually, following keeping static at room temperature for 15 min and centrifugation at 12,000 rpm for 15 min. The upper water phase was extracted, mixed with 0.5 ml isopropyl alcohol and placed at room temperature for 10–30 min, followed by centrifugation at 12,000 rpm for 10 min at 4℃ and discarding the supernatant to find RNA precipitation at the bottom of tube. Next, l ml 75% ethanol was added and the centrifuge tube was gently shaken to suspend the precipitation. After centrifugation at 8,000 rpm for 5 min at 4℃, the supernatant was discarded, and the specimens were dried at room temperature or underwent vacuum drying for 5–10 min. Then, 20 μl DEPC was used to dissolve the precipitation and the RNA concentration was determined. The primers used in this study were synthesized by Takara company in Dalian (Table 1). The reverse transcription was carried out as per the instructions of complementary DNA (cDNA) reverse transcription Kit (K1622; Beijing Ya An Da Biotechnology Co. Ltd., Beijing, China). The reaction conditions were as follows: 42℃ for 30–50 min (reverse transcription reaction) and 70°C for 5 min (reverse transcriptase inactivation reaction). The reverse transcriptional cDNA was diluted to 50 ng/μl, following a subsequent fluorescence quantitative PCR for reserve. The reaction amplification system was 25 μl, adding 2 μl each time and put into a fluorescence quantitative PCR instrument (ViiA 7; Zhongshan University Da'an Gene Co. Ltd., Guangzhou, China) for detection. The reverse transcription reaction conditions were as follows: predenaturation at 95℃ for 4 min, 30 cycles of denaturation at 95℃ for 30 s, and annealing at 57℃ for 30 s and extension at 72℃ for 30 s. With the total cDNA as template, the U6 gene was used as the internal reference of LEF1-AS1 and miR-330-5p, while glyceraldehyde-3-phosphate (GAPDH) was the internal reference of LEF1, E-cadherin, N-cadherin, and vimentin. The relative quantitative method ( ) was used to calculate the relative transcription level. The formula was as follows: ΔΔCt = ΔCt model group−ΔCt normal group, where ΔCt = Ct target gene−Ct internal reference. is the relative transcription level of the target gene (Ayuk, Abrahamse, & Houreld, 2016). The experiment was repeated three times.
| Gene | Sequence (5′–3′) |
|---|---|
| LEF1-AS1 | F: TTTGTGTGGCCTGGACTCTC |
| R: AACCCCTGGGACACAAACTG | |
| miR-330–5p | F: GGGACACAGGGCCAGAGAC |
| R: CAGTGCGTGTCGTGGAGT | |
| LEF1 | F: AGAACACCCCGATGACGGA |
| R: GAGGGTCCCTTGTTGTAGAGG | |
| E-cadherin | F: TGCCCAGAAAATGAAAAAGG |
| R: GTGTATGTGGCAATGCGTTC | |
| N-cadherin | F: CAACTTGCCAGAAAACTCCAGG |
| R: ATGAAACCGGGCTATCTGCTC | |
| Vimentin | F: GAGAACTTTGCCGTTGAAGC |
| R: GCTTCCTGTAGGTGGCAATC | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| GAPDH | F: ACCCAGAAGACTGTGGATGG |
| R: TCTAGACGGCAGGTCAGGTC |
- Note. F: forward; GAPDH: glyceraldehyde-3-phosphate; LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; miR: microRNA; R: reverse; RT-PCR: reverse-transcription quantitative polymerase chain reaction.
2.13 Western blot analysis
The transfected cells in each group were selected and added with protein lysate to lyse for 30 min at 4℃, while shaking every 10 min, followed by centrifugation at 12,000 rpm for 20 min at 4℃ with the lipid layer discarded. The protein concentration of the supernatant of each sample was determined using the bicinchoninic acid kit (20201ES76; Yeasen Biotechnology Co. Ltd., Shanghai, China). After quantitation based on different concentrations, the protein was transferred to polyvinylidene fluoride membrane by the wet transfer method after polyacrylamide gel electrophoresis, followed by sealing with 5% BSA for 1 hr at room temperature. Then, the following rabbit anti-human antibodies were added and incubated at 4℃ overnight in a rocking bed, followed by washing with Tris-buffered saline tween (TBST) three times, 5 min per time. The rabbit anti-human antibodies were LEF1 (ab22884, 1:2,000), E-cadherin (ab40772, 1:20,000), N-cadherin (ab76057, 1:1,000), and vimentin (ab92547, 1:2,000; Abcam Inc., Cambridge, MA). The HRP-labeled sheep anti-rabbit IgG (ab6721, 1:1,000; Abcam, Cambridge, UK) diluent was added and incubated for 1 hr at room temperature, followed by washing with TBST three times, 5 min per time. Then the developer was added to develop. ImageJ 1.48u software (National Institutes of Health) was used for protein quantitative analysis based on the ratio of the gray value of each protein and the gray value of GAPDH. The experiment was repeated three times.
2.14 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay
After transfection for 48 hr, the cells were collected and counted. After counting, the cells were seeded in a 96-well plate with a cell density of 3 × 103–6 × 103 cells in each well (100 μl; with six repeating wells). Three time points including 24, 48, and 72 hr were set to carry out the following experiments. Each well was added with 20 μl of 5 mg/ml MTT solution and then incubated for 2 hr at 37℃, followed by terminating culture. Then the supernatant was aspirated and 150 μl of dimethylsulfoxide was added to each well. The optical density (OD) values at 570 nm were measured using an enzyme-linked immunosorbent detector (NYW-96M; Beijing Nuo Ya Wei Instrument Co. Ltd., Beijing, China). Each experiment was repeated three times. The time points including 24, 48, and 72 hr were taken as the abscissa, the OD value as the ordinate, and cell viability curve was plotted.
2.15 Transwell assay
Matrigel matrix (40111ES08; Shanghai Yi Sheng Biotechnology Co. Ltd., Shanghai, China; Matrigel:DMEM = 1:2) diluted by precooling serum-free DMEM was settled in the Transwell cell culture apical chamber (3413; Beijing Unique Biotechnology Co. Ltd., Beijing, China), followed by incubating at 37℃ for 4–5 hr to be solidified. Then, 100 μl of serum-free medium was used to dilute the transfected cells to prepare the cell suspension with a concentration of 1 × 106 cells/ml, followed by inoculation. Next, 500 μl DMEM nutrient solution containing 20% fetal bovine serum was added into the lower chamber with three repeating wells in each group. After culturing for 24 hr with 5% CO2 at 37℃, the Transwell chamber was washed with PBS two times, fixed with 5% glutaraldehyde, stained with 0.1% crystal violet for 5 min at 4℃, followed by rinsing with PBS two times. Cotton balls were used to wipe the surface cells. Photographs of the cells were obtained from an inverted fluorescence microscope (TE2000; Nikon Company, Beijing, China), randomly selecting five fields (×200), with the average value as the number of the cells through the chamber in each group. Each experiment was repeated three times.
