Tcf3-activated lncRNA Gas5 regulates newborn mouse cardiomyocyte apoptosis in diabetic cardiomyopathy
Abstract
Diabetic cardiomyopathy can cause cardiac dysfunction and eventually lead to heart failure and sudden death. Long noncoding RNA (lncRNA) Gas5 has been reported to play a function in cardiomyocyte. Here we studied the function of Gas5 on newborn mouse cardiomyocyte (NMC) apoptosis to detect its molecular mechanism. High-glucose treatment was implemented to induce the apoptosis of NMC in this study. And terminal deoxynucleotidyl transferase dUTP nick-end labeling assay, JC-1 assay, and flow cytometry analysis were conducted to know about the apoptosis of NMC when Gas5 and Tcf3 were silenced. Meanwhile, RNA pull-down assay and luciferase reporter assay were conducted to verify the binding of RNAs. Finally, rescue assay was implemented to evaluate the influence on apoptosis situation affected by competing endogenous RNA pathways. Tcf3 was found to bind to the Gas5 promoter to activate the expression of Gas5. Meanwhile, Gas5 and Tcf3 were both found to promote the apoptosis of NMC. Also, mmu-miR-320-3p could bind to Gas5 and Tcf3. Moreover, the Gas5/miR-320-3p/Tcf3 pathway was found to modulate the apoptosis of NMC. In conclusion, Tcf3-activated lncRNA Gas5 regulates NMC apoptosis in diabetic cardiomyopathy.
1 INTRODUCTION
The characteristics of diabetes are hyperglycemia and endogenous insulin aberrant regulation, and diabetes has been studied to be connected to increasing risk of heart failure.1, 2 It has been found that diabetic cardiomyopathy is a kind of myocardial disease, but it is not caused by hypertension or coronary artery disease, which cause much risk of developing heart failure.3 Even though many characteristics of diabetic cardiomyopathy have been reported, the inner molecular mechanism of diabetic cardiomyopathy should be further studied.
Long noncoding RNAs (lncRNAs) have been recently studied in detail. lncRNAs aer more than 200 nucleotides in length and without the function of coding protein.4 Playing important function in many biological processes, lncRNAs can modulate the biological behavior of various cells, such as cancer cell and myocardial cell.5-8 For example, lncRNA TINCR and lncRNA H19 have been reported to regulate cardiomyocyte apoptosis in diabetic cardiomyopathy.9, 10 Meanwhile, the lncRNA growth arrest-specific 5 (GAS5) has been shown to be associated to cardiac diseases. For example, lncRNA GAS5 can modulate the fibrosis of cardiac fibroblasts.11 And lncRNA GAS5 can regulate oxidative stress in the heart.12 However, the function of lncRNA GAS5 in diabetic cardiomyopathy is still not clear. Thus, in this study, we are going to fully investigate the progression of the lncRNA GAS5 in diabetic cardiomyopathy.
lncRNAs can function as competing endogenous RNAs not only in cancer cells but also in myocardial cells to modulate the RNA expression.11, 13, 14 lncRNAs can sponge to microRNA (miRNA) and play the role of lncRNA-miRNA-messenger RNA (mRNA) to regulate the progression of cells.13, 15 For example, the lncRNA APF can modulate the autophagy in myocardial infarction through adjusting the expression of miR-188-3p.16 lncRNA CARL can modulate apoptosis of cardiomyocytes via regulating the expression of miR-539 and PHB2.17
In this study, we are going to explore the mechanism associated to lncRNA GAS5 for discovering a better treatment for diabetic cardiomyopathy.
2 MATERIALS AND METHODS
2.1 Cell culture and treatment
Newborn rat cardiomyocyte (NRC) and newborn mouse cardiomyocyte (NMC) were isolated from the 1- to 2-day old Sprague-Dawley rats (Charles River Laboratories, Boston, MA) as described previously.18 Cell samples of NRC and NMC were cultured in Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA) containing 1% penicillin/streptomycin antibiotics in 5% CO2 at 37°C. Cells were treated with 30 mmol/L high glucose for 0, 12, 24, and 48 hours to induce cell apoptosis, with nontreated cells as control.
2.2 RNA isolation and quantitative real-time polymerase chain reaction
One milliliter of TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) was added to the culture medium for extracting total RNA, then reverse transcription was achieved with SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) for complementary DNA synthesis. The Power SYBR Green Master mix (Applied Biosystems, Carlsbad, CA) was applied for preparing the polymerase chain reaction (PCR) reaction system, followed by 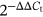 method. GAPDH or U6 served as an endogenous control.
method. GAPDH or U6 served as an endogenous control.
