Circular RNAs as prognostic and diagnostic biomarkers in renal cell carcinoma
Abstract
Background
Circular RNAs (circRNAs) play pivotal roles in proliferation, apoptosis, migration, and invasion of renal cell carcinoma (RCC) cells. This study is aimed to systematically summarize the current evidence regarding the clinical implications of circRNAs in RCC patients.
Methods
A systematic search in PubMed, Embase, and Web of Science was performed until January 1, 2022. The correlation between the expression of circRNAs and clinicopathological, prognostic, and diagnostic features of RCC was evaluated using the meta-analysis.
Results
Ultimately, 41 studies with 3485 RCC patients were included in this study: 26 studies for clinicopathological features, 31 studies for prognosis, and eight studies for diagnosis. Altered expression of circRNAs was significantly associated with clinicopathological characteristics of RCC, including tumor size, tumor stage, lymph node metastasis, distant metastasis, and TNM stage. The tumor promoter circRNAs were associated with reduced overall survival (OS) (Hazard Ratio (HR) = 1.98, 95% confidence interval [CI] 1.68–2.34) and disease/progression/recurrence-free survival (DFS/PFS/RFS) (HR = 2.34, 95% CI 1.85–2.97). Contrarily, the tumor suppressor circRNAs were linked with better OS (HR = 0.49, 95% CI 0.40–0.60) and DFS/PFS/RFS (HR = 0.40, 95% CI 0.28–0.59). The pooled sensitivity and specificity of circRNAs for RCC diagnosis in tissue samples were both 0.84. These results in fluid samples (serum and urine) were 0.78 and 0.69, respectively.
Conclusion
CircRNAs can serve as promising diagnostic and prognostic biomarkers for RCC.
1 INTRODUCTION
In adults, renal cell carcinoma (RCC) is the most prevalent type of kidney cancer, which leads to more than 175,000 deaths annually in the world, with an age-standardized global incidence of 4.5 per 100,000 persons.1-3 Partial and radical nephrectomy is the standard of care for patients with localized clear cell RCC. However, 30% of patients have metastatic lesions at initial diagnosis, and more than 25% eventually develop metastasis after the operation.4 In patients with metastatic RCC, available therapies often fail to inhibit the tumor growth or achieve complete remission effectively, with estimated patients' overall survival (OS) remaining less than 1 year.5 Therefore, precise estimation of recurrence risk after nephrectomy is essential for personalized follow-up and finding patients who benefit from specific targeted therapies.6 Unfortunately, biomarkers for early diagnosis and monitoring of RCC are not available thus far, and current prognostic evaluations are based on conventional properties such as tumor stage, size, and grade, offering restricted predictive accuracy for clinical outcomes.7 A better understanding of the molecular mechanisms involved in RCC could lead to finding novel molecular biomarkers for both diagnosis and prognosis assessment.
Renal cell carcinoma tumorigenesis is a highly complex process involving various dysregulations in genetic and epigenetic pathways.8, 9 Non-coding RNAs (ncRNAs) are a part of epigenetic alternations investigated in the last decade. It has been revealed that numerous ncRNAs are involved in cancer development and progression.10-12 Because of high specificity and easy detection in the tissues, serum, and body fluids, exploring the potential application of ncRNAs as diagnostic, prognostic, and novel therapeutic targets in cancer is currently an emerging area of interest.13, 14 CircRNAs are single-stranded, closed-loop RNAs widely expressed in the human genome and are distributed in several malignancies.15 Formerly, circRNAs were categorized as ncRNAs because of their conserved structure; however, recent investigations have revealed the translation of some circRNAs.16 CircRNAs regularly function through interacting with microRNAs (miRNAs), a type of ncRNAs that regulate gene expression post-transcriptionally.17 CircRNAs also play pivotal roles in modulating transcription and splicing and are highly stable and evolutionary conserved with tissue-specific expression patterns, making them valid biomarkers for diagnostic and prognostic evaluations.18, 19
Although circRNAs have been demonstrated to be involved in various malignancies, including breast cancer, lung adenocarcinoma, colon cancer, and hepatocellular carcinoma, their overall pathophysiological contribution to RCC is still largely unknown.20-22 Increasing evidence has demonstrated that circRNAs play critical roles in RCC cell proliferation, apoptosis, migration, and invasion.23-25 A comprehensive understanding of circRNAs may contribute to developing new diagnostic and prognostic biomarkers and novel therapeutic targets for RCC in the future. The present study is aimed to systematically summarize the current evidence regarding the clinical implications of circRNAs in RCC patients.
2 MATERIALS AND METHODS
This systematic review and meta-analysis was conducted and reported in compliance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement and a registered protocol.26, 27 The ethical and Institutional Review Board (IRB) approvals were obtained for each of the included studies; therefore, no additional approvals were required for this study.
2.1 Search strategy
A systematic search in PubMed, Embase, and Web of Science was performed using the keywords [“Renal Carcinoma” OR “Kidney neoplasm” OR “Renal cancer”] AND [“RNA, Circular”] until January 1, 2022, without any language or study type restrictions. Furthermore, the references within the included studies and review articles were manually evaluated to find additional results. The detailed search strategy in each electronic database is described in Supporting information.
2.2 Study selection
After removing duplicate results, two experienced investigators in the fields of cancer biology and circRNAs independently reviewed the titles and abstracts of the remaining articles to choose those studies addressing the significance of circRNAs in RCC. The studies without a clear description of circRNAs in RCC, alongside the retracted articles, were excluded. At this point, the full texts of the selected studies were independently assessed for inclusion by two investigators based on the following eligibility criteria. The discrepancies were resolved by discussion with a third author.
