miR-144 suppresses cell proliferation and migration in colorectal cancer by targeting NRAS
Xuemei Ji, Yanqing Liu, and Xiaoming Kao contributed equally to this study.
Abstract
Colorectal cancer (CRC) is a type of malignant cancer that has become particularly prevalent worldwide. It is of crucial importance to CRC treatment that the underlying molecular mechanism of CRC progression is determined. The NRAS gene is an important small G protein that is involved in various biological processes, including cancers. NRAS is an oncogene in many neoplasms but its function and regulation in CRC have seldom been investigated. In this study, it was uncovered that the NRAS protein was significantly upregulated in CRC tissues. According to a bioinformatics prediction, we identified that miR-144 may target NRAS to suppress its expression. In vitro experiments indicated that miR-144 decreased NRAS expression in different CRC cell lines (SW480, LoVo, and Caco2). By inhibiting NRAS, miR-144 repress SW480 cell proliferation and migration. Moreover, miR-144 decelerated the growth of SW480 xenograft tumors in vivo by targeting NRAS. In summary, our results identified a novel miR-144-NRAS axis in CRC that could promote the research and treatment of CRC.
1 INTRODUCTION
Colorectal cancer (CRC) has stood top among the most noticeable malignancies worldwide due to its high heterogeneity and poor prognosis. According to clinical reports, there are 1.2 million newly diagnosed patients with CRC and 608 700 deaths due to CRC annually. Currently, CRC-caused death ranks fourth among all cancer-related death all over the world.1 Although CRC has been intensively investigated at the molecular level,2-4 the precise mechanisms that participate in CRC initiation and development remain complicated and obscure.
Various genetic and epigenetic variations in colorectal epithelial cells are the fundamental causes for the occurrence and progression of CRC. Oncogenes (eg, c-Myc5 and HuR 6 activation and tumor suppressor genes (eg, TIA17 and PDCD48) silencing play crucial roles during CRC carcinogenesis. Ras proteins, which are molecular modulators, mainly control cell growth by transducing signals from receptor tyrosine kinases localized on the cytomembrane to intranuclear transcription factors and cell cycle proteins.9, 10 Ras proteins have two physiological states, namely, an activated (GTP-bound) form and an inactivated (GDP-bound) form. In addition, a family of guanine nucleotide exchange factors, such as SOS1, SOS2, and RASGRF,11 catalyze a change in Ras to its active form, and Ras-GAPs play an opposing role on Ras proteins though their GTPase activity.12 Neuroblastoma rat-sarcoma homolog (NRAS) is the archetypical member of the RAS superfamily and has been recently extensively investigated as an oncogene. A mutation in the NRAS gene leads to the malignant proliferation and metastasis of cells and it has a high frequency in hematological tumors, reaching 27.3%.13 However, NRAS mutations are much rarer than KRAS or BRAF variation in colorectal cancer, and the incidence rate is only 2%-4%.14, 15 This is the reason why the roles of NRAS in metastatic colorectal cancer have been much less investigated. A recent study revealed that NRAS mutation cases had a higher incidence of distant metastases and a shorter metastasis-free interval, suggesting the potential oncogenic function of NRAS in CRC.16 However, the exact role of NRAS and its regulation in CRC should be elucidated.
MicroRNAs (miRNAs) (consisting of 19-23 nucleotides) are a class of conserved noncoding RNA molecules, mainly posttranscriptionally regulating gene expression. MicroRNAs interact with the 3′-untranslated regions (3′UTRs) of mRNAs to promote mRNA cleavage and induce translational repression according to the principle of complementary base-pairing.17, 18 In previous studies, miRNAs have gained intensive attention in numerous human diseases, including various neoplasms.19 Emerging evidence has suggested that most miRNAs are considered oncomiRs or tumor-suppressive miRNAs based on their target mRNA(s) and/or overall effects on tumor development.19, 20 Aberrant miRNA production also participates in CRC occurrence, and some miRNAs even influence several fundamental molecular events, such as cell proliferation, differentiation, division, migration, invasion, and apoptosis.21 miR-144 has been reported to be significantly expressed at low levels in glioblastoma (GBM), and its decreased expression is related to a poorer prognosis in patients.22 In human epithelial cancers, miR-144 prevents cancer metastasis by binding to ADAMTS5 and ADAM10.23 miR-144 also impedes cell multiplication and promotes cell apoptosis in prostate cancer by targeting CEP55.24 Cai et al25 reported that miR-144 prevents the multiplication and migration of rectal cancer cells by reducing the ROCK 1 expression. However, studies of miR-144 function in CRC are limited. Thus, the expression status, specific roles, and potential principles of miR-144 in CRC need to be further elucidated.
