The Bacterial Species Behind the Wound and Their Antibacterial Resistant Pattern: A Three-Year Retrospective Study at St. Dominic Hospital, Akwatia, Ghana
Funding: The authors received no specific funding for this work.
ABSTRACT
Background
Wound infections are often underestimated issues that can lead to chronic illnesses, and since the introduction of antibiotics, wound complications have become less common. However, due to the increased and irrational use of these antibiotics, the resistance in the bacterial isolates has become very common. This has led to reduced treatment options, delay in wound healing, and high treatment costs. This study aimed to investigate bacterial wound infections and their antibiotic resistance at St. Dominic Hospital, Ghana.
Methods
A total of 517 records of wound swab culture and susceptibility testing, and patient demographics from 2020 to 2022 were collected from the microbiology unit of St. Dominic Hospital in the Eastern Region of Ghana. The data were entered into Microsoft Excel 2019, cleaned, and exported into IBM SPSS v26 for the statistical analysis. p < 0.05 was considered statistically significant for all analyses.
Results
The overall prevalence of bacteriological agents causing wound infection in individuals who visited the St. Dominic Hospital from 2020 to 2022 was 70.21% (363/517), with S. aureus 79/363 (21.76%) being the most abundant isolate. Out of the 79 S. aureus isolated, 40 (50.63%) and 39 (49.37%) were resistant to ampicillin and cephalexin, respectively. More than 50% of the predominant Gram-negative isolate, K. pneumoniae, were resistant to clindamycin 45/72 (62.50%) but susceptible to levofloxacin 70/72 (97.22%), cefotetan 69/72 (95.83%), and chloramphenicol 67/72 (93.06%).
Conclusion
Antibacterial susceptibility patterns revealed significant resistance trends, particularly among Gram-negative isolates, emphasizing the urgent need for prudent antibiotic use and ongoing surveillance to combat resistance.
1 Background
A wound occurs when there is physical disruption of the skin, which is one of the main barriers to the development of bacterial pathogen infections in interior tissue. Infection can result once microorganisms get through the skin barrier [1].
The prevalence of wound infections varies significantly across different studies and populations. Wound infections are among the most prevalent hospital-acquired infections, contributing to 70%–80% of fatalities [2]. A systematic review focusing on surgical wound infections reported a pooled prevalence of 3.3%, with higher rates observed in males compared to females [3]. In a general surgery setting, a cross-sectional study found a prevalence of 4.7% with factors such as prolonged hospitalization and immunodeficiency being significant risk factors [4]. In contrast, a study in Nigeria indicated a much higher overall prevalence of 64.8%, influenced primarily by age, with S. aureus identified as the predominant pathogen [5]. Another Nigerian study over 5 years reported a prevalence of 70.1%, again highlighting S. aureus as a common aetiological agent [6]. In Northern Ghana, the prevalence was 32.3%, with significant associations found with gender and diabetes [7]. Other significant causative bacteria include E. coli, P. aeruginosa, and K. pneumoniae, which collectively contribute to the polymicrobial nature of these infections [8].
Antibiotic resistance in wound infection is a critical concern, with a concerned prevalence of methicillin-resistant Staphylococcus aureus (MRSA) and extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae [9]. Korol et al. [10] in a study indicates MRSA is an increasingly important pathogen that causes more than 50% of S. aureus hospital-acquired infections in the United States and Europe, and presents challenges to treatment due to multiple antibiotic resistance. In another study in Ethiopia by Tsige, Tadesse [11], the isolated MRSA showed complete resistance to penicillin (100%), followed by erythromycin and ciprofloxacin (61.5%) and cotrimoxazole and gentamicin (53.8%).
Also, Namburi, Sisira [12] and Hasan in Bangladesh [8] in their studies reported that more than 50% of the isolates from wound culture were resistant to the commonly prescribed antibiotics such as ampicillin and penicillin, while fluoroquinolones and amikacin showed better efficacy against these pathogens. The emergence of polyresistant strains, particularly A. baumannii, further complicates treatment options, as these pathogens often show limited susceptibility to available antibiotics [13]. This resistance not only prolongs the duration of infections but also increases morbidity and mortality rates among infected patients [14].