2.3 Cell transfection
The designed short hairpin RNAs (shRNAs) for Tcf3 and Gas5, along with negative control (NC)-shRNAs, were available from GenePharma (Shanghai, China) for gene silencing using transfection kit Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The pcDNA3.1-Tcf3 and pcDNA3.1-NC, as well as miR-320-3p mimics/inhibitor and NC mimics/inhibitor were all procured from GenePharma for 48 hours of cell transfection.
2.4 Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was implemented as per the standard protocol of the EZ-ChIP Kit (Millipore, Billerica, MA). The cross-linked chromatin samples were sonicated for immunoprecipitation with anti-Tcf3 antibody or control anti-immunoglobulin G (IgG) antibody (Millipore). After adding magnetic beads, DNA enrichment was assayed.
2.5 Luciferase reporter assay
Gas5 promoter was PCR amplified for cloning into luciferase reporter pGL3 vector (Promega, Madison, WI) and then cotransfected with Tcf3 overexpression or silencing plasmids. Besides, the Gas5 or Tcf3 fragments covering the miR-320-3p wild-type (WT) and mutated (Mut)-binding sites were cloned into pmirGLO vector (Promega), named as Gas5-WT/Mut and Tcf3-WT/Mut. All luciferase activities were monitored by the Luciferase Reporter Assay System (Promega).
2.6 Terminal deoxynucleotidyl transferase dUTP nick-end labeling assay
The terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay was implemented using the standard method of One-Step TUNEL Apoptosis Assay Kit (Beyotime, Shanghai, China). All cell nuclei were detected by culturing in 4′,6-diamidino-2-phenylindole (DAPI) solution. TUNEL-positive cells were measured by a fluorescence microscope (NIKON, Tokyo, Japan).
2.7 JC-1 assay
Cell apoptosis was also monitored via the change in mitochondrial transmembrane potential (ΔΨm). Cells in 96-well plates were cultured with 10 mM of JC-1 dye (Beyotime) for 30 minutes, followed by washing in the assay buffer. ΔΨm was analyzed by a fluorescent plate reader.
2.8 Flow cytometer assay
Flow cytometer assay was conducted for the detection of cell apoptotic rate utilizing the protocol of Annexin V-labeled with 7-aminoactinomycin D and phycoerythrin (BD Biosciences, San Jose, CA). After double-staining for 15 minutes in the dark, apoptotic cells were analyzed with FACSCalibur flow cytometer (BD Biosciences).
2.9 Western blot
Total protein extracts from cells were prepared to treat with electrophoresis on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein samples were then shifted to polyvinylidene difluoride membranes and cultured with 5% skimmed milk. Primary antibodies against loading control GAPDH and Bax, Bcl-2, cleaved caspase-3, total caspase-3, as well as their relative horseradish peroxidase-marked secondary antibodies were all obtained from Abcam (Cambridge, MA) and applied after dilution. All samples were subjected to the enhanced chemiluminescence fluorescence test kit finally for detecting protein density.
2.10 Subcellular fraction
On the basis of the method of PARIS Kit (Ambion, Austin, TX), subcellular fraction analysis was carried out. After RNA isolation and purification, the expression level of Gas5 was assayed, with GAPDH as cytoplasmic control and U6 as nuclear control.
2.11 Fluorescence in situ hybridization
The designed RNA fluorescence in situ hybridization (FISH) probe specific to Gas5 was available from RiboBio (Guangzhou, China) and utilized in light of the user manual. DAPI dye was applied for counterstaining cell nuclei. Images of stained cells were taken by the NIKON microscope.
2.12 RNA pull-down
The miR-320-3p sequences covering the WT and MT Gas5 or Tcf3 binding sites were severally synthesized and biotinylated into their relative Bio-miR-320-3p-WT/Mut probes. The prepared protein extracts were used to mix with these probes and beads 1 hour. The relative enrichment of Gas5 or Tcf3 was analyzed by quantitative real-time (qRT)-PCR.
2.13 Statistical analyses
Data were all exhibited as the average of independent biotriplicates and results were shown as the means ± standard deviations. GraphPad Prism V5.0 was applied for processing statistical analysis via the Student t-test or one-way analysis of variance, with a p value below .05 representing a significant level.