2.16 Scratch test
After incubating the transfected cells in a 37℃ and 5% CO2 incubator for 24 hr, 10 μl head of the transfer liquid gun was used to scratch on monolayer cells with the shape of the Chinese character “—.” After washing with PBS three times, the serum-free medium was added and incubated for 24 hr, and the culture medium was washed away. Following washing with PBS for three times, cell migration at 0 and 24 hr was observed under an inverted microscope. Three images from each group were obtained and the relative distance between cells on the two sides of the scratches was measured. The relative migration distance of cells was obtained by the distance difference being divided by 2, and the cell relative mobility was calculated. The formula was as follows: cell relative mobility = relative migration distance/distance from scratch edge to scratch midline at 0 hr. Each experiment was repeated three times.
2.17 Xenograft tumors in nude mice
A total of 60 male BALB/c nude mice (5–7 weeks, 22–24 g, purchased from Shanghai Ling Chang Company, Shanghai, China) were selected and fed in the experimental animal center of Hsiang-Ya Medical College before the experiment (experimental animal certificate was No. 159). The feeding environment was no specific pathogen (SPF), comfortable temperature and environment, aseptic feed and drinking water, each 12 hr of day and night, and 7-day adaptive feeding.
After culturing the transfected cells for 48 hr, the cells in each group which were in a good growth state and in the logarithmic growth period were obtained to prepare a 5 × 107 cells/ml cell suspension. The 0.20 ml cell suspension was inoculated with an 1 ml syringe on the left side of the BALB/cnu/nu nude mice. (Each group of transfected cells was inoculated with six nude mice.) After inoculation, all nude mice were kept in the laminar mask in the SPF level animal house for observation. The data (volume and quality) were recorded at 7, 14, 21, and 28 days after inoculation. The short diameter (a) and long diameter (b) of the tumor were recorded by a vernier caliper, and the tumor volume was calculated based on the formula π(a2b)/6. The measurement of tumor volume and quality was repeated three times.
2.18 Statistical analysis
All data were processed by using SPSS 21.0 statistical software (IBM Corp., Armonk, NY). The measurement data were expressed by mean ± standard deviation. One-way analysis of variance was used for comparisons among multiple groups. p < 0.05 was indicative of significant statistical difference.
3 RESULTS
3.1 LEF1-AS1 and LEF1 are highly expressed and miR-330-5p is poorly expressed in PCa
To determine differentially expressed lncRNA and mRNA, the PCa chip GSE55945 was analyzed. The results showed that LEF1-AS1 and LEF1 were highly expressed in PCa (Figure 1a). The results of correlation analysis showed that there was a positive correlation between LEF1-AS1 and LEF1 (Figure 1b). Next, positive expression of E-cadherin, N-cadherin, and vimentin in tissues was detected by immunohistochemistry. The results showed that (Figure 1c,d) in the control group, the positive expression rate of E-cadherin was 70.69%, the positive expression rate of N-cadherin was 17.24%, and the positive rate of vimentin was 34.48%; in the PCa group, the positive expression rate of E-cadherin was 29.31%, the positive expression rate of N-cadherin was 82.76%, and the positive rate of vimentin was 65.52%. The results above indicated that compared with the control group, the positive expression rate of E-cadherin in the PCa group significantly decreased, while the positive expression rate of N-cadherin and vimentin increased significantly (all p < 0.05).
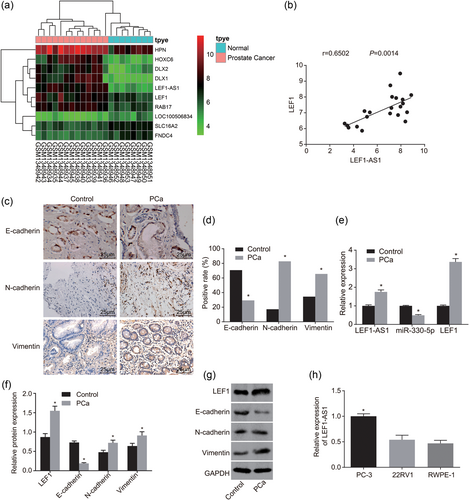
LEF1-AS1 and LEF1 are highly expressed, miR-330-5p is poorly expressed in PCa tissues and there is EMT occurring in PCa. (a) Chip GSE55945 screening image, the abscissa denotes the sample number, the vertical axis represents the name of the differential gene, the histogram on the upper right is color gradation, the color change from top to bottom represents the value from large to small in microarray data, each rectangle corresponds to a sample value, each column represents the amount of expression of all genes in each sample, the dendrogram on the left represents the results of cluster analysis of different genes from different samples, the bar on the top represents the sample type, the box on the upper right represents the sample color reference, blue as the normal control sample, red as tumor sample; (b) correlation analysis results of LEF1-AS1 and LEF1 on chip GSE55945; (c) immunohistochemistry pictures (×400); (d) histogram of immunohistochemistry results; (e) RT-qPCR was used to determine the expression of miR-330-5p and the mRNA level of LEF1 and LEF1-AS1; (f) western blot analysis was used to determine the protein expressions of LEF1 and EMT; (g) protein band diagrams of LEF1 and EMT in PCa; (h) RT-qPCR was performed to screen the highly expressed LEF1-AS1 cell line. *p < 0.05 versus the control group or RWPE-1 cell line. EMT: epithelial–mesenchymal transition; GAPDH: glyceraldehyde-3-phosphate; LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; lncRNA: long noncoding RNA; miR: microRNA; PCa: prostate cancer; RT-qPCR: reverse-transcription quantitative polymerase chain reaction [Color figure can be viewed at wileyonlinelibrary.com]
RT-qPCR was performed to measure the expression of LEF1-AS1, miR-330-5p, and LEF1 in tissues. The RT-qPCR results (Figure 1e) showed that compared with the control group, the PCa group had significantly increased LEF1-AS1 and LEF1 expression, while significantly decreased miR-330-5p expression (p < 0.05).
