Tenascin cytotactin epidermal growth factor-like repeat binds epidermal growth factor receptor with low affinity
Abstract
Select epidermal growth factor (EGF)-like (EGFL) repeats of human tenascin cytotactin (tenascin C) can stimulate EGF receptor (EGFR) signaling, but activation requires micromolar concentrations of soluble EGFL repeats in contrast to subnanomolar concentrations of classical growth factors such as EGF. Using in silico homology modeling techniques, we generated a structure for one such repeat, the 14th EGFL repeat (Ten14). Ten14 assumes a tight EGF-like fold with truncated loops, consistent with circular dichroism studies. We generated bound structures for Ten14 with EGFR using two different approaches, resulting in two distinctly different conformations. Normal mode analysis of both structures indicated that the binding pocket of EGFR exhibits a significantly higher mobility in Ten14–EGFR complex compared to that of the EGF–EGFR complex; we hypothesized this may be attributed to loss of key high-affinity interactions within the Ten14–EGFR complex. We proved the efficacy of our in silico models by in vitro experiments. Surface plasmon resonance measurements yielded equilibrium constant KD of 74 µM for Ten14, approximately three orders of magnitude weaker than that of EGF. In accordance with our predicted bound models, Ten14 in monomeric form does not bind EGFR with sufficient stability so as to induce degradation of receptor, or undergo EGFR-mediated internalization over either the short (20 min) or long (48 h) term. This transient interaction with the receptor on the cell surface is in marked contrast to other EGFR ligands which cause EGFR transit through, and signaling from intracellular locales in addition to cell surface signaling. J. Cell. Physiol. 211: 748–758, 2007. © 2007 Wiley-Liss, Inc.
EGFR belongs to the ErbB family of Type I receptor tyrosine kinases, and plays an integral role in regulating cellular functions (Wells, 1999, 2000). Active EGFR signals from the cell surface and intracellularly as it is internalized; intracellular signaling is qualitatively distinct from surface signaling and likely promotes proliferation over migration (Haugh et al., 1999a,b; Pennock and Wang, 2003). The activity of EGFR is shut off by dephosphorylation when unliganded, and over a longer term by lysosomal degradation secondary to internalization. Thus, persistence and subcellular localization of receptor occupancy would impact cellular response from EGFR activation.
EGFR is activated by the (EGF)-like (EGFL) family of soluble ligands which includes EGF, transforming growth factor alpha (TGFα), heparin-binding EGF, amphiregulin, and a number of virally encoded factors (Citri and Yarden, 2006). These peptides are characterized by an EGFL domain consisting of a sequence distribution of six cysteines that form disulfide bridges, giving them a tight and closely packed structure (Carpenter and Cohen, 1990). They bind the extracellular (EC) domains I and III of EGFR with very high affinity (Kim et al., 2002); the physiological affinities of EGF and TGFα for EGFR are in the very low nanomolar range (Wells, 1999). EGFL domains are present in other proteins families, including a number of extracellular matrix (ECM) proteins (tenascin, fibrillin 1, del1, laminin, thrombospondin 1), and are arranged typically as an array of sequential EGFL repeats (Hohenester and Engel, 2002). These EGFL repeats have disulphide bonds similar to EGF, and this intra-molecular cross-linking of the cysteines is essential for function (Zanuttin et al., 2004). With few exceptions, little is known about the function of these matrix EGFL repeat domains.
Human tenascin C is an ECM protein re-expressed during normal tissue regeneration, and implicated in tumor progression (Tsunoda et al., 2003). Interestingly, its expression profile coincides with active cell migration and proliferation, properties similar to those elicited by EGFR activation (Chen et al., 1994a; Jones and Jones, 2000). It is a hexabrachion with an array of 84 full and 6 half EGFL repeats, a fibronectin-type III array and a terminal fibrinogen globe (Jones and Jones, 2000). Recently, we demonstrated that select EGFL repeats of human tenascin C (e.g., the 14th repeat, Ten14), when presented in soluble form, can signal through EGFR in a receptor-dependent fashion (Swindle et al., 2001). However, micromolar concentrations of Ten14 were required to elicit responses comparable to those observed with EGF in the nanomolar range. A similar function has been reported for EGFL repeats from laminin V (Schenk et al., 2003).
The finding that EGFL repeats can signal through a classical receptor such as EGFR invited attention on a new class of receptor ligands, matrikines (Schenk and Quaranta, 2003; Tran et al., 2004) that are encoded as part of larger matrix components. The significantly lower affinities of these ligands would reflect the matrix-constrained situation of their physiological environment, in which limited diffusion and multimeric presentation would result in avidities approximately three orders of magnitude greater than individual soluble affinity constants. However, how this low affinity binding would be accomplished at the submolecular level has evaded explanation.
We hypothesized that the low affinity of Ten14 for EGFR is driven by weak inter-residue contacts with the receptor due to deletions and substitutions of key residues in the EGFL-binding domain of Ten14 required for tight binding, resulting in a more flexible mode of interaction that could accommodate a constraining environment. EGFR binding of ligands is usually accompanied by conformational changes in the complex that accommodate/optimize the interactions in the bound form (De Crescenzo et al., 2000). Our postulate assumes an enhancement in this type of conformational flexibility and its persistence even after binding.
Structural analysis of the complex showed that though Ten14 lacks the C-terminal loop present in EGF and TGFα found to be important for high affinity interaction with EGFR (Kramer et al., 1994), other structural contacts are established that may be sufficient for activation of receptor. Accordingly, a much weaker KD and increased mobility for the Ten14-EGFR interaction is observed as compared to EGF. As a result, Ten14 is neither internalized nor degraded over short- and long-term signaling via EGFR, and leads to compartmentalization of EGFR at the cell surface. This may lead to altered biochemical and biophysical signaling responses downstream of the receptor. An effort into characterizing the interaction of EGFL repeats with cell surface receptors has not been undertaken before, and elucidation of mechanistics of EGFL repeat-mediated signaling will allow for a more complete understanding of this new class of low-affinity growth factors embedded within the ECM.