Eligibility criteria:
I. The study consisted of adult (above 18 years of age) patients with diagnosed RCC based on the histopathological investigation;
AND
II. The correlation between the expression of at least one circRNA and clinicopathological, prognostic, or diagnostic features of RCC was evaluated;
AND
III. Adequate data were reported for extraction or calculation of odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) for clinicopathological features, hazard ratios (HRs) and 95% CIs for prognostic endpoints, and sensitivity and specificity for the diagnostic outcomes.
The non-original studies (reviews, letters, and commentaries), conference abstracts, and studies without available data for predefined outcomes were excluded.
2.3 Data extraction and outcome definition
The following data were extracted from all the included studies: the first author name, publication year, country, sample size, circRNAs and their expression profile, detection sample, method of detection, and cutoff points (and the number of patients with high-level and low-level expression). The circRNAs were categorized into two groups based on their expression profile and overall impact on RCC progression: (1) Tumor promoter (upregulated), and (2) Tumor suppressor (downregulated).28 The data extraction process was independently performed by two authors and double-checked by a third investigator to ensure accuracy.
The clinicopathological features were defined as: (1) Age (older vs. younger); (2) Gender (male vs. female); (3) Estimated glomerular filtration rate (eGFR) (<60 vs. ≥60 ml/min/1.73 m2); (4) Tumor size (larger vs. smaller); (5) Tumor grade (III + IV vs. I + II); (6) T stage (III + IV vs. I + II); (7) Lymph node metastasis (yes vs. no); (8) Distant metastasis (yes vs. no); and (9) TNM stage (III + IV vs. I + II). The cutoff values for age ranged from 50 to 70 years among the included (the majority of which used the cutoff value of 60 years). Regarding tumor size, the cutoff values ranged from 3 to 7 cm (most of the included studies used the cutoff value of 5 cm to classify the tumors based on their size). In line with previous meta-analyses, we defined older and younger groups for age and smaller and larger groups for tumor size according to the categories of the included studies.28, 29 The number of patients in high-and low-expression groups of the circRNAs for these endpoints, and p-values for the chi-squared test comparing the high- and low-expression groups were gathered.
Two endpoints were assumed for the prognosis of RCC: 1. OS, and 2. Disease-free survival (DFS), or Progression-free survival (PFS), or Recurrence-free survival (RFS). As DFS, PFS, and RFS address similar outcomes, they were considered one endpoint, DFS/PFS/RFS.30 The follow-up duration, analysis method (univariate or multivariate), and effect sizes (HRs and 95% CIs) were extracted. According to the previous methods, the HRs and 95% CIs were estimated based on provided data and Kaplan–Meier survival curves if they were not reported directly.31
The sensitivity, specificity, and area under the receiver operating characteristic (ROC) curve (AUC) of the circRNAs for detecting RCC were regarded as diagnostic endpoints. According to the ROC curves of various circRNAs, the number of true-positive, false-positive, true-negative, and false-negative cases were calculated.
2.4 Quality assessment
The methodological quality of studies included in the prognostic analysis was investigated using the Newcastle-Ottawa scale (NOS), which consists of three main domains (population selection, comparability, and outcome assessment) with a total score of nine.32 The risk of bias within the diagnostic studies was evaluated with the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool, comprising four categories (patient selection, index test, reference standard, and flow and timing) and a maximum score of seven.33 Two investigators independently completed this process, and a meeting was arranged in case of inconsistency.
2.5 Statistical analysis
All the analyses were conducted in Stata (version 14.2; Stata Corp) with p-value <0.05 indicating statistical significance. For clinicopathological features, pooled ORs and 95% CIs were calculated for the tumor promoter and tumor suppressor circRNAs. Concerning the prognostic endpoints, pooled HRs and 95% CIs were used to explore the association between the expression of tumor promoter and tumor suppressor circRNAs and the survival of patients with RCC. In this regard, separate analyses were performed to pool HRs obtained from univariate and multivariate analyses. In multivariate analysis, the results were adjusted for age,34-39 gender,34-36, 38, 39 tumor grade,34, 36-42 tumor/clinical stage,34-37, 39-44 tumor size,36, 40, 43, 44 metastasis,34, 35, 41-43 and surgical margin.40 Sensitivity analysis was also performed to evaluate each study's impact on the pooled HRs by omitting one study at a time from the meta-analysis. Regarding RCC diagnosis, pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and AUC of the circRNAs were calculated. Separate analyses were performed to ascertain the diagnostic accuracy of circRNAs detected in tissue samples and body-fluid (serum and urine) samples.
The statistical heterogeneity was evaluated by Cochrane's Q test (p-value <0.05 indicating heterogeneity) and the Higgins' I-squared test (I2 > 50% signifying heterogeneity).45 Random-effect models were implemented to pool effect sizes if the statistical heterogeneity was high (I2 > 50%); otherwise, fixed-effect models were utilized. Publication bias was investigated by visual assessment of funnel plots and Egger's test for prognostic meta-analysis, and Deeks' funnel plot asymmetry test for diagnostic meta-analysis with p-values <0.05 demonstrating the presence of publication bias.