This study was reported that NRAS expression is upregulated in CRC and promotes the multiplication and migration of tumor cells. miR-144 functions as a CRC-suppressive miRNA through regulating NRAS. By suppressing NRAS expression, miR-144 inhibits CRC cell multiplication and migration and slows CRC xenograft tumor growth.
2 MATERIALS AND METHODS
2.1 CRC patient tissues
CRC and adjacent noncancerous tissues were obtained from 13 patients who underwent surgical resection of CRC at Jinling Hospital (Nanjing, China). Both the CRC specimens and adjacent normal specimens were confirmed by pathologic diagnosis. All patients’ samples in this study were obtained from consentient individuals and the collectors of these specimens complied with the protocols authorized by the Ethics Committee of Nanjing University. Clinical specimens were instantly frozen in nitrogen canister after resection and conserved at−80°C. The clinicopathological data of 13 CRC patients is shown in Table 1.
| Case | Age | Gender | Tumor subtype | Pathological stage |
|---|---|---|---|---|
| 1 | 61 | Female | Rectal adenocarcinoma | IIIB |
| 2 | 64 | Male | Sigmoid adenocarcinoma | IIA |
| 3 | 61 | Male | Colonic | IIIA |
| adenocarcinoma | ||||
| 4 | 55 | Female | Rectal adenocarcinoma | IIA |
| 5 | 71 | Male | Colonic | IIIB |
| adenocarcinoma | ||||
| 6 | 47 | Male | Colonic | IIA |
| adenocarcinoma | ||||
| 7 | 76 | Male | Colonic | IIIA |
| adenocarcinoma | ||||
| 8 | 51 | Male | Colonic | IIIB |
| adenocarcinoma | ||||
| 9 | 71 | Male | Colonic | IIIA |
| adenocarcinoma | ||||
| 10 | 63 | Male | Rectal adenocarcinoma | IIA |
| 11 | 77 | Male | Rectal adenocarcinoma | IIB |
| 12 | 62 | Male | Colonic | IIIA |
| adenocarcinoma | ||||
| 13 | 67 | Male | Colonic | IIIB |
| adenocarcinomaAbbreviation |
- Abbreviation: CRC, colorectal cancer.
2.2 Cell culture
Human CRC cell lines (SW480, LoVo, and Caco2) were ordered from the SIBCB (Shanghai, China).
2.3 RNA isolation and real-time quantitative reverse transcription PCR (qRT-PCR)
Total RNA from CRC cells or tissue samples was isolated by using Trizol reagent (Invitrogen, CA). The procedure and analysis of qRT-PCR experiments are following methods described in a previous study.7
2.4 Antibodies
Antibodies used to be probed for NRAS and GAPDH were ordered from Abcam (ab77392; Abcam, CA) and Santa Cruz Biotechnology (sc-25778; Santa Cruz, CA), respectively.
2.5 RNA oligos and NRAS vector
MiR-144 mimic, miR-144 inhibitor, and NRAS siRNA (including negative control RNAs) were ordered from GenePharma (Shanghai, China). NRAS overexpression vector was constructed from Genescript (Nanjing, China). RNA and proteins were isolated 48 hours after transfection.
2.6 Luciferase assay
pMIR-REPORT vector was constructed to carry a 570-bp fragment of NRAS 3′UTR with presumed miR-144 target site from Genescript. To confirm combining selectivity, NRAS 3′UTR sequences binding to the miR-144 seed region were changed from ATACTGT to TATGACA, and then the mutant sequences were inserted into identical same types of vector (Genescript). β-galactosidase (β-gal) expression plasmid (Ambion) was applied to verifying the efficiency of transfection.