Materials and Methods
Structure Prediction for Ten14
Homology modeling and ab initio techniques were used to predict the 3D structure of Ten14. The chains corresponding to the active conformations of TGFα (PDB code IMOX-chain C (Ogiso et al., 2002)) and EGF (PDB code 1IVO-chainC (Garrett et al., 2002)) were chosen as templates. Sequence analyses for Ten14, TGFα and EGF were first performed using CLUSTALW (Pearson and Lipman, 1988). Model structures were obtained using three servers—Robetta (Kim et al., 2004), ESyPred3D (Lambert et al., 2002) and Swiss-Model (Schwede et al., 2003). Four queries were submitted to Robetta: Ten14 without a template (ab initio), Ten14 with TGFα as the template, Ten14 with EGF as the template and Ten14 with TGFα–EGFR as the template. In all, 25 models were obtained—ten for Ten14 without template and five each for the other three prediction queries with templates. For predictions with ESyPred3D and Swiss-Model, one model was obtained from each server with TGFα as template, resulting in a total of 27 models for Ten14. Distance root-mean-square deviations (dRMSD) between each model and the known TGFα and EGF structures were then calculated The Ten14 model with the lowest dRMSD was chosen for docking with EGFR.
Circular Dichroism (CD) Measurements
CD spectra for Ten14 were recorded in 10 mM phosphate buffer, pH 7.4, using an AVIV 202 series CD spectrophotometer (Lakewood, NJ) held at 25°C with a thermostatically controlled cell holder in a fused quartz cell with a path-length of 0.1 cm. For protein concentration of 0.18 mg/ml, ten spectra measured every 1 nm in the far UV-region (185–280 nm) were averaged. CD spectra were subjected to subtraction from buffer blank, normalization and smoothing, using the AVIV data manipulation software. Analysis of the data was carried out with the program CDSSTR, which used seven reference datasets, and is available through the DICHROWEB web server at www.cryst.bbk.ac.uk/cdweb/html/home.html (Lobley et al., 2002; Whitmore and Wallace, 2004).
Docking of Ten14 With EGFR
Ten14 was docked onto EGFR in two binding conformations. EGF-EGFR and TGFα–EGFR crystal structures were used as reference. For Structure I, we used the results from “Consensus” server (Prasad et al., 2003, 2004), which indicated that residues 21 through 31 in Ten14 had good overlap with active EGF and TGFα. Hence, we superimposed the co-ordinates of residues numbered 21 through 31 from Ten14 (the model with lowest dRMSD was chosen) onto corresponding residues in TGFα, in addition to matching the corresponding cysteines in Ten14 and TGFα. The remaining residues were transposed so as to maintain the overall structure of Ten14. For Structure II, Ten14 was docked to EGFR in a manner so as to allow for the anchoring interaction of Leu12 of Ten14 with the hydrophobic pocket in domain I of EGFR, determined by Leu14, Leu69, and Leu98 of receptor. This transposition of the entire Ten14 structure also led to a favorable interaction between Arg19 of Ten14 and Asp354 of EGFR (a residue forming a key salt bridge with EGF and TGFα). Finally, with only the backbone atoms of Ten14 and all atoms of EGFR fixed, the docked structure was minimized using CHARMM. Structures I and II for Ten14-EGFR were then analyzed using the Gaussian network model (GNM) and “FastContact.”
Structure Evaluation
The binding dynamics of the ligands complexed with EGFR were analyzed using the iGNM database and server (http://ignm.ccbb.pitt.edu/). iGNM generates residue mobilities in different modes of motion accessible near native conditions. We examined the most cooperative (lowest frequency) modes for EGFR structure alone (chain A from 1 MOX), and for the complexes of EGFR with EGF (chains A and B from 1IVO), with TGFα (chains A and B from 1 MOX), and with Ten14 (Structures I and II). The slow mode fluctuations for receptor alone and with ligand were visualized using both color-coded ribbon diagrams and mobility distribution curves (eigenvectors) as a function of residue index.
The key residues in ligand and receptor that contribute towards favorable and unfavorable interactions were identified using the software “FastContact” (Camacho and Zhang, 2005). This software identifies interactions between ligand and receptor residues that contribute maximally towards overall electrostatic and desolvation energies, and total binding energy. The resulting high affinity interactions between both structures of Ten14-EGFR were compared against those occurring in the EGF–EGFR and TGFα–EGFR complexes.
Surface Plasmon Resonance (SPR) Analysis
Ten14 binding kinetics was examined by SPR using the BIAcore 3000 system. Expression and purification of Ten14 was performed as described previously (Swindle et al., 2001). Recombinant human EGF (hEGF) (BD Biosciences, Bedford, MA) was used as the control analyte. The Ten14 and hEGF runs were performed on separate chips; each experimental series was repeated. For both runs, 25 µg/ml of recombinant EC domain of EGFR (EGFR-ED) (Research Diagnostic Inc., Flanders, NJ) in 10 mM sodium acetate (pH 5.0) was cross-linked to a CM5 sensor chip surface using the EDC/NHS coupling method (Amine coupling kit, BIAcore Inc., Uppsala, Sweden), resulting in immobilizations of ∼7,000 resonance units (RUs) of EGFR-ED for Ten14 studies and ∼2,500 RUs for hEGF studies. Separate flow cells from each chip derivatized without EGFR-ED were used as control. For Ten14, sterile 0.22 µm filtered phosphate buffered saline (PBS) was used as the running buffer and 10 mM Glycine-HCl, pH 3.0 (Regeneration kit, BIAcore Inc.) was used for regeneration of the chip surface. Increasing concentrations of Ten14 (1.88, 3.75, 7.5, 15, and 30 µM) or hEGF (0.03, 0.06, 0.125, 0.25, 0.5, 1, 3, and 10 µM) were then injected into their respective flow cells. For each concentration of Ten14, 30 min of association, dissociation and regeneration cycles were used, all at a flow rate of 5 µl/min. Increasing concentrations of human EGF was prepared in sterile HBS-EP buffer (BIAcore Inc.), and injected over the surfaces at flow rate of 30 µl/min, with a 10 min association pulse and 15 min of dissociation, without the need for a regeneration step. The sensograms obtained for each ligand concentration for all runs were graphed and analyzed for steady state binding using the BIAEvaluation software (BIAcore Inc.).