3 RESULTS
3.1 Search results and study selection
After duplicate removal, the systematic search within the databases yielded 233 results, and the manual search added one record (Figure 1). Based on the title and abstract review, 138 articles were excluded, and 95 studies were recruited for the full-text review. Among these, 54 studies were excluded due to the following reasons: non-original articles (n = 19), conference abstracts (n = 3), and studies without available data for any of the defined outcomes (n = 32). Ultimately, 41 studies consisting of 3485 patients with RCC were included in this study, among which 26 studies investigated clinicopathological features,34, 37-39, 42-44, 46-64 31 studies evaluated prognostic outcomes,23, 25, 34-44, 46, 48, 50, 51, 53-57, 63-71 and eight studies assessed the diagnosis of RCC.34, 40-42, 48, 58, 72, 73 The studies were published between 2017 and 2022 and were conducted in China (n = 38), Germany (n = 2), and Canada (n = 1). The majority of studies utilized quantitative real-time polymerase chain reaction (qRT-PCR) on RCC tissue samples for their investigation (two studies evaluated serum samples,58, 72 and one study evaluated urine samples73).
3.2 Clinicopathological features
Among 26 studies with a total of 2048 patients, the expression of six circRNAs (circ-0001451, circ-RAPGEF5, circ-AMOTL1L, circ-ESRP1, circ-CYP24A1, and circ-DVL1) was downregulated,34, 43, 57, 58, 62, 63 whereas 18 circRNAs (circ-001895, circ-001287, circ-HIPK3, circ-PDK1, circ-MYLK, circ-0085576, circ-PTCH1, circ-400,068, circ-NUP98, circ-001842, circ-RS-7 (two studies), circ-AKT1, circ-SDHC (two studies), circ-TLK1, circ-SNX6, circ-0054537, circ-CHST15, and circ-PPP6R3) were upregulated in the RCC tissue37-39, 42, 44, 46-56, 59-61, 64 (Table S1). The tumor suppressor (downregulated) circRNAs were associated with smaller tumor size (OR = 0.71 [95% CI 0.58–0.87]; p = 0.001), lower T stage (OR = 0.45 [95% CI 0.27–0.75]; p = 0.002), less lymph node metastasis (OR = 0.29 [95% CI 0.12–0.72]; p = 0.008), less distant metastasis (OR = 0.24 [95% CI 0.14–0.41]; p < 0.001), and lower TNM stage (OR = 0.36 [95% CI 0.24–0.56]; p < 0.001). The tumor promoter (upregulated) circRNAs were correlated with higher T stage (OR = 2.29 [95% CI 1.17–4.49]; p = 0.015), more lymph node metastasis (OR = 1.57 [95% CI 1.00–2.48]; p = 0.050), more distant metastasis (OR = 1.96 [95% CI 1.27–3.02]; p = 0.002), and higher TNM stage (OR = 2.26 [95% CI 1.55–3.30]; p < 0.001). No association was observed between the expression of circRNAs and age, gender, eGFR, or tumor grade (Table 1).
| Tumor promoter (upregulated) | Tumor suppressor (downregulated) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Studies (no.), Cases (no.) | OR | 95% CI | p-value | Heterogeneity (PQ, I2) | Studies (no.), Cases (no.) | OR | 95% CI | p-value | Heterogeneity (PQ, I2) | |
| Age (older/younger) | 20 studies, n = 1479 | 1.06 | 0.95–1.20 | 0.293 | PQ = 0.969, I2 = 0.0% | Six studies, n = 569 | 0.85 | 0.71–1.02 | 0.077 | PQ = 0.105, I2 = 45.1% |
| Gender (male/female) | 20 studies, n = 1479 | 1.06 | 0.99–1.14 | 0.109 | PQ = 0.908, I2 = 0.0% | Six studies, n = 569 | 1.07 | 0.95–1.21 | 0.259 | PQ = 0.381, I2 = 5.5% |
| eGFR (<60/≥60 mL/min/1.73 m2) | 2 studies, n = 174 | 1.13 | 0.76–1.69 | 0.537 | PQ = 0.290, I2 = 10.7% | – | – | – | – | – |
| Tumor size (larger/smaller) | 13 studies, n = 837 | 1.09 | 0.79–1.50 | 0.589 | PQ <0.001, I2 = 74.4% | Three studies, n = 406 | 0.71 | 0.58–0.87 | 0.001 | PQ = 0.650, I2 = 0.0% |
| Tumor grade (III + IV/I + II) | 17 studies, n = 1329 | 1.21 | 0.97–1.51 | 0.083 | PQ = 0.001, I2 = 61.0% | Three studies, n = 384 | 0.88 | 0.46–1.68 | 0.692 | PQ = 0.073, I2 = 61.7% |
| T stage (III + IV/I + II) | 8 studies, n = 616 | 2.29 | 1.17–4.49 | 0.015 | PQ <0.001, I2 = 76.1% | Three studies, n = 182 | 0.45 | 0.27–0.75 | 0.002 | PQ = 0.678, I2 = 0.0% |
| Lymph node metastasis (yes/no) | 12 studies, n = 929 | 1.57 | 1.00–2.48 | 0.050 | PQ <0.001, I2 = 76.0% | Two studies, n = 103 | 0.29 | 0.12–0.72 | 0.008 | PQ = 0.638, I2 = 0.0% |
| Distant metastasis (yes/no) | 14 studies, n = 1040 | 1.96 | 1.27–3.02 | 0.002 | PQ <0.001, I2 = 74.6% | Four studies, n = 430 | 0.24 | 0.14–0.41 | <0.001 | PQ = 0.952, I2 = 0.0% |
| TNM stage (III + IV/I + II) | 15 studies, n = 1175 | 2.26 | 1.55–3.30 | <0.001 | PQ <0.001, I2 = 77.6% | Three studies, n = 379 | 0.36 | 0.24–0.56 | <0.001 | PQ = 0.417, I2 = 0.0% |
- Abbreviations: CI, confidence interval; eGFR, estimated glomerular filtration rate; OR, odds ratio.
- Bold format indicate P < 0.05.