3 RESULTS
3.1 Tcf3 could bind to Gas5 promoter and activate the expression of Gas5
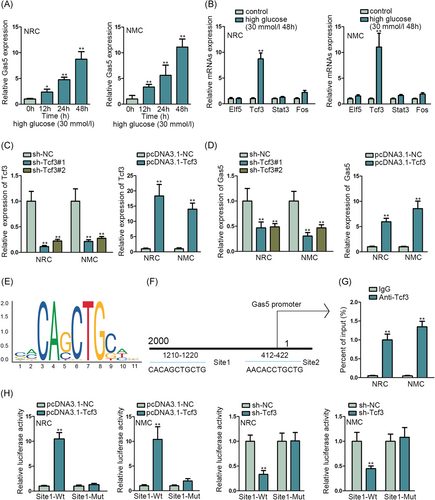
lncRNA GAS5 has been found to modulate the fibrosis of cardiac fibroblasts.11 And lncRNA GAS5 can regulate oxidative stress in the heart.12 Here we detected the expression of lncRNA Gas5 in NRC and NMC when treated with 30 mmol/L high glucose via qRT-PCR assay (Figure 1A). We found the expression of lncRNA GAS5 was upregulated with the hours of 30 mmol/L high-glucose treatment. And then, we considered transcriptional factors that can bind to the lncRNA promoter to activate the expression of lncRNA.19 After that, we detected the transcriptional activators such as Elf5, Tcf3, Stat3, and Fos according to the UCSC website. We implemented the qRT-PCR assay to assess the expression of them under the environment with high glucose of 30 mmol/L for 48 hours (Figure 1B). The results showed that the expression of Tcf3 was increased with 30 mmol/L high glucose. Next, we verified the inhibition efficiency and overexpression efficiency of Tcf3 in NRC and NMC by qRT-PCR assay (Figure 1C). Meanwhile, we investigated the expression of Gas5 when Tcf3 was inhibited or overexpressed by qRT-PCR assay (Figure 1D). We found that the expression of Gas5 was significantly decreased by Tcf3 inhibition and increased by Tcf3 overexpression. After that, we presented the sequence logo of Tcf3 and the binding site of the Tcf3 and Gas5 promoter (Figure 1E,F). Moreover, ChIP assay and luciferase reporter assay was conducted to verify the binding of Tcf3 and Gas5 promoter (Figure 1G,H). We found that Gas5 promoter was abundantly enriched in anti-Tcf3, and luciferase activity in wild binding site was increased by overexpressed Tcf3 and reduced by inhibited Tcf3, which proved the binding of Tcf3 and Gas5 promoter. In conclusion, Tcf3 could bind to the Gas5 promoter and activate the expression of Gas5.
3.2 Inhibited Gas5 and Tcf3 could reduce the apoptosis of NRC and NMC
For detecting the function of lncRNA Gas5 in NRC and NMC, we first evaluated the silence efficiency of Gas5 via qRT-PCR assay (Figure 2A). Next, we conducted a series of functional assays to detect the apoptosis situation of NRC and NMC when Gas5 was silenced. And we first treated high glucose (30 mmol/L) to induce the apoptosis of NRC and NMC. TUNEL assay, JC-1 assay, and flow cytometry analysis found that TUNEL-positive cells and cell apoptosis rate were both reduced by inhibited Gas5, and the JC-1 ratio was upregulated by silenced Gas5 (Figure 2B-D). Meanwhile, Western blot assay also found that Bax and cleaved caspase-3 protein concentration was decreased by Gas5 knockdown, and the Bcl-2 protein level was elevated by Gas5 inhibition (Figure 2E). In a word, Gas5 inhibition could reduce the apoptosis of NRC and NMC, which was elevated by high glucose (30 mmol/L).
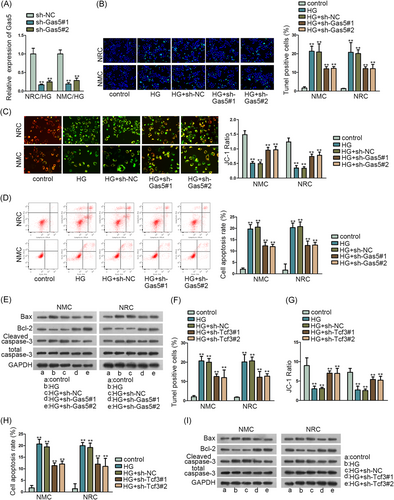
Meanwhile, as transcriptional activator Tcf3 has been reported to play a role in heart formation,20 we further detected its function in NRC and NMC. The apoptosis situation of NRC and NMC was delineated via TUNEL assay, JC-1 assay, flow cytometry analysis, and Western blot assay when Tcf3 was knocked down (Figure 2F-I). High glucose (30 mmol/L) was used to induce the apoptosis of NRC and NMC. And we found the apoptosis of NRC and NMC could be decreased by Tcf3 knockdown also. Both TUNEL-positive cells and cell apoptosis rates were decreased and the JC-1 ratio was increased by Tcf3 inhibition. Moreover, Bax and cleaved caspase-3 protein concentrations were decreased and the Bcl-2 protein level was elevated by silenced Tcf3. In conclusion, inhibited Gas5 and Tcf3 could reduce the apoptosis of NRC and NMC.