Internalization and Ligand Depletion Assays
For all in vitro experiments, NR6WT fibroblasts expressing ∼100,000 human EGFR/cell were cultured and maintained as described previously, and quiesced in medium containing 0.5% dialyzed serum (Wells et al., 1990; Chen et al., 1994a). Depletion assays were performed by incubating quiesced NR6WT cells in medium containing various concentrations of murine EGF (mEGF) or Ten14. Cells were lysed and growth factor concentrations determined for time 0 and 48 h via immunoblotting using anti-Xpress™ antibody (Invitrogen, Carlsbad, CA). To measure ligand internalization, quiesced NR6WT cells were incubated in binding buffer at 37°C. I125-EGF (0.6 nM) and I125-Ten14 (100 nM) were introduced for varying time points at 37°C. After washing, cell-surface associated ligand was removed with stripping buffer (HCl, pH 2.0). Cells were then lysed with 1 N NaOH. Internalization was measured as counts per minute (CPM) on a Packard 5005 Cobra Gamma Counter. To measure EGF receptor internalization and degradation, mEGF (1 nM) and Ten14 (2 µM) were introduced into the media at various time points at 37°C. Samples were collected and total EGFR levels were determined using immunoblotting utilizing the monoclonal EGFR antibody (BD Transduction Labs, San Jose, CA). Equal loading was assured using the anti-GAPDH antibody (Abcam Inc., Cambridge, MA). Relative densitometric values were derived with NIH image shareware v1.63 and Adobe Photoshop. Each experiment was repeated at least twice.
Immunofluorescence Assays
To assess localization of active EGFR, 10,000 NR6WT cells quiesced on glass coverslips and treated with increasing concentrations of mEGF (10 nM, 1 nM) or Ten14 (1 µM, 0.1 µM, 0.01 µM) for 30 min. After washing with cold PBS, cells were fixed in 3% paraformaldehyde for 30 min and lysed for 30 min with buffer containing 1% Triton X-100, 1 mM PMSF and 1 µg/ml aprotonin, followed by blocking in 5% BSA. In order to assess the localization of total versus phosphorylated EGFR, cells were incubated for 3 h at room temperature in a mixture of rabbit polyclonal anti-EGFR antibody (Santa Cruz Biotech., Santa Crux, CA) and mouse monoclonal phospho-EGFR antibody (Upstate, Chicago, IL), both at a final concentration of 5 µg/ml. After a brief wash in PBS containing 0.5% BSA, coverslips were incubated in a mixture of Alexa Fluor 647 anti-mouse secondary antibody, Alexa Fluor 488 anti-rabbit secondary antibody (both at 1 µg/ml) and 25 µg/ml propidium iodide at room temperature for 30 min. In order to assess the co-localization of ligand and receptor, slips treated with 1 µM Ten14 or 10 nM mEGF were incubated with a mixture of mouse monoclonal anti-Xpress™ antibody (Invitrogen) and rabbit polyclonal anti-EGFR antibody, followed by appropriate secondary antibodies. After a last wash, the slips were washed and mounted onto glass slides using gelvatol. After overnight drying, the slides were imaged using a Zeiss Axioplan confocal laser-scanning microscope, with each one imaged simultaneously for all channels. Each image was scanned along the Z-axis in 7–10 sectional planes with 0.43 µm steps (512 × 512 pixels per sectional plane). Images were collected and analyzed using Adobe Photoshop ver. 6.0. All RGB images were first pasted onto a canvas and RGB levels were adjusted from 0 to 128 bits achieve an optimal signal to noise ratio. Individual channel images were then separated and pasted separately onto another canvas to display green, red, and composite images.
Results
Predicted Structure for Ten14 Conforms to Other EGFR Ligands
Ten14 shares low but adequate sequence homology with EGF (25%) and TGFα (32%) for homology modeling techniques (Fig. 1A). Of the 27 models generated, the best model was selected from dRMSD calculations based on inter-residue distances between Cα atoms of 11 conserved residues: Cys5, Pro6, Cys9, Gly13, Cys15, Cys20, Cys22, Gly25, Tyr26, Gly28, and Cys31 (numbers refer to residue number in Ten14 sequence; corresponding residues were selected from EGF and TGFα as shown in Fig. 1A). The dRMSD value for the selected model was less than 1 Å when compared to EGF and TGFα (Supplemental Table 1). This structure was essentially recapitulated when we fixed the conserved amino acids and minimized the energies (data not shown), providing validation of the homology model. Ten14 is predicted to be composed of an anti-parallel β-hairpin encompassing the six backbone cysteines and an N- and C-terminal loop (Fig. 1B). This agrees well with the experimentally determined secondary structure for soluble Ten14 in CD studies, which indicates that soluble Ten14 is composed of 27% β-sheet and only 1% α-helix (Fig. 1C). As CD is particularly powerful for the prediction of α-helices (Johnson, 1990), our structural predictions for Ten14 are very compelling for lack of helical content. Also, binding geometry of the six cysteines of Ten14 agrees well with EGF and TGFα, in addition to residues in the beta-sheet region (Fig. 1B). As this binding is crucial for activity of EGFL repeats of tenascin C, we believe that the predicted Ten14 structure represents the functional form of the native soluble monomer (Zanuttin et al., 2004).