2.7 Cell proliferation assay
A total of 96-well plates were utilized to culture SW480 cells with 2 × 104 cell density each well. Cell counting kit-8 (CCK-8) reagent (Beyotime, Nanjing, China) were used to assay the cell proliferation index at different time points (every 12 hours, until an endpoint 60 hours) after transfection. The mean value of absorbance in triplicate was recorded as a final result at 450 nm wavelength.
A total of 48-well plates (Corning) were used to culture SW480 cells for EdU assay. The proliferation rate of CRC cells was estimated by the aid of an EdU assay kit (RiBoBio, Guangzhou, China) at 80% cells confluency after transfection. The protocol of the EdU assay kit was abided by but the nucleus staining materials were changed to obtain more distinct images. Specifically, the nucleus dye was DAPI (Beyotime) instead of Hoechst 33342 (provided by kit). Then, all stained cells were photographed by microphotography (BX51 Olympus, Japan).
2.8 Cell migration assay
The migration phenomenon of CRC cells was detected in 24-well millipore plates and the bottom of transwell chambers were covered with an 8-μm pore membrane (Millipore). The chambers were inverted in Petri dishes and 1% gelatin was added to the bottom of the membrane. The transfected CRC cells were collected after 24 hours and then resuspended in RPMI-1640 without FBS. The upper chamber was added with the resuspended cells approximately 2 × 104 cells each well and the lower compartment was loaded with 500 μL RPMI-1640 (20% FBS). The whole plates were placed in a common humid incubator with 5% CO2 at 37°C for 1 day. Then, migrant cells were fixed by 4% paraformaldehyde for 25 minutes (RT), followed by staining using 0.1% crystal violet for 20 minutes at RT.
2.9 Mice experiments
Tumor xenografts were established with male SCID mice about 4-week-old. At the beginning ofestablishment of the mice model, SW480 cells were treated with different vector tools to upregulate miR-144 and/or NRAS. Specifically, lentivirus infected alone SW480 cells to elevate miR-144 level, and NRAS vector was transfected alone the CRC cells to increase expression of NRAS. The rest of SW480 cells were cotransfected with the LV-miR-144 and the NRAS vector. Subsequently, SCID mice were subcutaneously injected with the above SW480 cells infected and/or transfected and each mouse was carried 3 × 104 cells. Five mice were divided into one group. The mice were killed to separate grafted tumors from them after 30 days and the weights of neoplasm were measured immediately after resection. Then, parts of the tissues were applied to extracting purpose RNA and protein, and the rest of samples were fixed for making pathological slides with 4% paraformaldehyde for 1 day The fixed tissues were stained with hematoxylin and eosin (H&E) or used to perform immunohistochemical staining for NRAS and Ki-67.
2.10 Statistical analysis
Representative images of the Western blot and cell migration experiments were selected from the acquired images that were obtained from at least three independent experiments. Triplicate experiments were strictly conducted, including RT-qPCR, luciferase reporter assay, and proliferation assay. Numerical data were demonstrated as the mean ± standard error of mean. For statistical significance of differences analysis, the Student t-test was used. P < .05 was identified as differences had statistical significance.
3 RESULTS
3.1 CRC tissues exhibit upregulated NRAS protein level
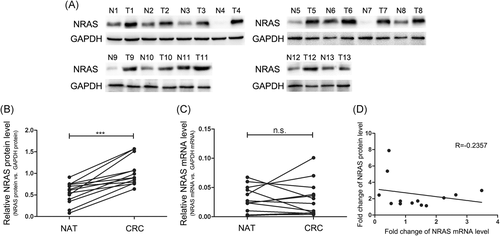
First, we investigated NRAS expression levels in 13 paired CRC tissues. Significantly increased NRAS protein level in CRC tissues (CRC) contrasted with normal adjacent tissues was observed (Figure 1A and 1B). Then, we analyzed the levels of NRAS mRNA expression in the same CRC samples and normal adjacent samples and identified irregular changes in the NRAS mRNA expression level between the tumor tissues and the adjacent nontumor tissues (Figure 1C). Pearson's correlation scatter plot was used to analyze the correlation between the differences in the levels of NRAS protein and mRNA expression. As shown in Figure 1D, there was little correlation between the protein and mRNA expression. The inconsistency between the changes in NRAS protein and mRNA expression suggested that NRAS expression was controlled by a posttranscriptional mechanism.