3.3 Prognosis
In total, 31 studies, including 2906 patients, investigated the prognostic endpoints (two studies reported the results for three circRNAs,40, 41 and each of the remaining 29 studies examined only one circRNA23, 25, 34-37, 39, 42-44, 46, 48, 50, 51, 53-57, 63, 65-71). These studies evaluated 30 distinct circRNAs (three circRNAs were investigated in more than one study: 1. Circ-EGLN3,23, 40, 41, 69 2. Circ-HIPK3,48, 67 and 3. Circ-RS-744, 70). The maximum follow-up duration was in the range of 40 to 240 months (Table 2). Based on NOS, the quality of included studies ranged from six to nine (from a total score of nine) with a median score of seven (Figure S1A).
| Study, year | circRNA (s) | Country | Detection method | Detected sample | Cutoff | Expression related to poor prognosis | Case (No.) | Outcomes | Max. follow-up (months) | HR availability | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wang, K. 2017 | circ-HIAT1 | China | qRT-PCR | Tissue | Median | Downregulated | 40 | OS | 40 | Directly | 6 |
| Wang, G. 2018 | circ-0001451 | China | qRT-PCR | Tissue | Median | Downregulated | 52 | OS | 50 | Directly | 9 |
| Zhou, B. 2018 | circ-PCNXL2 | China | qRT-PCR | Tissue | Median | Upregulated | 63 | OS | 60 | Indirectly | 7 |
| Chen, Z. 2019 | circ-001895 | China | qRT-PCR | Tissue | Median | Upregulated | 60 | OS | 133 | Indirectly | 7 |
| Franz, A. 2019 a | circ-EGLN3 | Germany | qRT-PCR | Tissue | Optimal | Downregulated | 99 | OS, RFS | 175 | Directly | 9 |
| circ-RHOBTB3 | Optimal | Downregulated | OS, RFS | Directly | |||||||
| circ-NOX4 | Optimal | Downregulated | OS, RFS | Indirectly | |||||||
| Lin, L. 2019 | circ-EGLN3 | China | qRT-PCR | Tissue | Median | Upregulated | 80 | OS | 60 | Indirectly | 7 |
| Chen, L. 2020 | circ-0001368 | China | qRT-PCR | Tissue | Median | Downregulated | 64 | OS | 60 | Directly | 9 |
| Chen, Q. 2020 | circ-RAPGEF5 | China | qRT-PCR | Tissue | NR | Downregulated | 245 | OS, RFS | 80 | Directly | 9 |
| Han, B. 2020 | circ-HIPK3 | China | qRT-PCR | Tissue | Median | Upregulated | 50 | OS | 60 | Indirectly | 7 |
| Lai, J. 2020 | circ-HIPK3 | China | qRT-PCR | Tissue | Median | Upregulated | 48 | OS | 60 | Indirectly | 7 |
| Li, J. 2020, 1 | circ-MYLK | China | qRT-PCR | Tissue | Relative expression | Upregulated | 71 | OS | 60 | Indirectly | 7 |
| Li, J. 2020, 2 | circ-TLK1 | China | qRT-PCR | Tissue | NR | Upregulated | 60 | OS, DFS | 66 | Indirectly | 7 |
| Li, W. 2020 | circ-CSNK1G3 | China | qRT-PCR | Tissue | NR | Upregulated | 64 | OS | 150 | Indirectly | 7 |
| Li, Wei 2020 | circ-PRRC2A | China | qRT-PCR | Tissue | Median | Upregulated | 516 | OS | 120 | Directly | 9 |
| Liu, G. 2020 | circ-0085576 | China | qRT-PCR | Tissue | ROC analysis | Upregulated | 76 | OS, DFS | 60 | Directly | 9 |
| Liu, H. 2020 | circ-PTCH1 | China | qRT-PCR | Tissue | Median | Upregulated | 39 | OS | 40 | Indirectly | 6 |
| Yu, R. 2020 | circ-NUP98 | China | qRT-PCR | Tissue | Median | Upregulated | 78 | OS, DFS | 60 | Indirectly | 7 |
| Yue, Y. 2020 | circ-101,341 | China | qRT-PCR | Tissue | Median | Upregulated | 60 | OS | 60 | Indirectly | 7 |
| Zeng, J. 2020 | circ-001842 | China | qRT-PCR | Tissue | Median | Upregulated | 97 | OS | 60 | Indirectly | 7 |
| Zhao, Y. 2020 | circ-RS-7 | China | qRT-PCR | Tissue | Median | Upregulated | 87 | PFS | 90 | Indirectly | 7 |
| Zhu, Q. 2020 | circ-AKT1 | China | qRT-PCR | Tissue | Median | Upregulated | 70 | OS | 60 | Indirectly | 7 |
| Cen, J. 2021 | circ-SDHC | China | qRT-PCR | Tissue | Median | Upregulated | 140 | OS | 99 | Directly | 9 |
| Frey, L. 2021 b | circ-EHD2 | Germany | qRT-PCR | Tissue | Optimal | Upregulated | 101 | OS, PFS | 240 | Directly | 9 |
| circ-NETO2 | Optimal | Downregulated | OS, PFS | Directly | |||||||
| circ-EGLN3 | Optimal | Downregulated | OS, PFS | Directly | |||||||
| Lv, Q. 2021 | circ-AGAP1 | China | qRT-PCR | Tissue | NR | Upregulated | 33 | OS | 150 | Directly | 7 |
| Zhang, G. 2021 | circ-EGLN3 | China | qRT-PCR | Tissue | Relative expression | Upregulated | 43 | OS | 60 | Indirectly | 7 |
| Huang, K. 2021 | circ-SNX6 | China | qRT-PCR | Tissue | Median | Upregulated | 81 | OS, PFS | 72 | Directly | 9 |
| Chen, Z. 2021 | circ-PPP6R3 | China | qRT-PCR | Tissue | Median | Upregulated | 96 | OS | 66 | Indirectly | 7 |
| Gao, L. 2021 | circ-AMOTL1L | China | qRT-PCR | Tissue | Median | Downregulated | 51 | OS | 60 | Indirectly | 7 |
| Gui, C. 2021 | circ-CHST15 | China | qRT-PCR | Tissue | Median | Upregulated | 175 | OS, PFS | 144 | Directly | 9 |
| Mao, W. 2021 | circ-RS-7 | China | qRT-PCR | Tissue | Median | Upregulated | 85 | OS | 80 | Directly | 9 |
| Wu, X. 2022 | circ-CYP24A1 | China | qRT-PCR | Tissue | Relative expression | Downregulated | 82 | OS | 145 | Indirectly | 7 |
- Abbreviations: circRNA, circular RNA; DFS, disease-free survival; HR, hazard ratio; NOS, Newcastle-Ottawa scale; NR, not reported; OS, overall survival; PFS, progression-free survival; qRT-PCR, quantitative real-time polymerase chain reaction; RFS, recurrence-free survival; ROC, receiver operating characteristic.
