NR2F1-AS1 regulated miR-423-5p/SOX12 to promote proliferation and invasion of papillary thyroid carcinoma
Abstract
Papillary thyroid carcinoma (PTC) is an aggressive histological subtype of thyroid carcinoma (THCA), whose occurrence rate is high. The participation of long noncoding RNAs in the pathologies of cancers has attracted significant attention during the past decades. The purpose of the current study is to investigate the role of NR2F1 antisense RNA 1 (NR2F1-AS1) in PTC. The expression of NR2F1 in THCA samples was analyzed by bioinformatics tool gene expression profiling interactive analysis. Levels of NR2F1-AS1, microRNA-423-5p (miR-423-5p), and SRY-box 12 (SOX12) were evaluated by a quantitative reverse transcription-polymerase chain reaction and Western blot. The impact of NR2F1-AS1 on PTC cell proliferation and invasion was assessed by Cell Counting Kit-8, EdU, and Transwell invasion assays. The interactions among NR2F1-AS1, miR-423-5p, and SOX12 were determined by RNA immunoprecipitation and luciferase reporter assays. Consequently, we found that NR2F1-AS1 and SOX12 levels were elevated in PTC, whereas miR-423-5p was downregulated in PTC cells. Functionally, NR2F1-AS1 silence led to reduced proliferation and invasion of PTC cells. Mechanistically, NR2F1-AS1 interacted with miR-423-5p to induce SOX12 expression in PTC cells. In conclusion, the present study firstly stated that NR2F1-AS1 regulated miR-423-5p/SOX12 to promote proliferation and invasion of PTC, indicating NR2F1-AS1 as a potential novel target for the molecular-targeted therapy of PTC.
1 INTRODUCTION
Papillary thyroid carcinoma (PTC), is the dominant type of thyroid cancer, and the most prevalent malignancy of the endocrine system.1 Recent decades have witnessed the rapid increase of the occurrence rate in PTC.2, 3 Patients with PTC usually have satisfactory outcomes, however, the 5-year survival rate of patients with PTC at advanced stage remains dismal.4 Hence, to further improve the prognosis of PTC, the precise mechanism of PTC progression still needs to be better understood.
Long noncoding RNAs (lncRNAs) are a family of conserved RNAs consisting of more than 200 bases and incapable of coding proteins.5 Increasing lncRNAs have been discovered during the past years, and the biological role of lncRNAs in cancers and diseases has become an intriguing research hot spot.6 A growing number of lncRNAs are identified as biomarkers in PTC. For example, lncRNA HOXA-AS2 facilitated PTC progression through regulating miR-520c-3p/S100A4 pathway7; lncRNA CNALPTC1 accelerated proliferation and migration of PTC through sponging miR-30 family8; and lncRNA UNC5B-AS1 aggravated proliferation, migration, and invasion in PTC cells.9 NR2F1 antisense RNA 1 (NR2F1-AS1) has been shown by a previous study to aggravate tumor growth and contribute to oxaliplatin resistance in hepatocellular cancer (HCC).10 Nevertheless, the function of NR2F1-AS1 in PTC remains unclear.
MicroRNAs (miRNAs) are known as single-strand, short (about 22 base pairs) RNAs, regulating gene expression posttranscriptionally via targeting 3′-untranslated region (3′-UTR) of messenger RNAs (mRNAs) and resulting in the degradation of mRNA or suppression the translation of functional proteins.11 It has been known that miRNAs could be competitively sponged by lncRNAs and prevented from silencing target genes in cancers,12-14 including in PTC.15 MiR-423-5p has been reported to be a suppressor of several cancers, such as osteosarcoma,16 brain metastasis of lung adenocarcinoma,17 nasopharyngeal carcinomas,18 as well as, colon cancer.19 However, the regulatory function of miR-423-5p in PTC and its association with NR2F1-AS1 have never been explored.
Sex determining region Y-box (SOX) proteins are a family of transcription factors, which regulate the activation of gene expression through recognizing and targeting the SOX target sequence A/TAACAA/T.20, 21 SOX12 is a newly identified member of the SOX protein family.22 Recent studies have shown that SOX12 is involved in carcinogenesis and the development of a variety of cancers. For instance, SOX12 promoted metastasis of gastric cancer through inducing MMP7 and IGF1.23 MiR-138 targeted SOX12 to inhibit proliferation, invasion, and migration in ovarian cancer.24 Silencing SOX12 repressed proliferation and metastasis of lung cancer cells.25 However, never has SOX12 been associated with PTC, and its relation with miR-423-5p and NR2F1-AS1 remains unknown.