3.2 miR-144 potentially targets NRAS
Because of miRNA is an essential posttranscriptional modulator of gene expression, we supposed some miRNAs regulate NRAS protein levels in CRC tissues. Online software Targetscan was utilized to predict possible miRNAs that regulate NRAS. miR-144 was identified as a potential candidate. The presumed combination between miR-144 and the NRAS 3′UTR is shown in Figure 2A. miR-144 seed region (2-8 bases of the mature miRNA) interacted with the target site in accordance with perfect base-pairing. Besides the miR-144 target sequence at the NRAS 3′UTR was highly conserved in diverse species (Figure 2A).
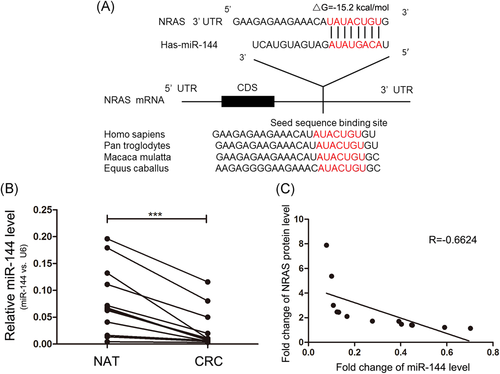
Next, we investigated whether there exists inverse correlation between miR-144 and NRAS levels in CRC samples. It was discovered that miR-144 expression levels were consistently lower in CRC samples compared with paired normal samples (Figure 2B). Furthermore, we identified the changes of the miR-144 expression levels were inversely correlated with those of the NRAS by using Pearson's correlation analysis (Figure 2C). Thus, NRAS was a candidate target of miR-144 in CRC.
3.3 miR-144 directly regulates the NRAS expression posttranscriptionally
NRAS expression levels were tested in CRC cell line SW480 after miR-144 alteration (increase or decrease). Figure 3A shows the efficient alteration of the miR-144 expression levels in SW480 cells by miR-144 mimic and inhibitor. As anticipated, NRAS expression dramatically decreased after miR-144 upregulation, whereas miR-144 inhibitor promoted the NRAS expression in SW480 cells (Figure 3B and 3C). Two other CRC cell lines (LoVo and Caco2) were used to strengthen the accuracy of these results (Figure 3A-C).
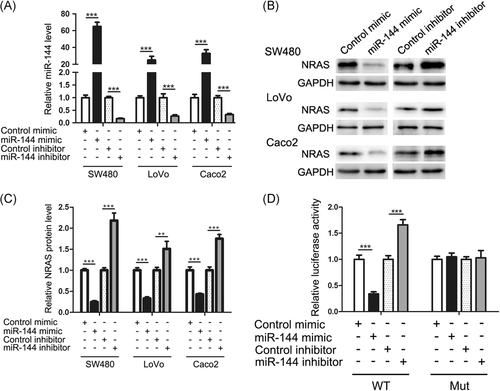
Next, to explore whether miR-144 prohibits NRAS expression through binding to the target site in the NRAS 3′UTR, we constructed a reporter plasmid to connect the NRAS 3′UTR sequence including the hypothesized miR-144 target site with the downstream location of the firefly luciferase gene. Subsequently, SW480 cells were transfected with the plasmid accompanied by the miR-144 mimic, miR-144 inhibitor, control mimic, or control inhibitor. As hypothesized, the upregulation of miR-144 expression led to nearly 70% decrease in luciferase signal contrasted to cells dealt with negative control RNAs, whereas miR-144 knockdown led to an approximate 60% increment in reporter activity compared with control cells (Figure 3D). The point mutations were subsequently induced into the miR-144 target region of the NRAS 3′UTR in a mutant plasmid. The plasmid was constructed to abrogate miR-144 target ability. The activity of mutant reporter was not affected by the upregulation or downregulation of the miR-144 expression (Figure 3D). These results indicated that target region is responsible for the interaction of miRNA-mRNA.