- a The circ-EGLN3 was upregulated in tumor tissue compared to the normal tissue; however, its downregulation (based on the cutoff value) was associated with poor prognosis. The cutoff points were identified by the software X-tile.
- b The circ-NETO2 and circ-EGLN3 were upregulated in tumor tissue compared to the normal tissue; however, their downregulation (based on the cutoff value) was linked with poor prognosis. The cutoff points were determined using the Cutoff Finder algorithm.
For the OS in univariate analysis, 23 studies with tumor promoter (upregulated) circRNAs (consisting of 2186 patients) and eight studies with tumor suppressor (downregulated) circRNAs (including 734 patients) entered the meta-analysis. The tumor promoter circRNAs (circ-PCNXL2, circ-001895, circ-HIPK3, circ-MYLK, circ-TLK1, circ-CSNK1G3, circ-PRRC2A, circ-0085576, circ-PTCH1, circ-NUP98, circ-101,341, circ-001842, circ-AKT1, circ-SDHC, circ-EHD2, circ-AGAP1, circ-SNX6, circ-PPP6R3, circ-CHST15, and circ-RS-7) were associated with reduced OS (HR = 1.98 [95% CI 1.68–2.34]; p < 0.001), while the tumor suppressor circRNAs (circ-HIAT1, circ-0001451, circ-RHOBTB3, circ-NOX4, circ-0001368, circ-RAPGEF5, circ-NETO2, circ-AMOTL1L, and circ-CYP24A1) were linked with better OS (HR = 0.49 [95% CI 0.40–0.60]; p < 0.001) (Figure 2A). The results were consistent in the multivariate analysis both for tumor promoter (seven studies with 1174 patients; HR = 3.08 [95% CI 2.09, 4.52]; p < 0.001) and tumor suppressor (five studies with 561 patients; HR = 0.42 [95% CI 0.30–0.60]; p < 0.001) circRNAs (Figure 2B).
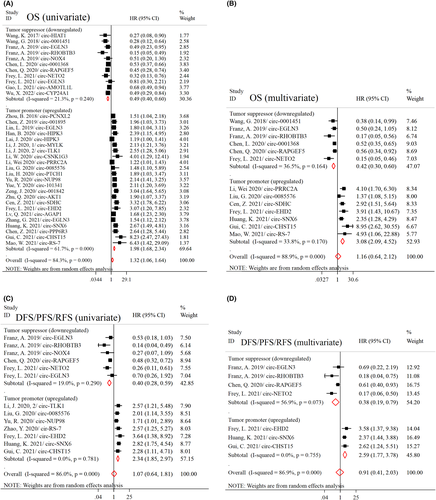
Pooling the HRs for the two circRNAs investigated in more than one study regarding OS in the univariate analysis did not reveal any significant association: 1. Circ-EGLN3 (four studies with 323 cases; HR = 1.11 [95% CI 0.64–1.93]; p = 0.714), and 2. Circ-HIPK3 (two studies with 98 cases; HR = 1.53 [95% CI 0.80–2.97]; p = 0.201) (Figure S2).
Regarding DFS/PFS/RFS in univariate analysis, 11 studies were included in the analysis (seven studies with 658 patients evaluating the tumor promoter and three studies with 445 patients assessing the tumor suppressor circRNAs). The expression of tumor promoter circRNAs was correlated with poor DFS/PFS/RFS (HR = 2.34 [95% CI 1.85–2.97]; p < 0.001), whereas the tumor suppressor circRNAs were associated with improved DFS/PFS/RFS (HR = 0.40 [95% CI 0.28–0.59]; p < 0.001) (Figure 2C). Similar findings were obtained from multivariate analysis for the tumor promoter (three studies with 357 cases; HR = 2.59 [95% CI 1.77–3.78]; p < 0.001) and tumor suppressor (three studies with 445 cases; HR = 0.38 [95% CI 0.19–0.79]; p = 0.010) circRNAs (Figure 2D).
Substantial statistical heterogeneity was detected in all pooled analyses; therefore, random-effect models were implemented. However, subgrouping the circRNAs based on their expression profile (tumor promoters and tumor suppressors) significantly reduced the heterogeneity in these analyses. No significant publication bias was detected according to the funnel plots and Egger's test (Figure S3). Sensitivity analysis revealed that the pooled results were stable after omitting each study from the analysis (Figure S4). Similar findings were obtained by limiting the studies to the Chinese investigations (Table S2).