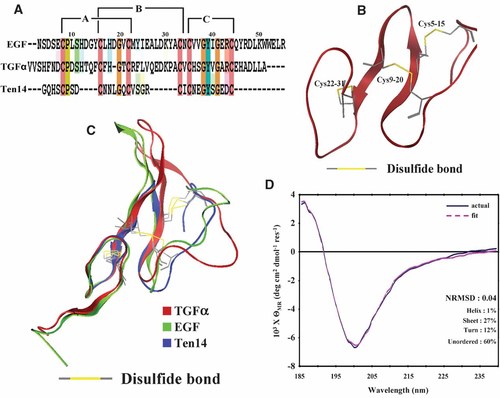
Structural modeling of Ten14. A: Multiple sequence alignment indicates that Ten14 has 25% sequence homology with EGF and 32% with TGFα; EGF and TGFα have 38% sequence homology. The six cysteines of Ten14 align with those of EGF and TGFα in addition to Pro6, Gly13, Gly25, Tyr26, and Gly28 of Ten14. Note, however, that the conserved arginine (Arg42 in TGFα) corresponds to an oppositely charged (aspartate) residue in Ten14. B: Ten14 is composed of an anti-parallel β-hairpin, with six cysteines forming the disulphide bridges in 1–3 (Cys5-Cys15), 2–4 (Cys9-Cys20), and 5–6 (Cys22-Cys31) orientation. C: Deletions at both N- and C-terminal regions make Ten14 significantly shorter compared to EGF and TGFα. Nevertheless, there is close overlap in the position of the six cysteines for all three structures. D: Analysis using CDSSTR yielded a best fit (pink dotted) for averaged Ten14 CD experimental spectra (blue) with a NRMSD of 0.04. Ten14 is composed mainly of β-sheet and β-turn, with negligible α-helix content, in excellent agreement with the modeled Ten14 structure.
Ten14 May Dock Onto EGFR in Alternative Structural Conformations
Ten14 binds to EGFR in or near the same region as EGF since Ten14 binding was competed by EGF, and an antibody that blocked EGF binding also blocked Ten14 (Swindle et al., 2001). As such, we modeled Ten14 binding to EGF/TGFα-binding pocket of EGFR (Fig. 2A). Rigid-body docking of Ten14 was performed using two methodologies based on distinct underlying principles, yielding two complexes (Structures I and II, Fig. 2). Structure I was generated on the intuitive assumption that if an unknown ligand shares sequence and structure homology with a known ligand in regions that directly interact with its receptor, the unknown ligand will most likely bind the receptor in a fashion similar to the known ligand. Hence, we first identified structurally conserved regions between Ten14, EGF and TGFα using the Consensus server (http://structure.bu.edu/cgi-bin/consensus/consensus.cgi). Consensus yields high-quality alignments for comparative modeling and identifies the alignment regions reliable for copying from a given template, even under low target-template identity. With maximum confidence on a scale of 0–9 (Supplemental Table 2), the Cα atoms of residues 20–31 of Ten14 were superimposed onto the corresponding TGFα residues in the ligand-binding pocket of TGFα–EGFR structure, with transposition of the other residues (Fig. 2B).
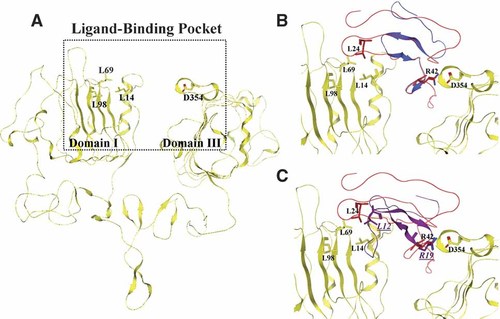
Docking of Ten14 with EGFR. A: Chain A of 1 MOX shows the ligand binding pocket of active EGFR. Leucines 14, 69, and 98 form an important ligand-dock site in domain I and Asp354 is involved in a crucial interaction with ligand in domain III. Both EGF and TGFα bind the receptor in this pocket. An additional interaction between Leu47 of TGFα with receptor has been shown in Supplemental Figure 1. B: Structure I of Ten14 (blue) docks to EGFR in the binding pocket. This docking was performed by overlap of the C-terminal region of Ten14 with the corresponding motif in TGFα (red). C: Structure II represents an alternative conformation, where Leu12 and Arg19 (underlined italicized) of Ten14 interact in a similar fashion as TGFα, forming contacts with the leucine pocket and Asp354 of EGFR, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
We modeled Structure II based on two observations: that the leading interaction responsible for molecular recognition (Rajamani et al., 2004) for both EGF and TGFα ligands corresponds to two ligand leucines binding to the same two hydrophobic pockets in EGFR, and that the leading salt bridge for both ligands involves Asp354 (Supplemental Fig. 1). These interactions were missing in Structure I. In Structure II, we were not only able to accommodate the single leucine of Ten14 (Leu12) in one of the hydrophobic pockets in EGFR, but the model complex also formed a salt bridge between Arg19 and Asp354 (Fig. 2C). Hence, the binding modes of EGF, TGFα, and Ten14 are quite homologous. We note that Structure II does not fulfill a second salt bridge present in EGF and TGFα (Supplemental Table 3), thus missing at least a third of the binding energy relative to these complexes.
GNM Analysis Suggests High Mobility of the EGFR–Ten14 Complexes
GNM calculations were performed to assess the collective dynamics of the ligand-receptor complexes. With GNM, the complex is modeled as an elastic network, the nodes of which are the α-carbons and the connectors (between all pairs of residues located within a cutoff distance of 8 Å) account for the equilibrium interactions that stabilize the native fold (Bahar et al., 1997; Bahar and Jernigan, 1998; Demirel et al., 1998; Bahar, 1999; Yang and Bahar, 2005). GNM has been used extensively in the past and collective dynamics predicted by the GNM, or elastic network normal mode analysis are insensitive to the detailed geometry, force field, or standard energy minimization protocols (Bahar and Rader, 2005; Ma, 2005). We used the iGNM web server for global mode analysis EGFR with or without ligand (Yang et al., 2005). Our analysis shows that the ligand binding pocket of EGFR between domains I and III is highly mobile in absence of ligand, allowing easy access and local rearrangements to accommodate ligand binding, as expected from experiments (De Crescenzo et al., 2000) (Fig. 3, Supplemental Fig. 2A). The ability of receptors to undergo conformational movements that facilitate substrate binding is consistent with our recent examination of intrinsic mobilities of proteins near their substrate binding sites (Tobi and Bahar, 2005). GNM analysis of collective dynamic for the TGFα–EGFR and EGF–EGFR confirmed that a very stable interaction is established for both EGF and TGFα with EGFR, that is, the mobilities of both ligands are significantly suppressed, indicating that both ligands bind with high affinity to the receptor as expected (Fig. 3, Supplemental Fig. 2B and C). However, in both Ten14–EGFR structures, Ten14 is found to be much more mobile in the bound form than classical EGFR ligands (Fig. 3, Supplemental Fig. 2D and E). As evidenced by the mobility plots for EGFR complexed with the ligands, differences exist in the mobility of residues near the ligand-binding pocket (Fig. 3) (Also see Supplemental Movies 1). The curves represent the normalized distribution of mobilities in the most cooperative (slowest) modes of motions. Regions near the ligand-binding pocket, (Boxes 1 and 2) are much more mobile in the Ten14-EGFR complexes (blue circles and purple line) compared to the TGFα (red diamonds) bound forms. Likewise, the terminal portions of the curves, detailing the mobility of the ligand residues, clearly indicate the significantly higher flexibility of Ten14 compared to TGFα for both structures, albeit the binding of Structure II to EGFR is more stable than Structure I.