3.3 miR-320-3p could bind to Tcf3 and Gas5 in NRC and NMC
As we detected that lncRNAs can act as a miRNA sponge and play the network of lncRNA-miRNA-mRNA to regulate the progression of cells,13, 15 we further detected the location of Gas5 by subcellular fraction assay and FISH assay (Figure 3A,B). The results indicated that Gas5 was mainly located in the cytoplasm of NRC and NMC. Thence we searched the possible miRNAs in ENCORI, and we detected the intersection of miRNAs that could bind to Gas5 and Tcf3 (Figure 3C). Mmu-miR-320-3p, mmu-miR-743a-3p, and mmu-miR-743b-3p were found out. Furthermore, the expression of miRNAs was assessed by qRT-PCR assay when NRC and NMC were treated with high glucose (30 mmol/L) (Figure 3D). The expression of miR-320-3p was significantly decreased in NRC and NMC treated with high glucose. After that, the binding sites Gas5 to miR-320-3p and Tcf3 to miR-320-3p were presented according to ENCORI (Figure 3E). The overexpression efficiency of miR-320-3p was detected by qRT-PCR assay (Figure 3F). Luciferase reporter assay and RNA pull-down assay were implemented, respectively, to verify the binding of Gas5 to miR-320-3p and Tcf3 to miR-320-3p (Figure 3G-J). Relative luciferase activity in Gas5 and Tcf3-WT was decreased significantly by miR-320-3p overexpression, and Gas5 and Tcf3 were both enriched in the pull-down of miR-320-3p WT. To further confirm the location of Gas5 in NRC and NMC treated with high glucose, we implemented subcellular fraction assay and FISH assay again (Figure 3K,L). We found that Gas5 was mainly located in the cytoplasm of NRC and NMC treated with high glucose. Also, the binding of Gas5 to miR-320-3p and Tcf3 to miR-320-3p were verified again by luciferase reporter assay and RNA pull-down assay in NRC and NMC treated with high glucose (Figure 3M-Q). Results indicated that miR-320-3p could bind to Tcf3 and Gas5 in NRC and NMC treated with high glucose. In conclusion, miR-320-3p could bind to Tcf3 and Gas5 in NRC and NMC.
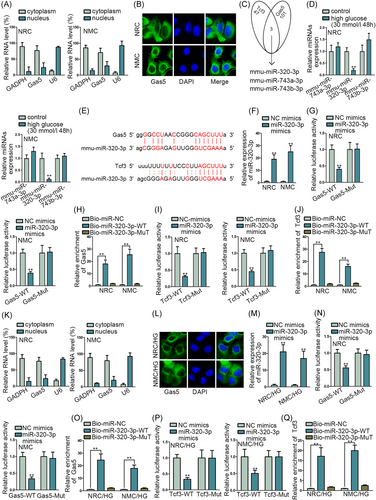
3.4 Gas5/miR-320-3p/Tcf3 axis could modulate the apoptosis of NRC and NMC
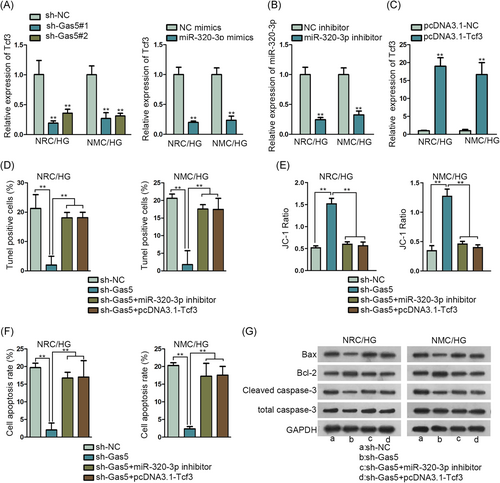
For detecting the function of the Gas5/miR-320-3p/Tcf3 axis in NRC and NMC, we implemented a series of rescue assay. First of all, we detected the expression of Tcf3 when Gas5 was inhibited or miR-320-3p was overexpressed in NRC and NMC treated with high glucose (30 mmol/L) (Figure 4A). And we found the expression of Tcf3 was significantly dropped down when Gas5 was silenced or miR-320-3p was overexpressed. Then, we implemented qRT-PCR assay to detect the inhibition efficiency of miR-320-3p and overexpression efficiency of Tcf3 in NRC and NMC treated with high glucose (Figure 4B,C). Furthermore, rescue assay of TUNEL assay, JC-1 assay, flow cytometry analysis, and Western blot assay was conducted in NRC and NMC treated with high glucose (Figure 4D-G). TUNEL assay found that Gas5 inhibition decreased the TUNEL-positive cells of NRC and NMC. However, the TUNEL-positive cells of NRC and NMC were countervailed by inhibited miR-320-3p and overexpressed Tcf3. Meanwhile, upregulated JC-1 ratio caused by Gas5 inhibition was increased again by knockdown of miR-320-3p and overexpressed Tcf3. Furthermore, flow cytometry analysis and Western blot assay also verified the decreased apoptosis of NRC and NMC by inhibited Gas5 could be offset by miR-320-3p inhibition and Tcf3 overexpression. In conclusion, the Gas5/miR-320-3p/Tcf3 axis could modulate the apoptosis of NRC and NMC.