Since antibiotics were introduced, complications from wounds have decreased. However, the frequent and indiscriminate use of broad-spectrum antibiotics has made bacterial resistance more common. This has resulted to reduced availability of treatment options, delay in wound healing, and increased treatment costs. This study seeks to identify the bacterial species causing wound infections and their antibiotic resistance patterns at St. Dominic Hospital in Ghana.
2 Methodology
2.1 Study Design
This study was carried out at St. Dominic Hospital in the Eastern Region of Ghana and utilised a retrospective study design. The study analysed archived laboratory results on wound culture and susceptibility tests spanning from January 2020 to December 2022.
2.2 Study Area
The study was conducted at St. Dominic Hospital. The hospital is located in Akwatia in the Eastern Region of Ghana. The hospital that was built and handed over to the Catholic Mission in 1960 now serves as the major health facility in the Akwatia, with Denkyembour as the district capital. The Denkyembour District is located at the south-western corner of the eastern region. It shares boundaries with Kwaebibirem and Akyemansa Districts to the north, West Akim Municipality to the south and Birim Central Municipality to the south-west according to Ghana Statistical Service (GSS) [15]. It falls between Latitude 7°. 30 W and 7°. 30 E and Longitude 1.30°N and 1.30°S. The hospital currently has organized wards, specialized clinics, public health and diagnostics units, among others. This makes the services provided in the facility extremely robust. St. Dominic Hospital is a member of the Christian Health Association of Ghana. It is under the national catholic health service. A collaborative relationship also exists between the hospital, district and regional health administrations. It has a bed capacity of 357 [16].
2.3 Study Participants
The study participants were patients who underwent wound culture and susceptibility testing between 2020 and 2022. The data on 517 participants were obtained from the hospital microbiology unit.
2.4 Inclusion Criteria
Archived results of wound culture and susceptibility testing were included in the study.
2.5 Exclusion Criteria
Archived wound testing results of 7 participants with incomplete and missing information (age, sex, department, isolate, and susceptibility results) were excluded from the study.
2.6 Sample Collection, Processing and Bacterial Identification
The wound was first cleaned with sterile normal saline. Each sample was collected by swabbing from the wound ground and edge using the Levine technique [17]. The samples were processed by inoculating them on chocolate agar, blood agar and McConkey agar and incubated appropriately. The inoculated blood and MacConkey agar were incubated in ambient air, while chocolate agar was incubated in CO2 environment. Identification was done by describing the colonial morphology on the agar plates, Gram staining, motility testing and by using biochemical tests including catalase, coagulase, oxidase, urease, citrate, indole and triple sugar fermentation tests. A novobiocin susceptibility test was also done to differentiate coagulase-negative Staphylococci species.
2.7 Antibacterial Susceptibility Testing
Antibacterial susceptibility testing, based on the local availability of the antibiotics, was performed using the Kirby Bauer disc diffusion method as described by Deku, Duedu [18], except that sterile normal saline was used instead of buffered phosphate saline, and the determination of the turbidity of the bacterial suspension using Densi CHEK plus densitometer was substituted with a macroscopic comparison with the 0.5 McFarland standard. The interpretation of the test result was done according to Clinical and Laboratory Standards Institute (CLSI) guidelines [19]. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as controls.
2.8 Data Collection
Data were collected at the Hospital Laboratory unit using a data extraction sheet. Demographics of patients with their wound culture and susceptibility results from 2020 to 2022 were retrieved. Data collected were later entered into Microsoft Excel 2019. Data were verified by double-checking the extracted data against the source. This was done by two individuals to ensure that possible errors were corrected. All assumptions of the various statistical analyses were tested and passed before the analysis was done.
2.9 Data Handling and Analysis
Data entered into the Microsoft Excel 2019 file were cleaned and exported into IBM Statistical Package for the Social Sciences Version 26.0 (Armonk, NY: IBM Corp). Descriptive statistics was used to calculate the overall prevalence of bacteriological agents of wound infection in the study population. Analysis was done based on subgroups such as age, sex, period and department using Pearson's chi-square analysis. p < 0.05 was considered statistically significant for all analyses.
2.10 Ethical Consideration
Ethical approval was obtained from the Research Ethics Committee of the University of Health and Allied Sciences (UHAS), Ghana, with reference number UHAS-REC A.3 [3] 23–24. All archived data for the study were kept undisclosed and used for the study only. Patients' consent was not sought for this study because it was retrospective.