The present study aimed to explore the biological influence and regulatory mechanism of NR2F1-AS1 in PTC.
2 MATERIALS AND METHODS
2.1 Cell culture
Human normal thyroid cells (Nthy-Ori 3-1), human PTC cells (CGTH-W3, BCPAP, K1, and TPC1), and human renal epithelial cell (293T) were purchased from the American Type Culture Collection (Manassas, VA). Cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) plus 10% fetal bovine serum (FBS; Invitrogen) under a humidified air condition with 5% CO2 at 37℃. Replacement of medium was done every 3 days.
2.2 Cell transfection
Lipofectamine 2000 (Invitrogen) was applied for conducting cell transfection, which lasted 48 hours as per its standard guide. For enhancing or inhibiting expression, K1 or TPC1 cells were transfected with various plasmids. Specific small interfering RNAs against NR2F1-AS1 (si-NR2F1-AS1#1, si-NR2F1-AS1#2, and si-NR2F1-AS1#3) or their corresponding negative control (NC) were formed by GeneChem (Shanghai, China), along with the pcDNA3.1 vector targeting SOX12 and the empty vector. Moreover, miRNA mimics, miRNA inhibitor, NC mimic, and NC inhibitor were acquired from GenePharma (Shanghai, China).
2.3 Quantitative real-time polymerase chain reaction
Trizol reagent (Invitrogen) was applied to isolate purified RNAs. Following synthesizing first-strand complementary DNA (cDNA) via the PrimeScript RT reagent Kit (TaKaRa, Tokyo, China), the cDNA was augmented by using SYBRPremix Ex Taq II (TaKaRa). The miR-423-5p was normalized to U6. NR2F1-AS1 or SOX12 was normalized to GAPDH via method.
2.4 Cell viability assay
Transfected K1 or TPC1 cells were planted into 96-well cell culture plates when concentration was up to 2000 cells per well. Cells were replaced with medium containing 10 μL Cell Counting Kit-8 (CCK-8) reagent (Beyotime, Shanghai, China) afterward. After 1 hour incubation, optical density absorbance at the wavelength of 450 nm was eventually monitored.
2.5 EdU analysis
K1 or TPC1 cells after transfection were added into 96-well plates as soon as the density reached 1 × 105 cells/mL. Cells were propagated for 1 day with a cover glass slide placed in the culture plate. Following incubation with EdU solution (Sigma-Aldrich, St. Louis, MO) for 4 hours, cells were fixed in methanol (Sigma-Aldrich) for 10 minutes. Instructions were strictly followed. Cardinal number was detected by a microscope (Leica, Wetzlar, Germany).
2.6 Cell invasion assay
Cell invasion capacity was explored by the Transwell assay. Briefly speaking, K1 or TPC1 cells after transfection maintained in 100 µL FBS-free media were planted into the top chambers precoated with Matrigel, while media with 20% FBS was plated to the lower chambers. Noninvasive cells were removed, while invasive cells were fixed by 4% paraformaldehyde (Sigma-Aldrich) and then stained by crystal violet solution (Sigma-Aldrich). Data were quantified via the ImageJ software.
2.7 Microarray analysis
A mirVanaTM RNA Isolation Kit (Ambion, São Paulo, Brazil) was utilized for the extraction of total RNA from cells. Then, cDNA was labeled with biotin after being synthesized from RNA. Biotinylated cDNA was subjected to purification, fragmentation, and hybridization. Microarrays were scanned by the Affymetrix Scanner 3000 7G (Affymetrix, Santa Clara, CA) after rinsing and staining. After that, microarray signals were evaluated via the Expression Console Software 1.41 (Affymetrix). Only proven sets with a P < .05 and fold change greater than 2.0 were included in the final gene list.
2.8 Luciferase reporter assay
The binding sites of NR2F1-AS1 on miR-423-5p, miR-3924, and miR-3617-5p or their corresponding NC were separately subcloned into the pmirGLO Dual-Luciferase vector and cotransfected with NC mimic, miR-423-5p mimic, miR-3924 mimic, and miR-3617-5p mimic into cells. Simultaneously, NR2F1-AS1-wild-type/mutant (WT/Mut) or SOX12-WT/Mut was cotransfected into 293T cells with miR-423-5p mimic or NC mimic. Forty-eight hours later, Dual-Luciferase Reporter Assay System (Promega, Madison, WI) was conducted.