3.4 Diagnosis
Eight studies, consisting of 604 RCC cases and 527 controls, investigated the diagnostic value of circRNAs for RCC detection34, 40-42, 48, 58, 72, 73 (Table 3). Among these, five studies utilized only RCC tissue samples,34, 40-42, 48 one study used both serum and tissue samples,72 one study investigated serum samples,58 and one study evaluated urine samples.73 The methodological quality of included studies was in the range of five to seven according to the QUADAS-2 (Figure S1B).
| Study, year | circRNA(s) | Country | Detection method | Detected sample | Expression | Case (No.) | Control (No.) | Sensitivity | Specificity | AUC | QUADAS-2 score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wang, G. 2018 | circ-0001451 | China | qRT-PCR | Tissue | Downregulated | 52 | 52 | 0.775 | 0.608 | 0.704 | 5 |
| Franz, A. 2019 | circ-EGLN3 | Germany | qRT-PCR | Tissue | Upregulated | 99 | 85 | 0.950 | 0.950 | 0.980 | 7 |
| circ-NOX4 | Downregulated | 0.910 | 0.710 | 0.810 | |||||||
| circ-RHOBTB3 | Downregulated | 0.720 | 0.910 | 0.820 | |||||||
| Han, B. 2020 | circ-HIPK3 | China | qRT-PCR | Tissue | Upregulated | 50 | 50 | 0.940 | 0.920 | 0.953 | 5 |
| Liu, G. 2020 | circ-0085576 | China | qRT-PCR | Tissue | Upregulated | 76 | 76 | 0.855 | 0.736 | 0.844 | 6 |
| Frey, L. 2021 | circ-EHD2 | Germany | qRT-PCR | Tissue | Upregulated | 101 | 81 | 0.525 | 0.852 | 0.757 | 7 |
| circ-NETO2 | Upregulated | 0.594 | 0.827 | 0.705 | |||||||
| circ-EGLN3 | Upregulated | 0.871 | 0.778 | 0.879 | |||||||
| Zheng, Z. 2021 | circ-PVT1 | China | qRT-PCR | Tissue | Upregulated | 90 | 90 | 0.911 | 0.922 | 0.930 | 6 |
| Serum | Upregulated | 60 | 40 | 0.933 | 0.675 | 0.860 | |||||
| Wang, Y. 2022 | circ-DVL1 | China | qRT-PCR | Serum | Downregulated | 60 | 66 | 0.833 | 0.681 | 0.808 | 5 |
| Peter, M. 2022 | circ-EGLN3 | Canada | qRT-PCR | Urine | Downregulated | 76 | 27 | 0.605 | 0.740 | 0.710 | 5 |
| circ-SOD2 | Urine | Downregulated | 0.618 | 0.666 | 0.680 |
- Abbreviations: AUC, the area under the curve; circRNA, circular RNA; qRT-PCR, quantitative real-time polymerase chain reaction; QUADAS-2, Quality Assessment of Diagnostic Accuracy Studies 2.
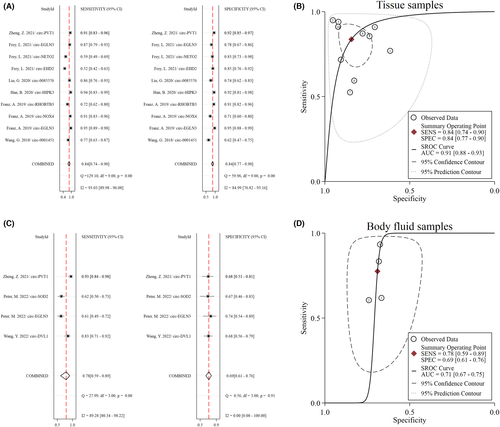
The pooled diagnostic performance of circRNAs for RCC based on tissue samples was as follows: sensitivity 0.84 (95% CI 0.74–0.90), specificity 0.84 (95% CI 0.77–0.90), PLR 5.4 (95% CI 3.40–8.30), NLR 0.19 (95% CI 0.11–0.33), DOR 28 (95% CI 12–66), and AUC 0.91 (95% CI 0.88–0.93) (Figure 3A,B). The pooled performance of circRNAs for RCC detection using body-fluid (serum and urine) specimens reached moderate accuracy (inferior to tissue samples): sensitivity 0.78 (95% CI 0.59–0.89), specificity 0.69 (95% CI 0.61–0.76), PLR 2.5 (95% CI 1.90–3.30), NLR 0.33 (95% CI 0.17–0.64), DOR 8 (95% CI 3–19), and AUC 0.71 (95% CI 0.67–0.75) (Figure 3C,D). Deeks' funnel plot asymmetry test detected no evidence of publication bias in these analyses (Figure S5).
4 DISCUSSION
A growing body of evidence indicated that circRNAs are highly stable and detectable molecules in body fluids and tissues and may act as valid biomarkers for cancer diagnosis and prognosis.46 The present study is a comprehensive systematic review and meta-analysis investigating the clinical significance of circRNAs in RCC in terms of clinicopathological characteristics, diagnosis, and prognosis. Our results included a total of 41 articles involving 3485 patients (31 studies for prognosis assessment and 26 and eight studies for clinicopathological and diagnostic assessments, respectively). According to the results of this study, the accuracy of circRNAs for RCC diagnosis was relatively high in tissue samples (sensitivity and specificity of 0.84) and moderate in fluid specimens (sensitivity of 0.78 and specificity of 0.69). Furthermore, altered expression of circRNAs was significantly associated with tumor characteristics, including tumor size, tumor stage, lymph node metastasis, distant metastasis, and TNM stage. Besides, increased expression of tumor promotor circRNAs and decreased expression of tumor suppressor circRNAs were associated with poor OS in RCC patients. These findings revealed that circRNAs could act as potential biomarkers for RCC diagnosis and prognosis assessment.