To measure protein expression of LEF1, E-cadherin, N-cadherin, and vimentin, western blot analysis was conducted. The western blot analysis results (Figure 1f,g) indicated compared with the control group, the PCa group had significantly decreased E-cadherin expression, while significantly increased expression of LEF1, N-cadherin, and vimentin (p < 0.05). The above results indicated that LEF1-AS1 and LEF1 were highly expressed in PCa tissues, and miR-330-5p was poorly expressed, and there was obvious EMT in PCa tissues.
RT-qPCR was used to screen cell lines with highly expressed LEF1-AS1. The RT-qPCR screening results (Figure 1h) showed compared with RWPE-1 cell line, there was no significant difference in the expression of LEF1-AS1 in the 22RV1 cell line (p > 0.05), while LEF1-AS1 expression in PC-3 cell line increased significantly (p < 0.05). Therefore, the PC-3 cell line was selected for the following experiments.
3.2 LEF1-AS1 is involved in the occurrence of EMT in PCa
An inverted microscope was used to observe cell morphology. The results of the inverted microscope showed the following (Figure 2a): the cells in the vector group were firmly adhered to the walls, the cells were polygonal, and few particles were visible in the cytoplasm. Compared with the vector group, in the LEF1-AS1 group, the cell wall was stronger and denser, and cells showed irregular polygons (p < 0.05); there was no significant difference in the si-NC group (p > 0.05); in the si-LEF1-AS1 group, cells showed round with irregular edges, increased particles were found in the cytoplasm, cell adhesion ability significantly decreased and suspended cells and cell debris significantly increased (p < 0.05). The above results showed that downregulation of LEF1-AS1 inhibited the activity of PCa cells.
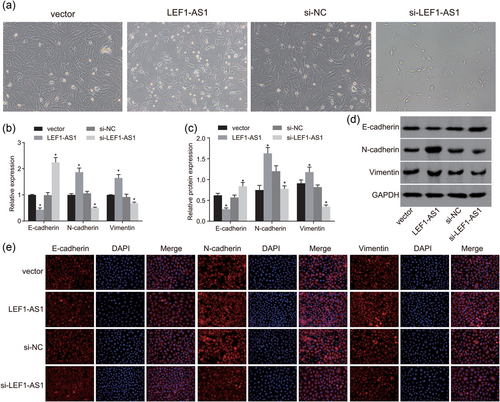
Downregulation of LEF1-AS1 inhibits the development of EMT in PCa. (a) An inverted microscope was used to observe cell growth morphology (×200); (b) RT-qPCR was conducted to measure mRNA expression of E-cadherin, N-cadherin, and vimentin; (c) western blot analysis was used to measure protein expression of E-cadherin, N-cadherin, and vimentin; (d) protein band diagrams of E-cadherin, N-cadherin, and vimentin; (e) immunofluorescence assay was used to detect the expression of E-cadherin, N-cadherin, and vimentin (×400); (f) fluorescence intensity of E-cadherin, N-cadherin, and vimentin. *p < 0.05 versus the vector group. DAPI: 4′,6-diamidino-2-phenylindole; EMT: epithelial–mesenchymal transition; GAPDH: glyceraldehyde-3-phosphate; LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; lncRNA: long noncoding RNA; miR: microRNA; NC: negative control; PCa: prostate cancer; RT-qPCR: reverse-transcription quantitative polymerase chain reaction [Color figure can be viewed at wileyonlinelibrary.com]
To measure mRNA expression of E-cadherin, N-cadherin, and vimentin, RT-qPCR was conducted. It was found that (Figure 2b) that compared with the vector group, the mRNA expression level of E-cadherin in the LEF1-AS1 group decreased significantly, while N-cadherin and vimentin mRNA expression increased significantly (p < 0.05); there was no significant difference in the E-cadherin, N-cadherin, and vimentin mRNA expression level in the si-NC group (p > 0.05); the si-LEF1-AS1 group had notably increased E-cadherin mRNA expression while significantly decreased N-cadherin and vimentin mRNA expression (p < 0.05).
To measure protein expression of E-cadherin, N-cadherin, and vimentin, western blot analysis was conducted. The results showed that (Figure 2c,d) compared with the vector group, the protein expression of E-cadherin in the LEF1-AS1 group decreased significantly, while N-cadherin and vimentin protein expression increased significantly (p < 0.05); there was no significant difference in E-cadherin, N-cadherin, and vimentin protein expression in the si-NC group (p > 0.05); the si-LEF1-AS1 group had notably increased E-cadherin protein expression while significantly decreased N-cadherin and vimentin protein expression (p < 0.05).
An immunofluorescence assay was used to detect the expression of E-cadherin, N-cadherin, and vimentin in each group. The results of the immunofluorescence assay showed the following (Figure 2e,f): E-cadherin, N-cadherin, and vimentin showed red fluorescence in the cells, and the nuclei were dyed blue by DAPI. Under Merge condition, E-cadherin, N-cadherin, and vimentin were mostly distributed in the cell membrane and cytoplasm. Compared with the vector group, the LEF1-AS1 group showed significantly reduced E-cadherin fluorescence, while significantly increased N-cadherin and vimentin fluorescence (p < 0.05); there was no significant difference in E-cadherin, N-cadherin, and vimentin fluorescence in the si-NC group (p > 0.05); the si-LEF1-AS1 group had notably increased E-cadherin fluorescence while significantly decreased N-cadherin and vimentin fluorescence (p < 0.05). The above results showed that downregulation of LEF1-AS1 inhibited the development of EMT in PCa.
3.3 LEF1-AS1 is involved in the proliferation, invasion, migration, tumor formation, and metastasis of PCa
An MTT assay was used to examine the cell proliferation ability in each group. The results showed that (Figure 3a) compared with the vector group, the LEF1-AS1 group had a significantly increased OD value after transfection for 48 and 72 hr (p < 0.05); no significant difference was found in the si-NC group (p > 0.05); the si-LEF1-AS1 group had a notably decreased OD value after transfection for 48 and 72 hr (p < 0.05).