3 Results
Table 1 provides an overview of the demographic characteristics of 517 study participants whose wound swab was taken for culture and susceptibility testing. Most of the study participants, comprising 118 out of the 517 (22.82%), fell within the 31–40 age group. Only 11 out of the 517 participants (2.13%), were under the age of 1 year. Most of the participants 398/517 (76.98%) were females and outpatients 279/517 (53.97%). The highest number of participants 238/517 (46.03%) sought medical care in the year 2022, followed by 187/517 (36.17%) in 2021, while 92/517 (17.79%) of the participants reported to the hospital in 2020.
| Variable indicator | Level | Number observed | % |
|---|---|---|---|
| Age | < 1 | 11 | 2.13 |
| 1–10 | 29 | 5.61 | |
| 11–20 | 24 | 4.64 | |
| 21–30 | 102 | 19.73 | |
| 31–40 | 118 | 22.82 | |
| 41–50 | 96 | 18.57 | |
| 51–60 | 64 | 12.38 | |
| 61–70 | 40 | 7.74 | |
| > 70 | 33 | 6.38 | |
| Sex | Female | 398 | 76.98 |
| Male | 119 | 23.02 | |
| Department | Inpatient | 238 | 46.03 |
| Outpatient | 279 | 53.97 | |
| Year | 2020 | 92 | 17.79 |
| 2021 | 187 | 36.17 | |
| 2022 | 238 | 46.03 |
Figure 1 shows the prevalence of bacteriological agents causing wound infection in the participants who visited the hospital during the year under consideration. There was bacterial growth in 363/517 (70.21%) of samples from the participants.
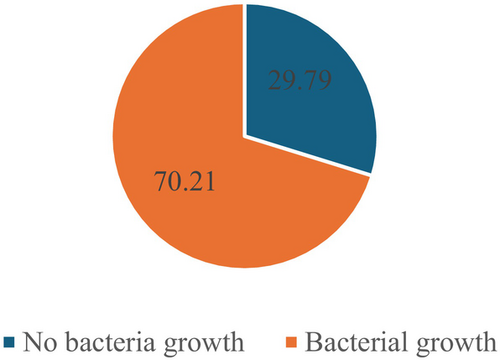
Figure 2 shows the proportion of the bacteriological agents of the various isolates from the wound swab of the participants who visited the hospital. The predominant isolate was S. aureus 79/363 (21.76%).
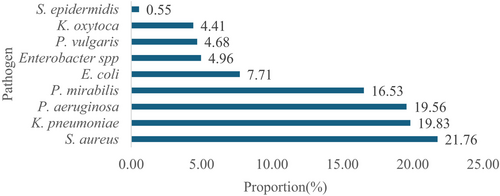
Table 2 provides an overview of the distribution of bacterial isolates stratified by the age groups of the study participants. The occurrence of S. aureus exhibited an upward trend as age advanced, reaching its peak in the 31–40 age group 19/79 (24.05%), while its lowest prevalence was observed in the < 1, 61–70, and > 70 age groups. K. pneumoniae showed a fluctuating distribution, with the highest percentage occurring in the 21–30 age group 21/72 (29.17%). No growth of K. pneumoniae was isolated in the swab from participants less than 1 year. P. aeruginosa also showed variability across age groups, with the highest proportion in the 31–40 age group 18/71 (25.35%) and the lowest in the < 1, 1–10 and 11–20 age groups 2/71 (2.82%). S. epidermidis was exclusively isolated in the 41–50 age group, exhibiting a prevalence of 100% (2/2). Generally, the highest proportion of bacterial isolates, accounting for 20.66% of the total isolates (75/363), was observed in the 31–40 age group, while the lowest proportion was identified in the < 1 age group, constituting only 1.65% (6/363). These distributions were however not statistically significant (p = 0.080).