2.9 RNA immunoprecipitationassay
For carrying out the RNA immunoprecipitation (RIP) assay, the EZ-Magna RIP Kit (Millipore, Billerica, MA) was employed on the basis of its specification. Cells were at the beginning lysed in complete RIP lysis buffer (Invitrogen). Subsequently, cell extract was treated with magnetic beads (Thermo Fisher Scientific, Waltham, MA) conjugated to an antibody against Ago2 (Millipore) or immunoglobulin G (Millipore). After incubating with proteinase K (Thermo Fisher Scientific) with shaking, the immunoprecipitated RNA was harvested. The NanoDrop spectrophotometer was applied so as to detect the density of RNA. Purified RNA was subjected to the quantitative real-time polymerase chain reaction.
2.10 Western blot analysis
K1 or TPC1 cells after transfection were lysed in radioimmunoprecipitation assay lysis buffer. Lysates were collected and then cleared via centrifugation. Next, proteins were transferred onto polyvinylidene fluoride membranes (Millipore) and treated at 4℃ with anti-SOX12 antibody (3 µg/mL, MA5-19999; Thermo Fisher Scientific) or anti-GAPDH antibody (1/20000, ab128915; Abcam, Cambridge) overnight, followed by incubating with secondary antibody. At last, membranes were visualized by the enhanced chemiluminescence system (Santa Cruz Biotechnology, Santa Cruz, CA).
2.11 Statistical analysis
All data in this study were shown as mean ± standard deviation. For comparing two or more groups, the Student t test or one-way analysis of variance were employed to carry out differences analysis. Differences were statistically significant when P < .05. GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA) was applied for statistical analyses. Each experiment was at least repeated three times.
3 RESULTS
3.1 Upregulation and tumor-promoting function of NR2F1-AS1 in PTC
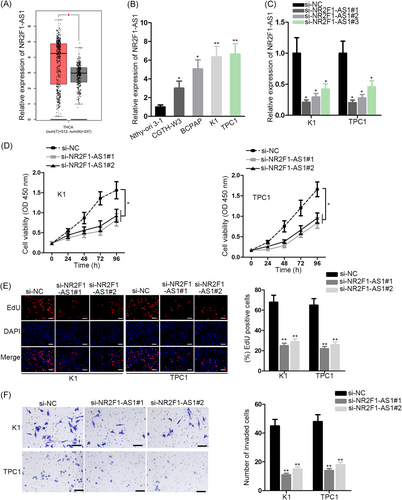
First, to probe the relation of NR2F1-AS1 with PTC, we analyzed its expression in PTC. With the application of gene expression profiling interactive analysis (http://gepia.cancer-pku.cn/), an online bioinformatics tool, we found that the NR2F1-AS1 level was upregulated in thyroid carcinoma (THCA) specimens than the normal specimens (Figure 1A). In addition, quantitative reverse transcription PCR (RT-qPCR) data revealed the increased expression of NR2F1-AS1 in PTC cell lines vs the normal cell line (Figure 1B). Since we observed that TPC1 and K1 cells expressed the highest NR2F1-AS1 level, we chose these two cell lines to knockdown the endogenous NR2F1-AS1 level. Results of RT-qPCR confirmed that si-NR2F1-AS1#1/2 silenced NR2F1-AS1 in both PTC cell lines more significantly (Figure 1C). Hence, we used si-NR2F1-AS1#1/2 to conduct loss-of-function assays to detect the impact of NR2F1-AS1 on the biological activities of PTC cells. Consequently, the proliferation of two PTC cell lines was impeded by the silence of NR2F1-AS1 (Figure 1D,E). Moreover, the invasive ability of PTC cells reduced as a result of NR2F1-AS1 knockdown (Figure 1F). Altogether, the data indicated that NR2F1-AS1 was upregulated in PTC and that silence of NR2F1-AS1 retarded proliferation and invasion of PTC in vitro.