4 DISCUSSION
It has been reported that diabetic cardiomyopathy can cause cardiac dysfunction and eventually lead to heart failure and sudden death.21 And the inner mechanism of diabetic cardiomyopathy has been reported before, which has proved that lncRNAs can play the modulating function in diabetic cardiomyopathy.22, 23 For example, KCNQ1OT1 can modulate the cell apoptosis of diabetic cardiomyopathy.23 lncRNA H19 reduces autophagy in diabetic cardiomyopathy by downregulating the expression of DIRAS3.24 And lncRNA MIAT can regulate the apoptosis of cardiomyocytes in diabetic cardiomyopathy via miR-22-3p.7 In our study, we scrutinized the role of lncRNA GAS5 in diabetic cardiomyopathy. Early studies have indicated that lncRNA GAS5 can modulate the fibrosis of cardiac fibroblasts.11 And lncRNA GAS5 can regulate the oxidative stress in the heart.12 In our study, we implemented high-glucose treatment to NRC and NMC and tested the expression of lncRNA GAS5 in NRC and NMC by qRT-PCR assay. And we found that lncRNA GAS5 expression was increased with the time of high-glucose treatment increasing. And then we detected the apoptosis of NRC and NMC by functional assays, such as TUNEL assay, JC-1 assay, flow cytometry analysis, and Western blot assay. Results indicated that increased apoptosis of NRC and NMC by high-glucose treatment was decreased by inhibited GAS5.
Transcriptional factors can bind to the lncRNA promoter to activate the expression of lncRNA.19 For example, transcription activator YY1 can activate the expression of lncRNA PVT1 in lung cancer.25 STAT3 can upregulate the expression of HOXD-AS1 in liver cancer.26 SOX2 activates the PVT1 expression to regulate the progression of breast cancer.19 In our study, we searched the transcription factors of GAS5 and tested their expression in NRC and NMC with high-glucose treatment. And we found that the expression of Tcf3 was significantly increased. After that, we detected the expression change of GAS5 by inhibiting Tcf3 and overexpressing Tcf3. And the expression of GAS5 was significantly changed by Tcf3 inhibition and Tcf3 overexpression. Also, ChIP assay and luciferase reporter assay detected the binding in Tcf3 and GAS5 promoter. Furthermore, functional assays also detected that increased apoptosis of NRC and NMC by high-glucose treatment was suppressed by Tcf3 inhibition.
lncRNAs act as miRNA sponge and play the network of lncRNA-miRNA-mRNA to regulate the progression of cells.13, 15 For example, lncRNA APF can modulate autophagy in myocardial infarction through adjusting the expression of miR-188-3p.11 In this study, we detected the miRNA that could bind to Tcf3 and GAS5. And we detected that miR-320-3p was downregulated in NRC and NMC on high-glucose treatment. Meanwhile, the rescue experiment detected the modulation of the Gas5/miR-320-3p/Tcf3 pathway in the apoptosis of NRC and NMC. In conclusion, Tcf3-activated lncRNA Gas5 regulates NMC apoptosis in diabetic cardiomyopathy. We hope our study can shed new light on the treatment of diabetic cardiomyopathy.
ACKNOWLEDGMENT
The authors would like to appreciate the supports of our experimenters.
CONFLICT OF INTERESTS
The author declares that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
DS was involved in study design, experiments, data interpretation, manuscript review. YJ, WH, and YY performed experiments, data interpretation, and analysis. FW, TW, and JT were involved in the investigation and drafting of the manuscript. All authors approved the final manuscript.