| Age (years) | < 1 | 1–10 | 11–20 | 21–30 | 31–40 | 41–50 | 51–60 | 61–70 | > 70 | p |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria isolates | 0.080 | |||||||||
| Staphylococcus aureus (n = 79) | 3 (3.80) | 9 (11.39) | 10 (12.66) | 13 (16.46) | 19 (24.05) | 11 (13.92) | 8 (10.13) | 3 (3.80) | 3 (3.80) | |
| Klebsiella pneumoniae (n = 72) | 0 (0.00) | 5 (6.94) | 4 (5.56) | 21 (29.17) | 10 (13.89) | 15 (20.83) | 7 (9.72) | 4 (5.56) | 6 (8.33) | |
| P. aeruginosa (n = 71) | 2 (2.82) | 2 (2.82) | 2 (2.82) | 11 (15.49) | 18 (25.35) | 11 (15.49) | 11 (15.49) | 7 (9.86) | 7 (9.86) | |
| P. mirabilis (n = 60) | 0 (0.00) | 2 (3.33) | 1 (1.67) | 3 (5.00) | 11 (18.33) | 13 (21.67) | 12 (20.00) | 9 (15.00) | 9 (15.00) | |
| E. coli (n = 28) | 0 (0.00) | 0 (0.00) | 1 (3.57) | 8 (28.57) | 6 (21.43) | 7 (25.00) | 3 (10.71) | 1 (3.57) | 2 (7.14) | |
| Enterobacter spp (n = 18) | 1 (5.56) | 1 (5.56) | 1 (5.56) | 4 (22.22) | 5 (27.78) | 2 (11.11) | 2 (11.11) | 2 (11.11) | 0 (0.00) | |
| P. vulgaris (n = 17) | 0 (0.00) | 0 (0.00) | 1 (5.88) | 5 (29.41) | 2 (11.77) | 6 (35.29) | 1 (5.88) | 1 (5.88) | 1 (5.88) | |
| K. oxytoca (n = 16) | 0 (0.00) | 1 (6.25) | 0 (0.00) | 4 (25.00) | 4 (25.00) | 3 (18.75) | 2 (12.50) | 2 (12.50) | 0 (0.00) | |
| S. epidermidis (n = 2) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 2 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Total (363) | 6 (1.65) | 20 (5.51) | 20 (5.51) | 69 (19.01) | 75 (20.66) | 70 (19.28) | 46 (12.67) | 29 (7.99) | 28 (7.71) |
- Note: Data are presented as frequencies with corresponding percentages in parentheses. p-value is significant at p < 0.05.
Table 3 shows the proportion of bacterial isolates stratified by gender, department and year of study. The table highlights the gender-based variations in bacterial isolates, with a higher total count of bacterial isolates in females 282/363 (77.69%) compared to males 81/363 (22.31%). This variation was statistically insignificant (p = 0.080). Furthermore, the table provides insights into the relationship between the bacterial isolates and both the department and the study period. From a statistical perspective, the analysis indicated a lack of significant association between the bacterial isolates and the department (p = 0.140). However, it did reveal a significant association between the bacterial isolates and the study period (p = 0.005).
| Gender | Department | Period | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial isolates | Female | Male | p | Inpatient | Outpatient | p | 2020 | 2021 | 2022 | p |
| 0.70 | 0.14 | 0.005 | ||||||||
| S. aureus (n = 79) | 61 (77.22) | 18 (22.78) | 32 (40.51) | 47 (59.49) | 17 (21.52) | 33 (41.77) | 29 (36.71) | |||
| K. pneumoniae (n = 72) | 56 (77.78) | 16 (22.22) | 36 (50.00) | 36 (50.00) | 19 (26.39) | 21 (29.17) | 32 (44.44) | |||
| P. aeruginosa (n = 71) | 52 (73.24) | 19 (26.76) | 26 (36.62) | 45 (63.38) | 11 (15.49) | 15 (21.13) | 45 (57.75) | |||
| P. mirabilis (n = 60) | 44 (73.33) | 16 (26.67) | 28 (46.67) | 32 (53.33) | 11 (18.33) | 30 (50.00) | 19 (31.67) | |||
| E. coli (n = 28) | 24 (85.71) | 4 (14.29) | 12 (42.86) | 16 (57.14) | 7 (25.00) | 5 (17.86) | 16 (57.14) | |||
| Enterobacter spp (n = 18) | 16 (88.89) | 2 (11.11) | 11 (61.11) | 7 (38.89) | 3 (16.67) | 11 (61.11) | 4 (22.22) | |||
| P. vulgaris (n = 17) | 15 (88.24) | 2 (11.76) | 12 (70.59) | 5 (29.41) | 3 (17.65) | 4 (23.54) | 10 (58.82) | |||
| K. oxytoca (n = 16) | 12 (75.00) | 4 (25.00) | 9 (56.25) | 7 (43.75) | 3 (18.75) | 4 (25.00) | 9 (56.25) | |||
| S. epidermidis (n = 2) | 2 (100.00) | 0 (0.00) | 0 (0.00) | 2 (100.00) | 1 (50.00) | 0 (0.00) | 1 (50.00) | |||
| Total (363) | 282 (77.69) | 81 (22.31) | 166 (45.73) | 197 (54.27) | 75 (20.66) | 123 (33.88) | 165 (45.46) | |||
- Note: Data are presented as frequencies with corresponding percentages in parentheses. p-value is significant at p < 0.05. p-value in bold is statistically significant.