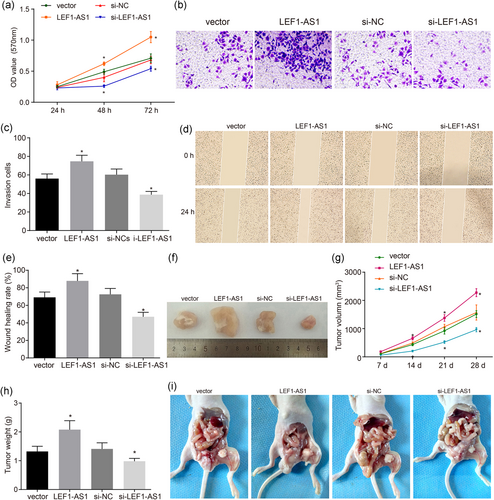
Downregulation of LEF1-AS1 inhibits the proliferation, invasion, migration, tumor formation, and metastasis of PCa. (a) OD value after transfection for 24, 48, and 72 hr; (b) the image of PCa cell invasion involved with LEF1-AS1 (×200); (c) histogram of PCa cell invasion ability involved with LEF1-AS1; (d) PCa cell migration ability involved with LEF1-AS1 (×100); (e) comparison of PCa cell migration involved with LEF1-AS1; (f) the image of PCa cell tumor formation in nude mice involved with LEF1-AS1; (g) tumor volume of PCa involved with LEF1-AS1; (h) histograms of tumor quality of PCa involved with LEF1-AS1; (i) the image of lymph node metastasis of nude mice with PCa involved with LEF1-AS1. *p < 0.05 versus the vector group. LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; lncRNA: long noncoding RNA; miR: microRNA; NC: negative control; OD: optical density; PCa: prostate cancer [Color figure can be viewed at wileyonlinelibrary.com]
A Transwell test was performed to examine cell invasion ability in each group. The results showed that (Figure 3b,c) compared with the vector group, the LEF1-AS1 group had a significantly increased cell invasion ability (p < 0.05); no significant difference was found in the si-NC group (p > 0.05); the si-LEF1-AS1 group had notably decreased cell invasion ability (p < 0.05).
A scratch assay was conducted to examine th cell migration ability in each group. The results showed that (Figure 3d,e) compared with the vector group, the LEF1-AS1 group had a notably elevated cell migration ability (p < 0.05); no significant difference was found in the si-NC group (p > 0.05); the si-LEF1-AS1 group had significantly reduced cell migration ability (p < 0.05).
The transplanted tumor model of nude mice was established to compare the tumor formation and metastasis in each group after transfection. Nude mice tumorigenicity test results showed (Figure 3f–i): no nude mice were found to have visceral organ metastasis, but the vector group, LEF1-AS1 group and si-NC group all had obvious para aortic lymph node metastasis (LNM). Compared with the vector group, the LEF1-AS1 group showed significantly increased cell tumorigenicity and LNM ability (p < 0.05); there was no significant difference in tumorigenicity and LNM ability in the si-NC group (p > 0.05); the si-LEF1-AS1 group had significantly reduced cell tumorigenicity and LNM ability (p < 0.05). The above results all indicated that downregulation of LEF1-AS1 inhibited the proliferation, invasion, migration, tumor formation, and metastasis of PCa.
3.4 Overexpression of miR-330-5p inhibits the occurrence of EMT in PCa
An inverted microscope was used to observe cell morphology. The results of the inverted microscope showed the following (Figure 4a): the cells in the blank and NC groups were firmly adhered to the walls, the cells were polygonal, and few particles were visible in the cytoplasm. Compared with the blank and NC groups, in the miR-330-5p inhibitor group, the cell wall was stronger and denser, and cells showed irregular polygons (p < 0.05); there was no significant difference in the LEF1-AS1 + miR-330-5p mimic and si-LEF1-AS1 + miR-330-5p inhibitor groups (p > 0.05); in the miR-330-5p mimic group, cells showed round with irregular edges, increased particles were found in the cytoplasm, cell adhesion ability significantly decreased and suspended cells and cell debris significantly increased (p < 0.05). The above results showed that upregulation of miR-330-5p inhibited the activity of PCa cells.
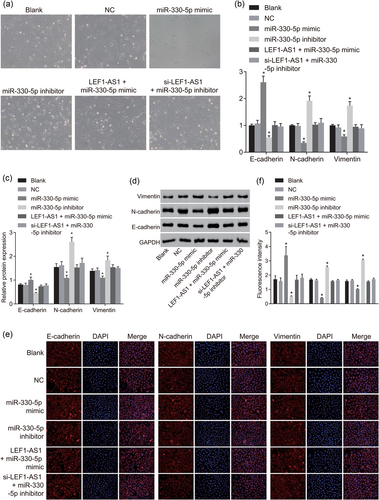
Upregulation of miR-330-5p inhibits the development of EMT in PCa. (a) An inverted microscope was used to observe cell growth morphology (×200); (b) RT-qPCR was conducted to measure mRNA expression of E-cadherin, N-cadherin, and vimentin; (c) western blot analysis was used to measure protein expression of E-cadherin, N-cadherin, and vimentin; (d) protein band diagrams of E-cadherin, N-cadherin, and vimentin; (e) immunofluorescence assay was used to detect the expression of E-cadherin, N-cadherin, and vimentin (×400); (f) fluorescence intensity of E-cadherin, N-cadherin, and vimentin. *p < 0.05 versus the blank and NC groups. DAPI: 4′,6-diamidino-2-phenylindole; EMT: epithelial–mesenchymal transition; GAPDH: glyceraldehyde-3-phosphate; LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; lncRNA: long noncoding RNA; miR: microRNA; NC: negative control; PCa: prostate cancer; RT-qPCR: reverse-transcription quantitative polymerase chain reaction [Color figure can be viewed at wileyonlinelibrary.com]
To measure mRNA expression of E-cadherin, N-cadherin, and vimentin, RT-qPCR was conducted. It was found that (Figure 4b) compared with the blank and NC groups, the mRNA expression of E-cadherin in the miR-330-5p mimic group increased significantly, while N-cadherin and vimentin mRNA expression decreased significantly (p < 0.05); the miR-330-5p inhibitor group had notably decreased E-cadherin mRNA expression while significantly increased N-cadherin and vimentin mRNA expression (p < 0.05); there was no significant difference in E-cadherin, N-cadherin, and vimentin mRNA expression in the LEF1-AS1 + miR-330-5p mimic and si-LEF1-AS1 + miR-330-5p inhibitor groups (p > 0.05).