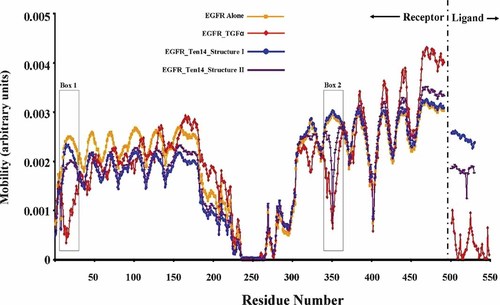
GNM analysis for Ten14 bound to EGFR suggests weak binding. The figure shows the distribution of fluctuations in the slowest modes for EGFR structure alone, and of the complexes TGFα–EGFR, Ten14–Structure I EGFR, and Ten14–Structure II EGFR. The curves represent the normalized mobility of each residue in the complex (Y-axis) as a function of residue index (X-axis). At the ligand-binding pocket entrance encompassing residues 10–40 (Box 1) and 350–370 (Box 2), EGFR alone (yellow squares) has considerably higher mobility than EGFR bound to TGFα (red diamonds), while the mobility in the Ten14-bound forms (blue circles and purple line) is comparable to the unbound forms. This comparison confirms that these regions involve stable and strong contacts with EGFR in the case of TGFα binding, which are either lost or weaker in the Ten14-bound form. Also, Ten14 ligand in both conformations is much more mobile than TGFα (right terminal portion of the curves). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Analysis of EGFR–Ten14 Interactions
We analyzed the interface between Ten14 and EGFR using “FastContact,” a program that computes the relative free energy of receptor and ligand residue contacts by summing electrostatic interactions and desolvation potentials that encapsulate hydrophobic interactions, the self-energy change upon desolvation and side chain entropy loss. Stronger interactions between ligand and receptor are manifested by lower (more negative) electrostatic and desolvation potentials. However, FastContact only accounts for inter-molecular contacts, and does not estimate differences in configurational entropy and thus introduces errors of up to 5 kcal/mol. Analysis of EGF–EGFR, TGFα–EGFR and Structures I and II of Ten14–EGFR complex yielded total electrostatic (ΔE) and desolvation energy potentials of −27.68 and −7.1 kcal/mol, respectively, for EGF–EGFR; −34.72 and −9.35 kcal/mol for TGFα–EGFR; −6.72 and 1.51 kcal/mol for Structure I, and −23.58 and −1.51 kcal/mol for Structure II of Ten14–EGFR. We emphasize that these are only relative energies—figuring in the error contributions arising from conformational entropy of the ligands and errors due to inherent shortcomings in the computation of binding energies using FastContact would significantly reduce the differences we observe with in silico binding energies, especially with EGF and TGFα. However, these estimates strongly suggest that Structure I, the model based on the same binding mode as EGF–EGFR, is not a good candidate for the complex. On the other hand, even after accounting for the aforementioned errors, Structure II recovers only a part (maybe about two thirds) of the affinity observed for EGF and EGFR, which may result in about a significant reduction in Ten14 affinity for EGFR with respect to EGF, consistent with the predicted decrement below. Detailed examination of the structures reveals that the highly attractive interactions between Asp345-Arg42 (Arg41 in EGF), Glu90-Lys29 (Lys28 in EGF), Lys464-Ala50 (Trp49 in EGF), and Arg125-Glu27 (Asp27 in EGF) between EGFR and the ligands EGF or TGFα are absent in the case of Ten14-Structure I (See Supplemental Tables 3A and B). However, for Structure I, an important salt bridge between Lys13 of the receptor and Glu29 of Ten14 is formed (Fig. 4A). This interaction is energetically more favorable for Ten14 as compared to those for EGF and TGFα (Supplemental Table 3). Also, favorable electrostatic interactions take place between Arg19 of Ten14 and Gln16 and Tyr45 of EGFR, shielding the hydrophobic Leu14 of EGFR and stabilizing the binding of Ten14 to EGFR (Fig. 4A). For Structure II, a number interactions are recapitulated in Ten14 that exist in EGF and TGFα, with Arg19 forming a highly favorable interaction with Asp354, and Glu29 interacting with Arg125, both of which lead to a tighter binding of Ten14 with EGFR in this conformation than Structure I (Fig. 4B). However, these interactions are not sufficiently stable to allow for tight binding of Ten14 to EGFR, evident from GNM analysis of Structure II (see Supplemental Fig. 2E).