Out of the 363 participants whose samples showed bacterial growth, 81 (22.31%) of the isolates were Gram-positive and the remaining 282 (77.69%) isolates were Gram-negative bacteria. The Gram-positive isolates were S. aureus 79/363 (21.76%) and S. epidermidis 2/363 (0.55%). The antibacterial susceptibility patterns of the Gram-positive bacterial isolates are presented in Table 4. Out of the 79 S. aureus isolated, 40 (50.63%) and 39 (49.37%) were resistant to ampicillin and cephalexin, respectively. However, the majority of these isolates were susceptible to ciprofloxacin 72/79 (91.14%), cefotaxime 72/72 (91.14%) and cefotetan 77/79 (97.47%). In contrast, all the isolates of S. epidermidis were resistant to erythromycin. About half of the Gram-positive isolates were resistant to ampicillin 41/81 (50.62%).
| Number of resistant pathogens to antimicrobial agents (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial isolates | CIP | AK/AM | PEN | CXM | CTX | CX | CTO | CN | CC | |
| S. epidermidis (n = 2) | 0 (0.00) | 0 (0.00) | 1 (50.00) | 1 (50.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| S. aureus (n = 79) | 7 (8.86) | 26 (32.91) | 13 (16.46) | 10 (12.66) | 7 (8.86) | 39 (49.37) | 2 (2.53) | 9 (11.39) | 19 (24.05) | |
| Total (81) | 7 (8.64) | 26 (32.10) | 14 (17.28) | 11 (13.58) | 7 (8.64) | 39 (48.15) | 2 (2.47) | 9 (11.11) | 19 (23.46) | |
| MRP | LEV | P | TET/TE | AMP | BA | E | ||||
| S. epidermidis (n = 2) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (50.00) | 1 (50.00) | 0 (0.00) | 2 (100.00) | |||
| S. aureus (n = 79) | 4 (5.06) | 7 (8.86) | 4 (5.06) | 22 (27.85) | 40 (50.63) | 1 (1.26) | 23 (29.11) | |||
| Total (81) | 4 (4.94) | 7 (8.64) | 4 (4.94) | 23 (28.40) | 41 (50.62) | 1 (1.24) | 25 (30.86) | |||
- Abbreviations: AK/AM = Amoxicillin–Clavulanate, AMP = Ampicillin, BA = Bacitracin, CC = Chloramphenicol, CIP = Ciprofloxacin, CN = Clindamycin, CTO = Cefotetan, CTX = Cefotaxime, CXM = Cefuroxime, CX = Cefoxitin, E = Erythromycin, LEV = Levofloxacin, MRP = Meropenem, P = Piperacillin, PEN = Penicillin, TET/TE = Tetracycline.
The Gram-negative isolates were K. pneumoniae (72), P. aeruginosa (71), P. mirabilis (60), E. coli (28), Enterobacter spp (18), P. vulgaris (17) and K. oxytoca (16). More than 50% of the predominant Gram-negative isolate, K. pneumoniae, were resistant to clindamycin 45/72 (62.50%) but were susceptible to levofloxacin 70/72 (97.22%), cefotetan 69/72 (95.83%) and chloramphenicol 67/72 (93.06%).