To measure protein expression of E-cadherin, N-cadherin, and vimentin, western blot analysis was conducted. The results showed that (Figure 4c,d) compared with the blank and NC groups, the protein expression of E-cadherin in the miR-330-5p mimic group increased significantly, while N-cadherin and vimentin protein expression decreased significantly (p < 0.05); the miR-330-5p inhibitor group had notably decreased E-cadherin protein expression while significantly increased N-cadherin and vimentin protein expression (p < 0.05); there was no significant difference in E-cadherin, N-cadherin, and vimentin protein expression in the LEF1-AS1 + miR-330-5p mimic and si-LEF1-AS1 + miR-330-5p inhibitor groups (p > 0.05).
An immunofluorescence assay was used to detect the expression of E-cadherin, N-cadherin, and vimentin in each group. The results of the immunofluorescence assay showed the following (Figure 4e): E-cadherin, N-cadherin, and vimentin showed red fluorescence in the cells, and the nuclei were dyed blue by DAPI. Under Merge condition, E-cadherin, N-cadherin, and vimentin were mostly distributed in the cell membrane and cytoplasm. Compared with the blank and NC groups, the miR-330-5p mimic group showed significantly increased E-cadherin fluorescence, while significantly reduced N-cadherin and vimentin fluorescence (p < 0.05); the miR-330-5p inhibitor group had notably decreased E-cadherin fluorescence while significantly increased N-cadherin and vimentin fluorescence (p < 0.05); there was no significant difference in E-cadherin, N-cadherin, and vimentin fluorescence in the LEF1-AS1 + miR-330-5p mimic and si-LEF1-AS1 + miR-330-5p inhibitor groups (p > 0.05). The above results showed that upregulation of miR-330-5p inhibited the development of EMT in PCa.
3.5 Upregulation of miR-330-5p suppresses the proliferation, invasion, migration, tumor formation, and metastasis of PCa
A MTT assay was used to examine cell proliferation ability in each group. The results showed that (Figure 5a) compared with the blank and NC groups, the miR-330-5p mimic group had a significantly decreased OD value after transfection for 48 and 72 hr (p < 0.05); the miR-330-5p inhibitor group had a notably increased OD value after transfection for 48 and 72 hr (p < 0.05); no significant difference was found in the LEF1-AS1 + miR-330-5p mimic and si-LEF1-AS1 + miR-330-5p inhibitor groups (p > 0.05).
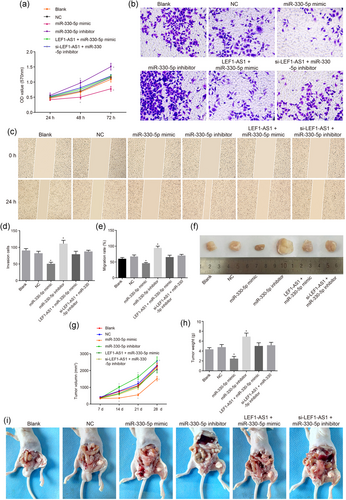
Upregulation of miR-330-5p inhibits the proliferation, invasion, migration, tumor formation, and metastasis of PCa. (a) OD value after transfection for 24, 48, and 72 hr; (b) the image of PCa cell invasion affected by miR-330-5p (×200); (c) histogram of PCa cell invasion ability affected by miR-330-5p; (d) histogram of PCa cell migration ability affected by miR-330-5p; (e) the image of PCa cell migration affected by miR-330-5p (×100); (f) the image of PCa cell tumor formation in nude mice affected by miR-330-5p; (g) tumor volume of PCa affected by miR-330-5p; (h) histograms of tumor quality of PCa affected by miR-330-5p; (i) the image of lymph node metastasis of nude mice with PCa affected by miR-330-5p. *p < 0.05 versus the blank and NC groups. LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; lncRNA: long noncoding RNA; miR: microRNA; OD: optical density; PCa: prostate cancer [Color figure can be viewed at wileyonlinelibrary.com]
A Transwell test was performed to examine cell invasion ability in each group. The results showed that (Figure 5b,d) compared with the blank and NC groups, the miR-330-5p mimic group had significantly decreased cell invasion ability (p < 0.05); no significant difference was found in the LEF1-AS1 + miR-330-5p mimic and si-LEF1-AS1 + miR-330-5p inhibitor groups (p > 0.05); the miR-330-5p inhibitor group had notably increased cell invasion ability (p < 0.05).
A scratch assay was conducted to examine cell migration ability in each group. The results showed that (Figure 5c,e) compared with the blank and NC groups, the miR-330-5p mimic group had notably decreased cell migration ability (p < 0.05); no significant difference was found in the LEF1-AS1 + miR-330-5p mimic and si-LEF1-AS1 + miR-330-5p inhibitor groups (p > 0.05); the miR-330-5p inhibitor group had significantly elevated cell migration ability (p < 0.05).
The transplanted tumor model of nude mice was established to compare the tumor formation and metastasis in each group after transfection. Nude mice tumorigenicity test results showed the following (Figure 5f–i): no nude mice were found to have visceral organ metastasis, but the blank group, NC group, miR-330-5p inhibitor group, LEF1-AS1 + miR-330-5p mimic group, and si-LEF1-AS1 + miR-330-5p inhibitor group all had obvious para aortic LNM. Compared with the blank and NC groups, the miR-330-5p mimic group showed significantly decreased cell tumorigenicity and LNM ability (p < 0.05); there was no significant difference in tumorigenicity and LNM ability in the LEF1-AS1 + miR-330-5p mimic and si-LEF1-AS1 + miR-330-5p inhibitor groups (p > 0.05); the miR-330-5p inhibitor group had significantly increased cell tumorigenicity and LNM ability (p < 0.05). The above results all indicated that upregulation of miR-330-5p inhibited the proliferation, invasion, migration, tumor formation, and metastasis of PCa.