3.4 miR-144 suppresses CRC cell proliferation by targeting NRAS
It was presumed that miR-144 inhibits the CRC tumorigenic process by suppressing NRAS expression. We first analyze the impact of miR-144 on CRC cell proliferation by utilizing CCK-8 and EdU assays. The upregulation of the miR-144 expression decreased SW480 proliferation; conversely, the downregulation of the miR-144 expression had the opposite influence SW480 proliferation (Figure 4A-C).
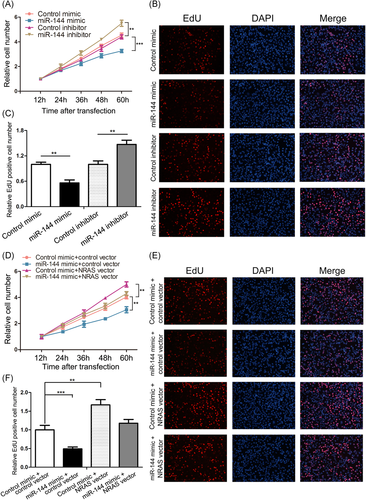
An important issue was whether the effects of miR-144 on CRC cell proliferation originated from NRAS suppression by miR-144 since a single miRNA could have different targets.26 Therefore, we examined the functions of NRAS on SW480 cell proliferation. NRAS protein expression was downregulated and upregulated using siRNA (si-NRAS) and an overexpression plasmid (NRAS vector), respectively. It is shown that effective downregulation and upregulation of NRAS expression in SW480 cells (Figure S1A and S1B). Consequently, NRAS overexpression promoted the multiplication of SW480 cells, whereas downregulation of NRAS expression inhibited on cell multiplication (Figure S1C-E). Therefore, miR-144 and NRAS affect cell proliferation in CRC. We subsequently verified whether the upregulation of NRAS expression is enough to remedy the antiproliferative functions of miR-144 in CRC cells. Some of the SW480 cells were transfected with the miR-144 mimic or NRAS vector and the others were cotransfected with the miR-144 mimic and NRAS vector. As supposed, SW480 cells cotransfected with the miR-144 mimic and NRAS vector had markedly higher proliferation rates compared with those cells dealt with the miR-144 mimic alone (Figure 4F).
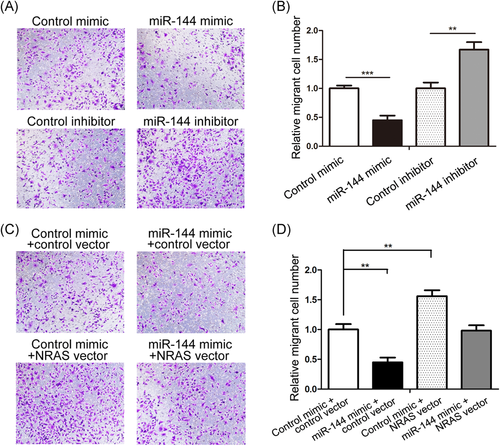
3.5 miR-144 suppresses CRC cell migration by targeting NRAS
We also checked the effect of miR-144 on CRC cell migration by performing the Transwell assay. The number of migratory cells was notably lower in SW480 cells after the miR-144 mimic transfection and higher in cells after the miR-144 inhibitor transfection inhibitor in contrast to control cells (Figure 5A and 5B). Then we validated that cell migration repression by miR-144 was due to the inhibition of NRAS. SW480 cells transfected with si-NRAS and the NRAS vector showed reduced and enhanced migration percentages, respectively (Figure S1F and S1G), suggesting that NRAS promotes SW480 cell migration. Then, a rescue experiment was performed in a similar manner as the proliferation experiment. SW480 cells cotransfected with the miR-144 mimic and NRAS vector exhibited a significantly higher migration rate than the control group (Figure 5C and 5D). Thus, the recovery of the ability of miR-144 to suppress cell migration by restoring NRAS demonstrated that miR-144 inhibits SW480 cell migration by targeting NRAS.