Close to 50% of P. aeruginosa isolates exhibited resistance to clindamycin 35/71 (49.30%). Interestingly, the isolates of Enterobacter spp. and K. oxytoca showed no resistance to chloramphenicol and cefotetan. Also, the isolate of P. vulgaris exhibited no resistance to chloramphenicol and cefotetan. Details of these results are presented in Table 5.
| Number of resistant pathogens to antimicrobial agents (%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial isolates | CRO | CHL | CTO | CAZ | CXM | CTX | CC | AK/AM |
| K. pneumoniae (n = 72) | 36 (50.00) | 5 (6.94) | 3 (4.17) | 12 (16.67) | 29 (40.27) | 33 (45.83) | 45 (62.50) | 8 (11.11) |
| P. aeruginosa (n = 71) | 10 (14.08) | 1 (1.41) | 2 (2.82) | 14 (19.72) | 10 (14.09) | 15 (21.13) | 35 (49.30) | 7 (9.86) |
| P. mirabilis (n = 60) | 14 (23.33) | 5 (8.33) | 4 (6.67) | 6 (10.00) | 13 (21.67) | 14 (23.33) | 34 (56.67) | 7 (8.75) |
| E. coli (n = 28) | 12 (28.57) | 1 (3.57) | 2 (7.14) | 7 (25.00) | 8 (28.57) | 10 (31.71) | 16 (57.14) | 1 (3.57) |
| Enterobacter spp. (n = 18) | 9 (50.00) | 0 (0.00) | 0 (0.00) | 6 (33.33) | 9 (50.00) | 13 (72.22) | 7 (38.89) | 3 (16.67) |
| P. vulgaris (n = 17) | 8 (47.06) | 0 (0.00) | 1 (5.88) | 2 (11.77) | 7 (41.18) | 5 (29.41) | 12 (70.59) | 1 (5.88) |
| K. oxytoca (n = 16) | 4 (25.00) | 0 (0.00) | 0 (0.00) | 3 (18.75) | 2 (12.50) | 2 (12.50) | 10 (62.50) | 3 (18.75) |
| Total (282) | 93 (32.98) | 12 (4.26) | 12 (4.26) | 50 (17.73) | 78 (27.66) | 92 (32.62) | 159 (56.38) | 30 (10.64) |
| MRP | TET | LEV | CIP | AMP | CN | |
|---|---|---|---|---|---|---|
| K. pneumoniae (n = 72) | 9 (12.50) | 8 (11.11) | 2 (2.78) | 20 (27.78) | 9 (12.50) | 25 (34.72) |
| P. aeruginosa (n = 71) | 4 (5.63) | 5 (7.04) | 0 (0.00) | 12 (16.90) | 4 (5.63) | 12 (16.90) |
| P. mirabilis (n = 60) | 7 (11.67) | 9 (15.00) | 1 (1.67) | 15 (25.00) | 12 (12.00) | 9 (15.00) |
| E. coli (n = 28) | 6 (21.43) | 5 (17.86) | 2 (7.14) | 8 (28.57) | 4 (14.29) | 6 (21.43) |
| Enterobacter spp. (n = 18) | 2 (11.11) | 2 (11.11) | 0 (0.00) | 6 (33.33) | 2 (11.11) | 7 (38.89) |
| P. vulgaris (n = 17) | 2 (11.77) | 1 (5.88) | 0 (0.00) | 5 (29.41) | 2 (11.77) | 6 (35.29) |
| K. oxytoca (n = 16) | 1 (6.25) | 3 (18.75) | 0 (0.00) | 3 (18.75) | 0 (0.00) | 6 (37.50) |
| Total (282) | 31 (10.99) | 33 (11.70) | 5 (1.77) | 69 (24.47) | 33 (11.70) | 71 (25.18) |
- Abbreviations: AK/AM = Amoxicillin–Clavulanate, AMP = Ampicillin, CAZ = Ceftazidime, CC = Clindamycin, CHL = Chloramphenicol, CIP = Ciprofloxacin, CN = Gentamycin, CRO = Ceftriaxone, CTO = Cefotetan, CTX = Cefotaxime, CXM = Cefuroxime, LEV = Levofloxacin, MRP = Meropenem, TET = Tetracycline.