3.6 LEF1 is involved in the proliferation, migration, invasion, and EMT of PCa
From the above results, we can see that both lncRNA LEF1-AS1 and miR-330-5p are involved in regulating the biological function of PCa. To further study whether LEF1 has an impact on the biological characteristics of PCa, we set up different groups to detect the mRNA and protein levels of LEF1 in each group by RT-qPCR and western blot analysis. The results showed that the mRNA and protein levels of LEF1 were significantly lower in the si-LEF1 group than in the si-NC group, while the mRNA and protein levels of LEF1 were significantly higher in the LEF1-AS1 + si-LEF1 group than in the si-LEF1 group (p < 0.05), and there was no significant difference in the mRNA and protein levels of LEF1 between the si-NC group and the si-LEF1 group (Figure 6a–c). First, the MTT assay showed that compared with the si-NC group, the OD value of cells in the si-LEF1 group was significantly decreased; compared with the si-LEF1 group, the OD value of cells in the LEF1-AS1 + si-LEF1 group was significantly increased (p < 0.05), and there was no significant difference in the proliferation ability of cells between the si-NC group and the si-LEF1 group (Figure 6d), indicating that after si-LEF1 transfection, LEF1-AS1 would reverse the effect of LEF1 on decreasing cell proliferation ability. Furthermore, we tested the effects of LEF1 on cell migration and invasion by scratch test and Transwell assay. The results showed that (Figure 6e–h) compared with the si-NC group, the ability of cell migration and invasion in the si-LEF1 group decreased significantly; compared with the si-LEF1 group, the ability of cell invasion in the LEF1-AS1 + si-LEF1 group increased significantly (p < 0.05), and there was no significant difference in the invasion ability of cells between the si-NC group and the si-LEF1 group.
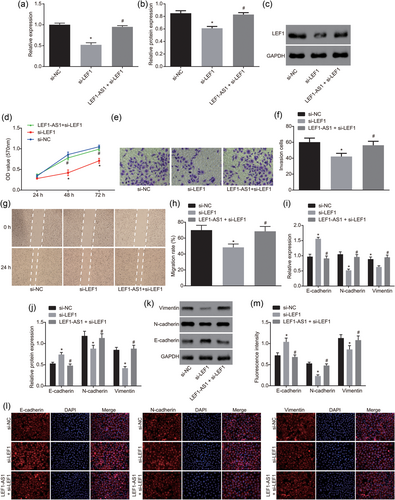
LEF1 is involved in the proliferation, migration, invasion, and EMT of PCa. (a) The relative mRNA expression of LEF1 was detected by RT-qPCR; (b, c) the protein level of LEF1 was detected by western blot analysis; (d) the proliferation of cells in each group was detected by MTT; (e and f) the invasion ability of cells in each group (×200) was detected by Transwell assay; (g and h) the migration ability of cells in each group (×100) was detected by scratch test; (i) mRNA expression of EMT markers was detected by RT-qPCR; (j and k) protein levels of EMT markers was detected by western blot analysis; (l and m) fluorescence intensity of EMT markers was detected by immunofluorescence. All the data were expressed by mean and standard deviation. T test was used between the two groups and one-way analysis of variance was used among the multiple groups. The experiment was repeated three times. *p < 0.05 versus the si-NC group; #p < 0.05 versus the si-LEF1 group. DAPI: 4′,6-diamidino-2-phenylindole; GAPDH: glyceraldehyde-3-phosphate; LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1, lymphoid enhancer-binding factor-1 antisense RNA 1; mRNA: messenger RNA; NC: negative control; OD: optical density; PCa: prostate cancer; RT-qPCR: reverse-transcription quantitative polymerase chain reaction [Color figure can be viewed at wileyonlinelibrary.com]
In addition, we detected the mRNA and protein levels of EMT-related markers by RT-qPCR and western blot analysis. Compared with the si-NC group, the mRNA and protein levels of N-cadherin and vimentin in the si-LEF1 group were significantly decreased, while the mRNA and protein levels of E-cadherin were significantly increased. Compared with the si-LEF1 group, the mRNA and protein levels of N-cadherin and vimentin in the LEF1-AS1 + si-LEF1 group were significantly increased, while the mRNA and protein levels of E-cadherin were significantly decreased (p < 0.05), and there was no significant difference in the levels of EMT-related markers between the si-NC group and the si-LEF1 group (Figure 6i–k). Immunofluorescence assay found that E-cadherin, N-cadherin, and vimentin showed red fluorescence in the cells and the nucleus was stained blue by DAPI. Under Merge condition, E-cadherin, N-cadherin, and vimentin were mainly distributed in the cell membrane and cytoplasm. Compared with the si-NC group, the fluorescence of E-cadherin in the si-LEF1 group was significantly higher, and the fluorescence of N-cadherin and vimentin were significantly lower (all p < 0.05); there was no statistical significance in E-cadherin, N-cadherin, and vimentin fluorescence in the LEF1-AS1 + si-LEF1 group (all p > 0.05). These results suggest that downregulation of LEF1 inhibits the development of EMT in PCa.
3.7 Downregulation of LEF1-AS1 or upregulation of miR-330-5p inhibits LEF1 expression
As shown in Figure 7a, lncRNA cell sublocation site analysis showed that LEF1-AS1 was concentrated in the nucleus and cytoplasm. The FISH experiment was used to analyze the location of LEF1-AS1 in cells. The results indicated that (Figure 7b) LEF1-AS1 did not show fluorescence in the NC probe group, and the nucleus was stained blue by DAPI. Under Merge condition, the nucleus showed blue fluorescence but no red fluorescence in LEF1-AS1. In the LEF1-AS1 probe group, LEF1-AS1 showed red fluorescence, and the nuclei were stained blue by DAPI. Under the condition of Merge, the two showed red and blue fluorescence together, and the red fluorescence displayed by LEF1-AS1 was in the nucleus. The above results showed that LEF1-AS1 was concentrated in the nucleus.