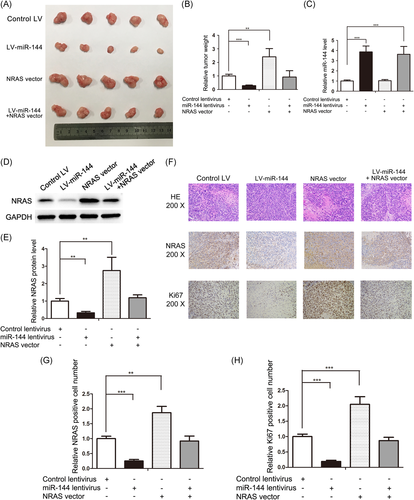
3.6 miR-144 inhibits mice CRC growth by targeting NRAS
A key problem to be solved was whether these effects of miR-144 and NRAS on CRC work in vivo. miR-144 lentivirus (LV-miR-144) and NRAS vector are used solely or combined in SW480. By subcutaneously implanting these SW480 cells into SCID mice, the CRC xenograft model was constructed. After sacrificing the mice, we observed a markedly decreased tumor weight in miR-144 overexpression group compared with the control group, whereas NRAS overexpression increased tumor weights (Figure 6A and 6B). Moreover, NRAS overexpression could abolish the growth-suppressing effects of miR-144 (Figure 6A and 6B), indicating that miR-144 inhibits tumor growth by silencing NRAS, which was collaborated by the miR-144 and NRAS levels in tumors (Figure 6C-E). H&E staining and immunohistochemical assays further demonstrated the antitumor growth role of miR-144 by suppressing NRAS (Figure 6F-H). These results were consistent with the in vitro results, which clearly validated the anti-CRC role of miR-144 by suppressing NRAS.
4 DISCUSSION
Colorectal cancer is a common tumor type in the digestive tract27 that seriously threatens human health due to its high morbidity in recent years.28 The molecular mechanisms of CRC pathogenesis mainly comprise genetic and epigenetic alterations that affect oncogenes or tumor suppressors expression. NRAS is a crucial tumor promoter in many cancers.29, 30 However, its potential role in CRC has not been elucidated. In our current study, the NRAS expression was found to be markedly upregulated in CRC tissues and promoted the proliferation and migration of CRC cells. The exact molecular mechanism responsible for the upregulated expression of NRAS was subsequently addressed.
As the fields of molecular biology and oncology develop, miRNAs are brought into the tumor suppressor or oncogene family. Many studies have suggested the various roles of miRNAs in the occurrence and development of CRC.31, 32 In this study, we elucidated that miR-144 expression is downregulated in CRC tissues, suppresses cell proliferation and migration in vitro and inhibits tumor growth in vivo. Using target prediction software,33 we identified NRAS as a potential target gene of miR-144. This hypothesis was demonstrated by Western blot analysis and a luciferase assay. By inhibiting NRAS, miR-144 repressed SW480 cell proliferation and migration. In addition, miR-144 decelerated SW480 xenograft tumor growth in SCID mice by targeting NRAS.
Nevertheless, there is a caveat that miR-144 is speculated to have targets other than NRAS, and NRAS may be modulated by other miRNAs besides miR-144. For example, NRAS is regulated by miR-34034 and miR-29,29 and miR-144 can simultaneously target CEP5524 and COX-2.35 Therefore, it is critical to check how essential the miR-144/NRAS is in the complex CRC oncogenesis pathways.
More notably, miRNAs have the potential to treat cancer.36, 37 Specifically, dysregulated miRNAs expression in cancers can be restored by various methods.38, 39 In addition to our results, another group has reported that miR-144 suppresses the initiation, progression, invasion, and recurrence of CRC tumors and that downregulation of the miR-144 expression is closely associated with a poor prognosis in patients with CRC and a decreased sensitivity of cancer cells to rapamycin.40 Therefore, targeting miR-144 may be a promising therapeutic way to treat CRC, which is worth further exploration.
In conclusion, this is the first study to show that miR-144 suppresses CRC tumorigenesis by directly targeting NRAS. This novel miR-144-NRAS axis could be a new therapeutic strategy for CRC.
ACKNOWLEDGMENT
This study was supported by grants from the National Natural Science Foundation of China (No. 81570506).
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.