4 Discussion
In this study, the prevalence of bacteriological agents causing wound infection in the participants was 70.21% (363/517). The proportion of the bacteriological agents isolated indicates significant insights into the prevalent bacterial species associated with wound infections during the study period. S. aureus emerged as the most dominant isolate, comprising approximately 21.76% (79/363) of the total isolates, suggesting its prominent role as a causative agent of wound infections among the population of the participants studied. On the other hand, S. epidermidis represented the least isolated species 2/363 (0.55%), implying its relatively minor contribution compared to other bacterial strains. The findings in this study are in agreement with other studies by various researchers in Egypt [20], Saudi Arabia [21], Italy [22], Ethiopia [23], Uganda [24] and Nepal [25]. In contrast, a study conducted in Nigeria [26] reported Pseudomonas spp as the most predominant bacterial isolate in that study.
The dominance of S. aureus in wound infections underscores its significant role as a major pathogen in hospital settings. This bacterium's ability to colonize the skin and medical equipment, coupled with its capacity to evade the host immune system, makes it a formidable challenge in wound care management [27]. In contrast, the low prevalence of S. epidermidis observed in this study may be attributed to its usual role as a skin commensal rather than a primary pathogen. However, S. epidermidis is known to act as an opportunistic pathogen, particularly in immunocompromised individuals or in cases involving indwelling medical devices [28]. This dual nature of S. epidermidis suggests that while it may not be a dominant pathogen in wound infections, its potential role should not be underestimated, especially in specific patient populations. These findings emphasize the importance of continued surveillance and the development of tailored strategies to manage and mitigate the burden of wound-associated infections.
A focus on controlling S. aureus through improved hygiene practices, sterilization of medical equipment and effective wound care protocols is essential. Additionally, understanding the dynamics of less prevalent species like S. epidermidis could provide insights into preventing opportunistic infections in vulnerable patients.
The study revealed 81 Gram-positive and 282 Gram-negative isolates. In agreement with this finding, studies done in Saudi Arabia [21], Ethiopia [23] and Italy [29] reported predominant Gram-negative isolates, while another study in Egypt [20] recorded more Gram-positives than Gram-negatives. Among the Gram-positive bacteria, S. aureus predominated with 79 cases, followed by S. epidermidis with 2 cases. On the other hand, K. pneumoniae was the most common Gram-negative isolate with 72 cases, followed closely by P. aeruginosa, 71 cases, and P. mirabilis, 60 cases. This finding is consistent with other studies done in Nigeria [30] and Egypt [20].
The study also reported on the prevalence of various bacteria across different age demographics. Based on age groups, S. aureus exhibited an increasing trend with advancing age, peaking in the 31–40 age group, while K. pneumoniae and P. aeruginosa showed fluctuating distributions across age groups. Interestingly, S. epidermidis was exclusively isolated in the 41–50 age group. Although the highest proportion of bacterial isolates was observed in the 31–40 age group, and the lowest in the < 1 age group, these distributions are not statistically significant. These age-related trends in bacterial isolates could be attributed to various factors. For instance, the increase in S. aureus with age might be linked to changes in skin integrity, which tend to occur as individuals age. Aging skin is more susceptible to microtrauma, reduced barrier function and changes in moisture levels, making it more vulnerable to colonization by bacteria like S. aureus [31]. Additionally, immune function tends to decline with age, a phenomenon known as immunosenescence, which could contribute to the higher prevalence of certain bacteria in older age groups [32]. The exclusive isolation of S. epidermidis in the 41–50 age group could also reflect age-related shifts in skin microbiota composition [33], possibly influenced by hormonal changes and environmental exposures over time [34].
On gender-based variations in bacterial isolates, the study recorded a higher total count in females compared to males. In contrast to this study's findings, a study in Nigeria [35] reported higher infection incidence among males than females. Similarly, a previous study in Rwanda [36] has reported significantly higher wound infection rates in males than females. The observed pattern of higher bacterial counts in females, although not statistically significant, aligns with the broader context of healthcare-seeking behaviour, where gender differences often emerge due to various sociocultural factors and could partially explain the higher incidence of bacterial species isolated in females. A study in Taiwan highlighted that women are more likely to engage in health-protective behaviours and seek medical care compared to men, who may delay seeking treatment due to sociocultural norms that emphasize masculinity and self-reliance [37]. Additionally, gender roles and expectations in certain societies may influence how men and women perceive and respond to illness, which could affect the timing and frequency of medical consultations and, consequently, the detection and management of infections [38, 39]. These differences could partly explain the variations in bacterial isolates observed in this study.