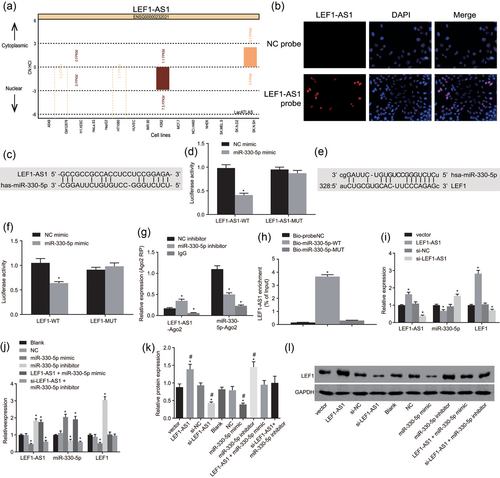
Downregulated LEF1-AS1 or upregulated miR-330-5p inhibits LEF1 expression. (a) Subcellular location website was used to analyze the location of LEF1-AS1 in cells; (b) FISH was used to detect the location of LEF1-AS1 in cells (×400); (c) the predictive binding site for miR-330-5p and LEF1-AS1–3′-UTR; (d) Wt-miR-330-5p/LEF1-AS1 cotransfection luciferase activity. *p < 0.05 versus the NC mimic group; (e) the predictive binding site for miR-330-5p and LEF1–3′-UTR; (f) Wt-miR-330-5p/LEF1 cotransfection luciferase activity; *p < 0.05 versus the NC mimic group; (g) RIP detects the combination of LEF1-AS1 and miR-330-5p with Ago2; *p < 0.05 versus the NC inhibitor group; (h) pull down test detects the enrichment of LEF1-AS1. *p < 0.05 versus the Bio-probe NC group; (i) the expression of LEF1-AS1, miR-330-5p, and LEF1 by regulating LEF1-AS1 expression; *p < 0.05 versus the vector group; (j) the expression of LEF1-AS1, miR-330-5p, and LEF1 by regulating miR-330-5p expression; *p < 0.05 versus the blank and NC groups; (k) western blot analysis was used to determine LEF1 protein expression; *p < 0.05 versus the vector group; #p < 0.05 versus the blank and NC groups; (l) protein band of LEF1. DAPI: 4′,6-diamidino-2-phenylindole; FISH: fluorescence in situ hybridization; IgG: immunoglobulin G; LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; lncRNA: long noncoding RNA; miR: microRNA; NC: negative control; PCa: prostate cancer [Color figure can be viewed at wileyonlinelibrary.com]
The target relationship between LEF1-AS1 & LEF1 and miR-330-5p was analyzed by using an online analysis software. There was a specific binding area between LEF1-AS1 and LEF1 gene sequences and miR-330-5p sequence. LEF1-AS1 was ceRNA of miR-330-5p (Figure 7c) and LEF1 was the target gene of miR-330-5p (Figure 7e). The luciferase report showed that LEF1-AS1 and LEF1 were the targets of miR-330-5p (Figure 7d,f). Compared with the NC mimic group, in Wt-miR-330-5p/LEF1-AS1 and Wt-miR-330-5p/LEF1 cotransfection group of miR-330-5p mimic transfection group, the luciferase signal decreased (p < 0.05); but there was no significant difference in mutant 3′-UTR luciferase activity (p > 0.05). The above results indicated that miR-330-5p can specifically bind LEF1-AS1 and LEF1 genes.
RIP test results showed (Figure 7g) that compared with the NC inhibitor group, in the miR-330-5p inhibitor group, the amount of LEF1-AS1 precipitated by the Ago2 antibody increased significantly while the amount of miR-330-5p decreased significantly (p < 0.05); in the IgG group, the amount of LEF1-AS1 and miR-330-5p precipitated by Ago2 antibody reduced notably (p < 0.05). The aforementioned results showed that LEF1-AS1 was the ceRNA of miR-330-5p.
A pull down test was used to detect the combination of LEF1-AS1 and miR-330-5p. Pull down test results showed the following (Figure 7h): LEF1-AS1 expression in the Bio-miR-330-5p-WT group was significantly higher than that in the Bio-probe NC group (p < 0.05), and Bio-miR-330-5p expression in the Bio-miR-330-5p-MUT group had no significant difference (p > 0.05). These results showed that Bio-miR-330-5p-WT promoted the enrichment of LEF1-AS1, but Bio-miR-330-5p-MUT cannot enrich LEF1-AS1. LEF1-AS1 competed for the binding sites of miR-330-5p and reduced the expression level of miR-330-5p.
RT-qPCR was performed to measure the expression of LEF1-AS1, miR-330-5p, and LEF1. The RT-qPCR results (Figure 7i,j) showed that compared with the vector group, the LEF1-AS1 group had significantly increased LEF1-AS1 and LEF1 expression while notably decreased miR-330-5p expression (all p < 0.05). There was no significant difference in the expression of each factor in the si-NC group (p > 0.05). In the si-LEF1-AS1 group, the expression of LEF1-AS1 and LEF1 decreased significantly, and the expression of miR-330-5p increased significantly (p < 0.05). Compared with the blank and NC groups, the miR-330-5p mimic group had remarkably reduced LEF1-AS1 and LEF1 expression while notably increased miR-330-5p expression (p < 0.05); the miR-330-5p inhibitor group showed significantly increased LEF1-AS1 and LEF1 expression while obviously decreased miR-330-5p expression (p < 0.05); the LEF1-AS1 + miR-330-5p mimic group had significantly increased LEF1-AS1 and miR-330-5p expression (p < 0.05) while no significant difference in LEF1 expression (p > 0.05); the si-LEF1-AS1 + miR-330-5p inhibitor group had significantly reduced LEF1-AS1 and miR-330-5p expression (p < 0.05) while no significant difference in LEF1 expression (p > 0.05). The above results suggested that LEF1-AS1 and miR-330-5p inhibited each other's expression so as to regulate the expression of LEF1.
Western blot analysis was performed to determine protein expression of LEF1. As shown in Figure 7k,l, compared with the vector group, the LEF1-AS1 group and miR-330-5p inhibitor group had significantly increased LEF1 protein expression (p < 0.05) while the si-LEF1-AS1 group and miR-330-5p mimic group had significantly decreased LEF1 protein expression (p < 0.05); there was no significant difference in LEF1 protein expression between other groups (p > 0.05). Compared with the blank and NC groups, the trend of each group was the same as above. The aforementioned findings indicated that downregulation of LEF1-AS1 or upregulation of miR-330-5p inhibited LEF1 expression.
4 DISCUSSION
PCa is the most incident male cancer in the world with risk factors of ethnicity, advanced age, and a genetic history of this cancer (Chang, Cui, & Song, 2018). It has been found that metastasis increases the risk for cancer death, and sequential and interrelated events are involved in cancer metastasis, in which miRs and EMT play important roles (C. Liu, Guan et al., 2015). EMT has been found to be a significant factor for the development of PCa (C. H. Liu, Tang et al., 2015). Besides that, lncRNAs are found to have great influence on the development and progression of cancers (Yang et al., 2014). In the present study, it was found that downregulated LEF1-AS1 could bind to miR-330-5p to inhibit the EMT, cell invasion and migration of PCa by suppressing LEF1 expression.