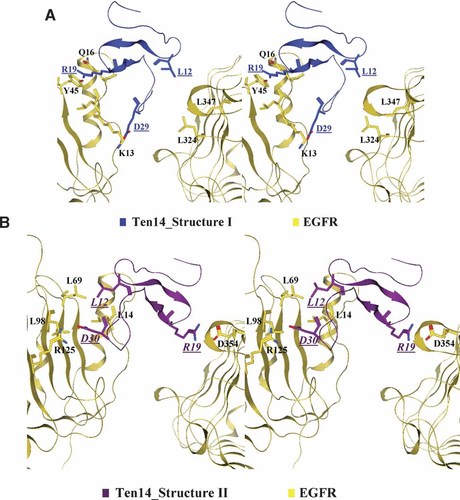
Interactions between Ten14 and EGFR. Stereo views of two important sets of interactions for both conformations of Ten14 (blue on top and purple on bottom) with EGFR (yellow ribbon) were generated using MOE. A: Interactions between Ten14-Structure I EGFR include a high-affinity salt-bridge between Lys13 of EGFR and Glu29 of Ten14. The side chains of the two residues are only 3 Å apart. This interaction is energetically much more favorable than the corresponding interaction between EGF and TGFα (See Supplemental Table 3A). Arg19 of Ten14 and Tyr45 and Gln16 of EGFR stabilize the binding of Ten14 to EGFR. This interaction buries Leu14 of EGFR and restricts it from solvent accessibility. Also, a potential interaction may be possible between Leu12 of Ten14 and a pocket formed on the receptor by Leu324 and Leu347. B: For Structure II of EGFR-bound Ten14, Leu12 of Ten14 sits in the hydrophobic pocket formed by Leu14, Leu69, and Leu98 of EGFR. This coupling leads to interaction of Arg19 with Ten14 with Asp354 of EGFR. Note also that Asp30 of Ten14 interacts with Arg125, a key residue for the TGFα–EGFR interaction (See Supplemental Table 3). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Ten14 EGFL Repeats Exhibit Ultra-Low Affinity for EGFR as Compared to EGF
Previous studies show that much higher concentrations of Ten14 are required to have biological and biochemical effects equivalent to EGF. This, coupled with the structural data indicating very weak binding, suggests that Ten14 may activate receptor in a manner distinct from other classical soluble ligands. A low affinity ligand such as Ten14 would follow a staccato mode of signaling, whereby it binds EGFR for a period sufficient to elicit signaling, but dissociates from the receptor before internalization. In order to confirm this, SPR analyses were performed with increasing concentrations of Ten14 and human EGF on sensor surfaces derivatized with the EC ligand-binding domain of EGFR (Fig. 5). We obtained a KD of 74 µM for Ten14, nearly a 1,000-fold higher than that of EGF (∼110 nM), which was similar to published values for EGF–EGFR interactions (Zhou et al., 1993; Brown et al., 1994; Domagala et al., 2000). Though we could not directly determine the association/dissociation rates for the interaction due to technical limitations pertaining to low fidelity of Ten14 to changes in buffer conditions, the KD values agree well with our observations in vitro and similar experiments performed with EGFR-ED (see discussion below). This unprecedented high KD for an EGFR–ligand interaction is in accordance with predictions of high mobility of Ten14 in the ligand-binding pocket of EGFR from structural modeling of the complex (Fig. 3).
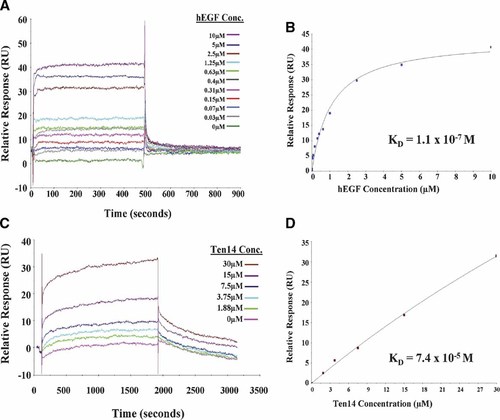
Surface plasmon resonance analysis of EGF/Ten14 binding to EGFR. A: Increasing concentrations of EGF (0.039, 0.078, 0.153, 0.31, 0.625, 1.25, 2.5, 5, and 10 µM) were run over CM5 surfaces derivatized without or with ∼2,500 RUs of extracellular domain (ED) of EGFR, using HBS-EP as running buffer. Sensograms were plotted against time using the BIAevaluate software package after subtraction from blank. B: Steady state RUs were plotted against concentration for each EGF level, and curves were fit for calculating the equilibrium dissociation constant KD using the “steady state” module in the BIAEvaluation software. We obtained a KD of 110 nM for EGF. C: Increasing concentrations of Ten14 (1.88, 3.75, 7.5, 15, and 30 µM) were run over surfaces derivatized without or with ∼7,000 RUs of EGFR-ED, using PBS as running buffer. Sensograms were plotted after blank subtraction. D: Sensograms were analyzed and a KD of 74 µM was obtained for Ten14, nearly a 1,000-fold higher than EGF. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Ten14 EGFL Repeats do not Undergo Internalization and Depletion
The much higher ligand mobility and KD for Ten14–EGFR interaction predicts an unstable binding unlikely to result in either ligand or receptor internalization/degradation. Specifically, we determined whether Ten14 was internalized and/or depleted in a manner similar to EGF when presented in soluble form to NR6WT murine fibroblasts overexpressing EGFR. Over a 48 h period, EGF concentrations of 10 and 1 nM were depleted from the medium; however, we saw no significant depletions of Ten14 at similar concentrations (Fig. 6A). In order to distinguish between the possibilities of Ten14 being recycled to cell surface as in the case of TGFα (French et al., 1995) versus rapid uncoupling from the receptor before internalization, we assessed internalization of radio-iodinated EGF and Ten14 ligand using a commonly derived protocol over a short time period that precludes recycling (Wiley et al., 1991). Although EGF was internalized dramatically, Ten14 was not (Fig. 6B). We also measured EGFR internalization under high levels of EGF and Ten14 to ensure that Ten14–EGFR binding did not persist to engage internalization and subsequent degradation. As expected, no significant Ten14-mediated degradation of EGFR was observed over extended time periods (Fig. 6C). Also, this in vivo data corroborated by the results we obtain for the Ten14–EGFR interaction in vitro (Fig. 5), where the high KD would drive rapid dissociation of Ten14. We conclude that Ten14 does not undergo receptor-mediated internalization, implying a rapid dissociation from the receptor leading to surface restricted activation of EGFR. Accordingly, immunofluorescent detection of active EGFR showed that upon treatment with EGF, significant co-localization of total and active EGFR was observed in internal compartments of the cell in the form of punctate dots (Fig. 6D). However, Ten14 treatment resulted in surface staining alone, with no active receptors in internal compartments.