The study also elucidates the relationship between bacterial isolates and both department and study period. While no significant association was found between bacterial isolates and department, an association was detected with the study period. These findings underscore the complex interplay of factors influencing bacterial prevalence, including age, gender, department and study period, with implications for clinical practice and further research in wound management and infection control.
The antimicrobial susceptibility patterns observed in this study highlight significant differences between Gram-positive and Gram-negative isolates, which have important implications for treatment and infection control practices. The number of S. aureus resistant to ampicillin and cephalexin, coupled with those susceptibility to ciprofloxacin, cefotaxime and cefotetan, suggests that the latter antibiotics may be more effective choices in managing infections caused by this organism. However, the resistance patterns observed in S. epidermidis, particularly to erythromycin, indicate the need for careful selection of antibiotics, considering the potential for treatment failure if resistance is not accounted for.
For Gram-negative isolates like K. pneumoniae, the resistance to clindamycin but susceptibility to levofloxacin, cefotetan and chloramphenicol points to the need for tailored antibiotic therapy based on susceptibility testing. The lack of resistance in some isolates, such as Enterobacter spp., K. oxytoca and P. vulgaris, to certain antibiotics, may present opportunities for effective treatment. Yet, it also underscores the necessity for ongoing surveillance to detect emerging resistance trends.
The distinct resistance profiles between Gram-positive and Gram-negative bacteria have significant implications for infection control practices. For instance, the significant number of isolates that exhibited resistance to commonly used antibiotics like ampicillin and clindamycin necessitate stricter antibiotic stewardship programs to prevent the further spread of resistant strains. In clinical settings, the choice of empirical therapy should be guided by local susceptibility data to ensure optimal outcomes. The variation in resistance patterns observed in this study compared to others could be attributed to factors such as differences in wound type, geographic location and patterns of antibiotic use and misuse, as highlighted in studies done in Ethiopia [23] and Italy [22]. These findings underscore the critical importance of prudent antibiotic use, tailored infection prevention and control measures and continued surveillance to combat the growing threat of antimicrobial resistance effectively.
Further research is warranted to explore the underlying factors contributing to these resistance patterns and to develop strategies to mitigate their impact on patient care. By integrating these insights into clinical practice, healthcare providers can better manage infections and reduce the burden of antimicrobial resistance in both hospital and community settings.
5 Conclusion
This study highlights the bacteriological agents causing wound infections at St. Dominic Hospital from 2020 to 2022, with a high prevalence of 70.21% (363/571). S. aureus was identified as the most dominant isolate, comprising 21.76% (79/363) of total cases, underscoring its critical role in wound infections and the necessity for targeted interventions against this pathogen. The study also observed a higher prevalence of Gram-negative bacteria, with K. pneumoniae and P. aeruginosa being the most common isolates. Antibacterial susceptibility patterns revealed significant resistance trends, particularly among Gram-negative isolates, emphasizing the urgent need for prudent antibiotic use and ongoing surveillance to combat resistance.
Author Contributions
John Gameli Deku, Francisca Esenam Goloe and Jonathan Maniye Nmoandor: conceptualization. Francisca Esenam Goloe, Vida Angmorkie Eshun and Eunice Agyei: methodology. John Gameli Deku: project administration. John Gameli Deku, Enoch Aninagyei, Richard Vikpebah Duneeh, Israel Bedzina and Kwabena Obeng Duedu: resources. Francisca Esenam Goloe, Vida Angmorkie Eshun and Eunice Agyei: investigation. John Gameli Deku, Francisca Esenam Goloe, Vida Angmorkie Eshun and Eunice Agyei: supervision. Francisca Esenam Goloe, Vida Angmorkie Eshun and Eunice Agyei: data curation. Israel Bedzina and Kwabena Obeng Duedu: formal analysis. John Gameli Deku, Israel Bedzina and Richard Vikpebah Duneeh: writing – original draft. John Gameli Deku and Enoch Aninagyei: writing – review and editing. All authors read and approved the final version of the manuscript.
Acknowledgements
The authors are grateful to the Management and Laboratory Staff of the St. Dominic Hospital for allowing us access to their data for this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data are available from the corresponding author upon satisfactory request.