Initially, one of our major results was that LEF1-AS1 and LEF1 were highly expressed while miR-330-5p was poorly expressed in PCa. LEF1-AS1 and LEF1 were the targets of miR-330-5p. Downregulation of LEF1-AS1 promoted miR-330-5p to inhibit LEF1 expression. And downregulation of LEF1 inhibits the development of EMT in PCa. LEF1, as a member of the lymphoid enhancer-binding factor/T-cell factor (LEF/TCF) family, interacts with nuclear β-catenin and functions as a critical transcriptional regulator of Wnt signaling pathway (Kobayashi & Ozawa, 2013). LEF1 was found to be necessary for upregulation of the mesenchymal marker vimentin independent of β-catenin activation (Nawshad, Medici, Liu, & Hay, 2007). Wnt is a complicated signaling pathway, which plays a pivotal role in the development and progression of many epithelial cancers such as PCa (Terry, Yang, Chen, Vacherot, & Buttyan, 2006). A previous study shows that LEF1, a transcriptional mediator of Wnt signaling pathway, is notably increased in ERG-high human PCa (Wu et al., 2013). Another study also indicated that LEF1 is highly expressed in androgen-independent PCa, and has the potential to be a biomarker for androgen-independent disease (Li et al., 2009). It has been reported that LEF1 high expression in PCa cells has a close relation to EMT and aggressive malignancy and miRs may be responsible for the phenotype through LEF1 overexpression (Liang, Li, Li et al., 2015). Moreover, it has been found that miR-330-5p is downregulated in pancreatic cancer and serves as a novel tumor suppressive miR, which can inhibit the development of pancreatic cancer (Trehoux et al., 2015). A recent study shows that miR-330-5p has downregulation in cutaneous malignant melanoma (CMM) and upregulation of miR-330-5p suppresses the proliferation and migration of CMM cells by regulating TYR and PDIA3 protein (Su, Zhou, Gan, & Zhang, 2016). Besides that, as previously reported, miR-26b is downregulated in some cancers including colon cancer, and miR-26b directly targets LEF1 as well as reduces LEF1 expression (Zhang et al., 2014). Another study also finds that miR-34a and miR-223 directly target and downregulate LEF1 (Rodriguez-Ubreva et al., 2014).
Additionally, this study also indicated that downregulation of LEF1-AS1 or upregulation of miR-330-5p inhibited EMT, invasion, migration, tumor formation, and metastasis of PCa. EMT is important to the metastasis of PCa and plays a critical part in deregulating the androgen axis (M. Wang, Liu et al., 2015). A former study shows that EMT is closely related to the cancer stem-like (CD44+) cell in the metastasis of PCa, which may lead to the failure of androgen deprivation treatment (Shang et al., 2015). Besides that, LEF1 plays a pivotal role in stem cell maintenance, organ development, and EMT through activation of EMT-related genes including N-cadherin, vimentin, and Snail. Abnormal LEF1 expression is associated with tumor formation, cell proliferation, migration, and invasion in cancers (Santiago, Daniels, Wang, Deng, & Lee, 2017). Increasing evidence demonstrates that LEF1-AS1 plays an important part in tumor formation and metastasis of colorectal cancer (Chen, Yu, Xu, & Shen, 2017). LEF1-AS1 expression is also reported to be remarkably upregulated in GBM and LEF1-AS1 silencing notably inhibits the progression of GBM, including cell proliferation and invasion (J. Wang et al., 2017). A previous study found that a new lncRNA called hepatocellular carcinoma-associated lncRNA, as a ceRNA, can regulate LAPTM4B expression through the competitive binding to common miRs, including miR-15a, miR-196a, and miR-196b, thereby promoting cell growth and metastasis of hepatocellular carcinoma (Xie et al., 2017). Moreover, a previous study showed that miR-330-3p is negatively related to EMT genes in melanoma (D. Wang, Li et al., 2015). It is also found that miR-330-5p exerts great influence on suppressing the proliferation and migration of keratinocytes through the regulation of Pdia3 (Kim, Yoo, Choi, & Yoon, 2015). Another study indicated that miR-330-5p directly targets and inhibits ITGA5 in GBM cells, and upregulation of miR-330-5p and downregulation of ITGA5 inhibit cell proliferation, invasion, and migration (Feng, Ma, Ji, Liu, & Hu, 2017). Evidence also shows that miR-34a and LEF1 cooperatively affect cell invasion of PCa and miR-34a has negative effect on the migration and invasion of PCa cells through LEF1 (Liang, Li, Daniels et al., 2015).
In summary, this study demonstrated that downregulation of LEF1-AS1 or upregulation of miR-330-5p suppressed EMT, cell invasion and migration of PCa by downregulating LEF1 expression (Figure 8). The obtained results suggest that LEF1-AS1 could be a potential predictive target for the treatment of PCa. However, due to the limited sample size and experimental conditions, further studies are needed to define the detailed mechanisms of LEF1-AS1 in treating PCa.
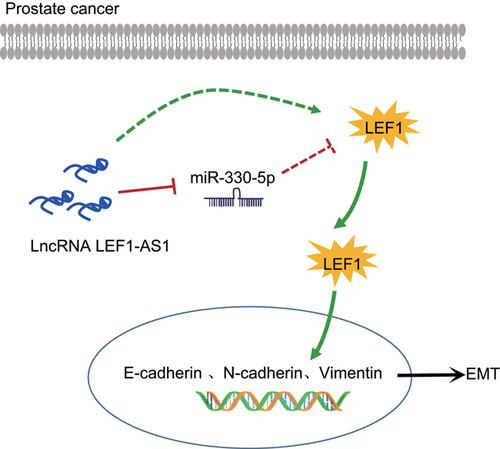
A schematic diagram depicts the molecular basis of LEF1-AS1/miR-330-5p/LEF1 in PCa. In PCa, the expression of LEF1-AS1 is upregulated, which could act as a ceRNA to sponge to miR-330-5p, thereby promoting the EMT process by activating LEF1. EMT: epithelial–mesenchymal transition; LEF1: lymphoid enhancer-binding factor-1; LEF1-AS1: lymphoid enhancer-binding factor-1 antisense RNA 1; lncRNA: long noncoding RNA; miR: microRNA; PCa: prostate cancer [Color figure can be viewed at wileyonlinelibrary.com]
ACKNOWLEDGMENTS
This project was supported by the Natural Science Foundation of Jiangsu Province (BK20151158), the Youth Medical Key Talent Project of Jiangsu Province (QNRC2016389, QNRC2016386), the Innovation Team Project of Xuzhou Central Hospital (XZB201607, XZB201610), the Key Research and Development Plan Project of Jiangsu Province (BE217635, BRA2017294). We would like to acknowledge the helpful comments on this paper received from our reviewers.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.