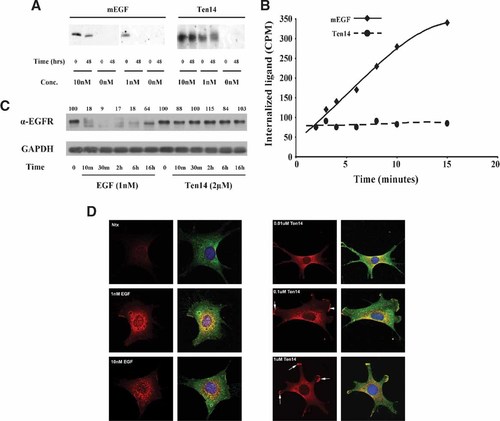
Ten14 activates EGFR on the cell surface without internalization. A: Following treatment of NR6WT cells for 48 h with Ten14 and mEGF, concentrations of growth factors in the supernatant were determined using the antibodies against ligands. Ten14 is not depleted from the medium over 48 h, compared to murine EGF. B: Over 20 min, minimal internalization of 125I-Ten14 is observed as compared to 125I-EGF. C: As opposed to EGF, Ten14 does not lead to degradation of EGFR over a 16 hr time period. D: With 1 and 10 nM EGF treatment, active EGFR (phospho-tyrosyl EGFR, red) is internalized into endosomal compartments, appearing as punctate blobs. Staining for total EGFR (green) shows this is the fate of the majority of cellular receptors. Internalization of active EGFR is accompanied by EGF internalization (data not shown). On the other hand, Ten14 causes localization of active EGFR solely at the cell surface with no internalization into intracellular compartments. All concentrations of Ten14 lead to activation and localization of EGFR into lammelipods (arrows). Also, Ten14 co-localizes with active receptor at the cell surface without internalization (data not shown). Blue stains for the nucleus.
Discussion
EGF-like repeats are found in many ECM proteins and have been implicated in signaling through EGFR (Swindle et al., 2001; Schenk et al., 2003). This creates a conundrum in that high affinity binding of a concatemerized tethered ligand to a growth factor receptor, resulting in ultra-high avidity, would not allow for internalization-mediated attenuation that is critical to prevent excess signaling and aberrant cell responses (Wells et al., 1990; Masui et al., 1991). Even dephosphorylation attenuation would be limited as the physical constraints coupled to the high affinity would results in highly persistent ligandation. As an answer to this confounding aspect, our data indicates that Ten14 may be a low affinity ligand with altered binding dynamics as compared to other soluble prototypical ligands such as EGF and TGFα. Thus, signaling from the tethered EGFL repeat would be attenuated by loss of ligandation and subsequent dephosphorylation.
We evaluate the binding (both in silico and in vitro) of the 14th EGFL repeat of tenascin C, even though at least three other repeats in the EGFL domain of tenascin C can potentiate signaling through EGFR (Swindle et al., 2001). We focus on Ten14 because we have previously optimized the purification and refolding process for Ten14, and, more importantly, the folding of the EGFL repeat 14 has been previously studied (Zanuttin et al., 2004).
Structural modeling of Ten14–EGFR complex was the first step towards understanding the basis of the low affinity of Ten14 for EGFR. Modeling was chosen over classical techniques such as X-ray crystallography due to limitations in the purification process of Ten14; Ten14 undergoes non-physiological protein aggregation at very high concentrations that are required for successful protein crystallization. Ten14 is also extremely sensitive to changes in pH and buffer conditions. Lastly, enzymatic de-glycosylation of EGFR, a required step for co-crystallization of EGFR with its ligands (Ogiso et al., 2002), may drive down the affinity of Ten14 further, as seen previously with EGFR (Soderquist and Carpenter, 1984; Wang et al., 2001). Alternative purification techniques are being explored, but these efforts lie beyond the scope of this communication. Modeling of Ten14–EGFR complex structure offers a reasonable alternative to crystallography. Structural models have been generated for a number of receptor-ligand complexes, with good correlation between the predicted model and the actual crystal structure (Paas et al., 2000). Also, templates for the Ten14–EGFR complex exist in the form of EGF–EGFR and TGFα–EGFR crystal structures (Garrett et al., 2002; Ogiso et al., 2002), an important consideration for structural modeling. These facts provided us with sufficient impetus to simulate the bound structure of Ten14 and EGFR and analyze the complex to identify important interactions between them. An important consideration for studying Ten14–EGFR interactions using structural modeling was the identification of the ligand binding region, as this would directly impact inferences drawn from detailed analysis of the resulting complex. We employed the following approaches to elucidate interactions between Ten14 and EGFR. The first approach relies on the fact that given sufficient sequence and structural homology, similar domains in distinct ligands may bind substrate in an identical manner (Rajamani et al., 2004). This is clearly evident with the EGF–EGFR and TGFα–EGFR complexes, where, despite their structural differences, significant overlap exists in the key ligand-motifs that interact with the receptor. Independently, we generated homology models of Ten14 based on the EGF and TGFα structures, and found that the top ranked models had a striking structural homology with the previously identified binding motifs. Based on this homology, we modeled a bound structure in a manner so as to satisfy the key molecular interactions, achieved by first fulfilling the hydrophobic requirements, followed by coupling of hydrophilic interactions resulting from the initial dock.