Abdominal computed tomography scan is the standard of choice in RCC diagnosis in patients with signs or symptoms of renal tumors, for example, unexplained hematuria74; however, in patients without any symptoms or patients with early developed tumors that cannot be recognized in imaging investigations, other diagnostic evaluations are warranted. Early diagnosis of RCC can effectively guide clinical treatment and significantly increase the radical nephrectomy rate, leading to better survival rates.75 In this regard, identifying specific biomarkers can help in the early detection of RCC patients. CircRNAs possess outstanding features as potential diagnostic biomarkers, including being highly stable and enriched in tissue samples, serum, and other body fluids.76 Regarding the diagnostic significance of circRNAs in RCC tissue samples, our results indicated a pooled sensitivity, specificity, and AUC of 0.84, 0.84, and 0.91, respectively, suggesting the potential role of circRNAs in RCC diagnosis. Furthermore, in body-fluid (serum and urine) specimens, circRNAs resulted in moderate accuracy with a sensitivity, specificity, and AUC of 0.78, 0.69, and 0.71, respectively. Thus, further studies are required to investigate the diagnostic accuracy of body fluid-augmented circRNAs (e.g., serum and urine samples) that are more likely to be future specimens of the RCC diagnosis. In this study, we separately pooled the data of tissue and body-fluid samples to avoid heterogeneity due to different sample types. Although the diagnostic performance of circRNAs is still limited for confirmation or exclusion of RCC, circRNAs may have significant advantages when combined with other biomarkers or clinical examinations, especially for diagnosing complex cases. Furthermore, combining two or more circRNAs might improve overall diagnostic value, which should be investigated in future multicenter and high-quality studies.
Regarding the clinicopathological significance of circRNAs in RCC, our results demonstrated that increased expression of several circRNAs, including circ-001895, circ-001287, circ-HIPK3, circ-PDK1, circ-MYLK, circ-0085576, circ-PTCH1, circ-400,068, circ-NUP98, circ-001842, circ-RS-7, circ-AKT1, circ-SDHC, circ-TLK1, circ-SNX6, circ-0054537, circ-CHST15, and circ-PPP6R3, is associated with higher tumor stage, more lymph node metastasis, more distant metastasis, and advanced TNM stage. On the other hand, decreased levels of six circRNAs (circ-0001451, circ-RAPGEF5, circ-AMOTL1L, circ-ESRP1, circ-CYP24A1, and circ-DVL1) were associated with smaller tumor size, lower tumor stage, absence of lymph node metastasis, less distant metastasis, and lower TNM stage. These results reveal that these circRNAs, with their differential expression patterns in RCC, can play pivotal roles in tumorigenesis and tumor progression and significantly correlate with clinicopathological features. For instance, circ-RAPGEF5 inhibits RCC migration, invasion, and proliferation by targeting the miR-27a-3p/TXNIP pathway.43 On the contrary, circ-HIPK3 promotes RCC tumor cell proliferation and metastasis through the miR-508-3p/CXCL13 signaling pathway,48 and circ-PDK1 promotes RCC tumor cell migration and invasion by regulating the miR-377-3P-NOTCH1 axis.49
Concerning the prognostic significance of circRNAs in RCC, our results revealed that both upregulated and downregulated circRNAs were significantly associated with survival outcomes (OS and DFS/PFS/RFS) in univariate and multivariate analysis, suggesting the substantial value of circRNAs as biomarkers for RCC prognosis. Among various circRNAs investigated in previous studies, upregulation of circ-CHST15 was associated with the worst OS with an HR of 8.23 (95% CI 2.47–27.43). Circ-CHST15 promotes EIF4EBP1 expression through sponging miR-125a-5p, subsequently resulting in the proliferation and migration of RCC cells.39 Upregulation of circ-RS-7 and circ-CSNK1G3 were also associated with poor OS with the HR of 6.43 (95% CI 1.42–29.09) and 4.01 (95% CI 1.29–12.41), respectively. Circ-RS-7 acts as a competing endogenous RNA for miR-139-3p inhibiting Transgelin degradation and promoting the proliferation and metastasis of RCC via the PI3K/AKT pathway.44 Circ-CSNK1G3 upregulates miR-181b and promotes tumor proliferation and metastasis through TIMP3-mediated epithelial to mesenchymal transition.71
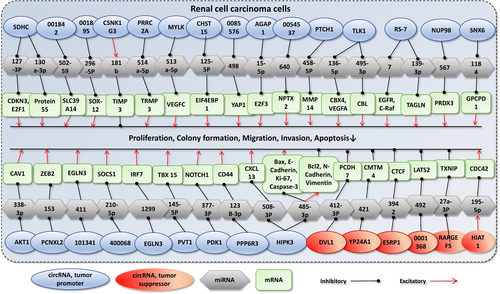
On the other hand, several circRNAs could act as tumor suppressors, and their high expression levels in tumor cells were significantly associated with better survival outcomes. Circ-RHOBTB3, circ-HIAT1, and circ-0001451 were tumor suppressor circRNAs with the lowest HRs (0.15, 0.27, and 0.28, respectively). There are several other circRNAs involved in RCC tumor progression. To better understand the molecular mechanisms of the circRNAs in RCC progression, we have summarized the identified molecular pathways in Figure 4. Taken together, we suggest that a combination of circ-CHST15, circ-RS-7, and circ-CSNK1G3, as tumor promoters with the largest HRs for OS (with pooled HR = 5.80 [95% CI 2.81–11.97]; p < 0.001) and circ-RHOBTB3, circ-HIAT1, and circ-0001451 as tumor suppressors with the smallest HRs for OS (with pooled HR = 0.23 [95% CI 0.13–0.42]; p < 0.001) can lead to a better prognostic evaluation in RCC patients. Further, well-designed studies are demanded to investigate the potential advantages of using a combination of these circRNAs for the prognosis assessment of RCC patients.