Both GNM and “FastContact” analysis of the models suggests that conformation of Structure II of Ten14 is much more stable in the EGFR binding pocket compared to Structure I (lower mobility of Structure II in GNM analysis of the complex (Fig. 3) and the lower ΔE of Structure II as compared to Structure I). Interestingly, Structure I provided us with a direct experimental test as it juxtaposed a non-conserved amino acid at the site of a salt bridge in EGF–EGFR and TGFα–EGFR complexes. We replaced Asp30 of Ten14 with a positively charged arginine corresponding with Arg41 in EGF (Arg42 in TGFα), and assessed its affinity for EGFR. If Structure I represents the true binding conformation of Ten14, the restoration of an important salt bridge between Ten14 and EGFR (by interaction of Arg30 in a D30R mutant of Ten14 with Asp354 of EGFR) should result in increased affinity of D30R for EGFR. This would also be reflected in the “FastContact” analysis of the D30R mutant complexed with EGFR, resulting in much lower ΔE as compared to Structure I of native Ten14, and much tighter binding. However, SPR measurements for the D30R mutant with EGFR-ED and “FastContact” analysis of the modeled D30R–EGFR complex yielded results showing no increase in affinity of the mutated form (data not shown). “FastContact” analysis suggests that Structure II loses a significant part of its affinity, which could result in a KD that is well within the range observed for Ten14 in vitro using SPR (Fig. 4B). Therefore, even though there is a lack of mutations predicted to increase affinity empirically, we believe Structure II most likely represents the true binding conformation of Ten14 for EGFR. However, mutational analysis of Structure II (in a manner similar to that for Structure I) with substitution of the key Arg19 to decrease affinity for EGFR further would be confounded by the fact that we are at the limits of detection of binding affinities by SPR, and any decrement would be indistinguishable from improperly folded ligands. Nevertheless, additional experiments are underway to validate the binding conformation of Structure II of Ten14 by assessing binding of Arg19 of Ten14 with Asp354 of EGFR using bi-functional crosslinking followed by receptor and ligand fragmentation and affinity purification. These technically daunting experiments lie beyond the scope of the present communication.
In order to assess if the low affinity is a direct result of altered binding dynamics of Ten14 to EGFR as compared to other soluble ligands, we performed SPR analysis using the ED domain of EGFR. SPR can be effectively used to predict kinetic binding parameters even in the micromolar levels (van der Merwe et al., 1994) as opposed to other biochemical techniques that are optimal only for studying high-affinity binding interactions. Our results indicate a KD of 110 nM for EGF, and though this is nearly two logs higher relative to measurements using live cells, it is in excellent agreement with similar experiments performed previously with EGFR-ED monomers (Zhou et al., 1993; Brown et al., 1994; Domagala et al., 2000). Also, the predicted KD for Ten14 is nearly 1,000-fold higher than EGF which agrees well with concentrations of ligand required to stimulate equivalent levels of activation of EGFR in vitro. As we could not directly measure the on-off rates of binding due to limitations arising from buffer considerations for Ten14, we attempted to verify our data with other independent techniques such as dynamic light scattering, but technical limitations hindered effective analysis of binding.
Receptor compartmentalization and trafficking are important aspects of regulation of EGFR-mediated cellular responses. Proliferation and differentiation are initiated by signaling cascades triggered at the cell surface and are maintained by signaling cascades that are functional in intracellular compartments (Haugh and Meyer, 2002). Cell migration though, seems to be a mainly cell surface signaling mediated phenomenon, and active EGFR in endosomal compartments contribute minimally to triggering PLCγ1 required cell migration (Chen et al., 1994b; Haugh et al., 1999a; Glading et al., 2001). In vitro experiments with EGF presented as a tethered ligand by coupling to a polymer matrix showed that EGF can promote cell migration as effectively as soluble EGF (Griffith, Wells, et al. personal communication). We also observe Ten14-mediated restriction of active EGFR at the cell surface over a range of ligand concentrations. We contend that Ten14, and possibly other select EGFL repeats of tenascin C, may play a similar role in physiological conditions, presenting itself as a two-dimensional matrikine ligand with low affinity for EGFR, leading to compartmentalization of receptor and steady activation of migratory cascades at the cell surface.
Based on this and previous studies, we posit that multiple EGFL repeats can potentially bind numerous EGFR as part of a signaling complex. In this context, EGFR signaling could be mediated by EGFL repeats being part of an intact tenascin C, or released as aggregates containing multiple EGFL-repeat subunits. In fact, we previously reported that simple dimerization of Ten14 stabilized the Ten14 EGFL-repeat interactions with EGFR (Swindle et al., 2001). The release of these subunits can potentially be mediated by the action of matrix metalloproteinases (MMPs) on tenascin C (Siri et al., 1995), and it has also been demonstrated in vivo for EGFL domains of laminin (Schenk et al., 2003). Thus, while the individual affinity would be low, the matrix constraints would increase the avidity; a similar situation is found with integrin binding sites (Carman and Springer, 2003). However, overall avidity for interactions with EGFR via multimeric ligand domains may increase or decrease based on the physical constraints imposed by being a component of a matrix protein and not a freely rotatable soluble monomeric ligand. Interestingly, we prefer Structure II binding precisely because it better allows for an integral EGFL repeat as part of intact tenascin C or an EGFL repeat domain to fit in the EGFR binding pocket.
Receptor binding to such matrix-constrained ligands would be enabled by the greater motility of the receptor–ligand interaction as we note, resulting in the lessened binding affinity and more transient occupation and activation profile. Such signaling may be relevant from the ECM standpoint, where a number of proteins, particularly tenascin C are found to be upregulated only during wound healing or tumor progression, both of which require potent activation of migratory signaling cascades (Zagzag et al., 2002; Ilunga et al., 2004; Juuti et al., 2004). Interestingly, an upregulation of MMPs is also observed concurrently with expression of tenascin C during numerous patho-physiological scenarios, characterized especially by instances involving potent cell migration (Jian et al., 2001; Cai et al., 2002; Kalembeyi et al., 2003). EGFL repeats of tenascin C may thus temporally and spatially activate select pathways downstream of EGFR, driven primarily by their low affinity for the receptor.
Acknowledgements
We thank Jonathan Steckbeck and the BIAcore facility at the University of Pittsburgh for help with using the machine. We thank Dr. Lee Wei Yang and other members of Dr. Ivet Bahar's and Dr. Linda Griffith's (MIT) laboratories. We also thank Dr. Richard Bodnar, Chris Shepard and Cecilia Yates for critical reading of the manuscript.