Although previous studies showed that several circRNAs are associated with survival outcomes of RCC patients, only circ-EGLN3, circ-HIPK3, and circ-RS-7 were investigated in more than one study. Circ-HIPK3 was investigated in two studies by Han et al.48 and Lai et al.67 Although both investigations indicated that upregulation of circ-HIPK3 in tumor tissue was significantly associated with poor OS in RCC patients, the meta-analysis of pooled data revealed no significant association between the expression level of circ-HIPK3 and OS, which may be due to the small sample sizes of these two studies (only a total of 98 patients). Circ-EGLN3 was evaluated in four previous studies.23, 40, 41, 69 This circRNA was upregulated in tumor tissue compared to the normal adjacent tissue in all four studies. In two studies based on the Asian population, Lin et al. and Zhang et al.23, 69 indicated that upregulation of circ-EGLN3 is significantly associated with poor OS, while two studies based on the German population indicated different findings; Franz et al.40 indicated that high expression level of circ-EGLN3 is significantly associated with increased OS and Frey et al.41 showed no significant association between the expression of circ-EGLN3 and OS. Pooled HR regarding the prognostic significance of circ-EGLN3 was not statistically significant. Future large-scale studies consisting of samples from diverse ethnicities are warranted to shed light on the prognostic significance of circ-EGLN3 and other circRNAs in patients with RCC, considering the potential differences in circRNAs' expression in different races and ethnicities. Circ-RS-7 was upregulated in RCC tissue samples in two studies and associated with poor PFS and OS in patients with RCC.44, 70
Our study had several strengths over previous reviews as a wide-ranging systematic review and meta-analysis regarding the potential clinical significance of circRNAs in RCC.77 We comprehensively included all the previous investigations in various aspects of diagnostic, prognostic, and clinicopathological characteristics of RCC and pooled the data to provide a better picture regarding the utility of circRNAs in patients with RCC. However, the following limitations merit consideration. Firstly, the number of studies and their sample sizes were less than desirable for diagnostic evaluations; only eight studies with 11 circRNAs were included in our analysis. Although we were able to conduct separate meta-analyses to examine the diagnostic value of different types of specimens (tissue and body-fluid samples), in the current literature, most studies used RCC tumor tissue samples to investigate the role of different circRNAs. Although these tumor tissue samples can provide valuable insights into the role of circRNAs in RCC, further research regarding circRNAs in more accessible samples, such as serum and urine, is warranted. Secondly, most of the study population consisted of Asian (Chinese) patients, which may have influenced our findings. Future studies on other races and ethnicities are warranted to assess the role of circRNAs as novel diagnostic and prognostic biomarkers in patients with RCC to obtain more definitive results. Thirdly, the HRs from 19 studies could not be directly extracted. For more comprehensive results, HRs and 95% CIs were estimated from the Kaplan–Meier survival curves based on the methods previously described by Tierney et al.31 Fourthly, none of the included studies in the diagnostic analysis examined the significance of circRNAs in differentiating metastatic from localized RCC. Future studies addressing the role of circRNAs in discriminating metastatic RCC from small localized tumors are needed, as there are substantial differences in disease profiles and prognosis of early and advanced cases. Fifthly, apparent heterogeneity existed between the included studies. We used a more conservative random-effect model to pool the effect sizes. After subgrouping based on the expression level of circRNAs, we found that altered expression might be the source of heterogeneity. Furthermore, the data on individual circRNAs appeared only once between the included studies, and few circRNAs appeared in more than one study, which could influence the statistical heterogeneity.
In summary, the present meta-analysis revealed that circRNAs could serve as promising diagnostic biomarkers for RCC, and aberrant expression of circRNAs is closely correlated with the clinicopathological and prognostic outcomes of patients with RCC. However, future well-designed, large-scale, and high-quality prospective studies, including patients with various ethnicities, are required to ascertain the clinical value of circRNAs in RCC.
AUTHOR CONTRIBUTIONS
Sina Rashedi involved in conceptualization; formal analysis; investigation; data curation, methodology, visualization, wrote the original draft, reviewed, and edited the manuscript. Mahta Mardani involved in conceptualization, data curation, investigation, wrote the original draft, reviewed, and edited the manuscript. Ali Rafati involved in conceptualization, data curation, wrote the original draft, reviewed, and edited the manuscript. Mohammad Mahdi Khavandi involved in methodology, formal analysis, reviewed, and edited the manuscript. Fatemeh Mohammadi involved in methodology, reviewed, and edited the manuscript. Salar Javanshir and Rojin Sarallah involved in data curation, reviewed, and edited the manuscript. Mahsa Dolatshahi, Mohammadmahdi Sabahi, and Sina Azadnajafabad involved in investigation, reviewed, and edited the manuscript. Hamed Tavolinejad involved in formal analysis, reviewed, and edited the manuscript. Nima Rezaei administered the project, supervised, reviewed, and edited the manuscript. All authors read and approved the final manuscript.
FUNDING INFORMATION
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are included in the manuscript and its supporting information.