3.2 NR2F1-AS1 acted as a sponge of miR-423-5p in PTC cells
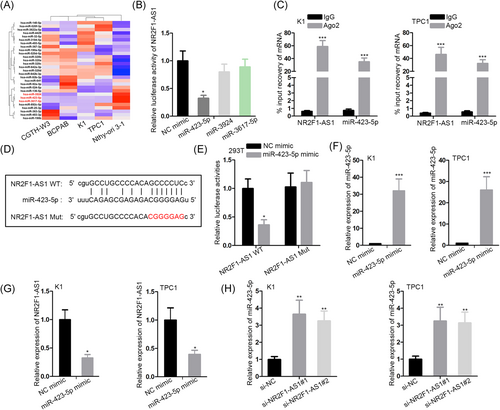
Next, the mechanism of NR2F1-AS1 in PTC was investigated. A myriad of studies have demonstrated that lncRNAs function in cancer development through acting as miRNA sponges.26, 27 Hence, we postulated that NR2F1-AS1 exerted its function in PTC through sponging miRNA. By searching Starbase3.0, we found 29 candidate miRNAs potentially targeted by NR2F1-AS1. To detect the involvement of these miRNAs in PTC, we examined their expression in cell lines by RT-qPCR. As shown in the heat map of Figure 2A, we found that miR-423-5p, miR-3924, and miR-3617-5p showed significant downregulation in all four PTC cells. Then, we detected the interplay of the three miRNAs with NR2F1-AS1, respectively. Luciferase activity of NR2F1-AS1 was overtly reduced by the ectopic expression of only miR-423-5p (Figure 2B). Therefore, we hypothesized that NR2F1-AS1 could sponge miR-423-5p in PTC. RIP analysis followed by RT-qPCR revealed that both NR2F1-AS1 and miR-423-5p were enriched in the immunoprecipitated products of Ago2 (Figure 1C). Besides, the miR-423-5p site on NR2F1-AS1 was obtained from Starbase3.0 (http://starbase.sysu.edu.cn/) and the site was mutated to generate NR2F1-AS1 Mut reporter (Figure 2D). Overexpression of miR-423-5p attenuated the luciferase activity of NR2F1-AS1 WT but not NR2F1-AS1 Mut (Figure 3E), indicating that miR-423-5p interacted with NR2F1-AS1 at the predicted site. Later, we confirmed the overexpression of miR-423-5p in TPC1 and K1 cells (Figure 2F). The expression of NR2F1-AS1 dropped with the presence of miR-423-5p overexpression in two PTC cell lines (Figure 2G). Also, miR-423-5p expression was induced upon the silence of NR2F1-AS1 (Figure 2H). Jointly, NR2F1-AS1 acted as a sponge of miR-423-5p in the PTC cells.
3.3 SOX12 was a target of NR2F1-AS1/miR-423-5p in PTC cells
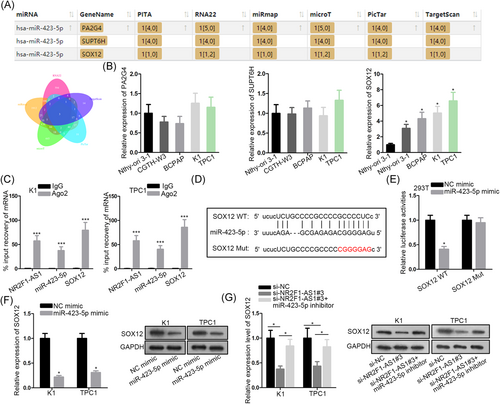
It is widely reported that by sponging miRNAs, lncRNAs could relieve downstream genes from the posttranscriptional suppression by miRNAs.26, 27 Thus, we set out to identify the downstream gene for NR2F1-AS1/miR-423-5p. The intersection of the searching results of six bioinformatics tools (PITA, RNA22, miRmap, microT, PicTar, and TargetScan) showed that three genes were putative targets for miR-423-5p, which were PA2G4, SUPT6H, and SOX12 (Figure 3A). Subsequently, we examined the expressions of three genes in cell lines. Results of RT-qPCR showed that only SOX12 was upregulated in PTC cell lines (Figure 3B). Therefore, we speculated that SOX12 was a target for miR-423-5p in PTC. RIP assay depicted that miR-423-5p was coimmunoprecipitated with NR2F1-AS1 and SOX12 with the Ago2 antibody (Figure 3C). Furthermore, binding sequences on SOX12 for miR-423-5p were predicted by Starbase3.0 and the sequences were substituted with the complementary sequences to generate SOX12 Mut reporter (Figure 3D). Overexpression of miR-423-5p caused a decrease of SOX12-WT rather than SOX12 Mut (Figure 3E). Besides, SOX12 mRNA and protein levels were reduced by a miR-423-5p mimic in PTC cells (Figure 3F). Moreover, SOX12 mRNA and protein expressions were reduced by NR2F1-AS1 silence and were restored by cotransfection of miR-423-5p inhibitor in PTC cells (Figure 3G). To sum up, SOX12 was a target of NR2F1-AS1/miR-423-5p in PTC cells.
3.4 NR2F1-AS1 accelerated proliferation and invasion of PTC cells through a SOX12-dependent way
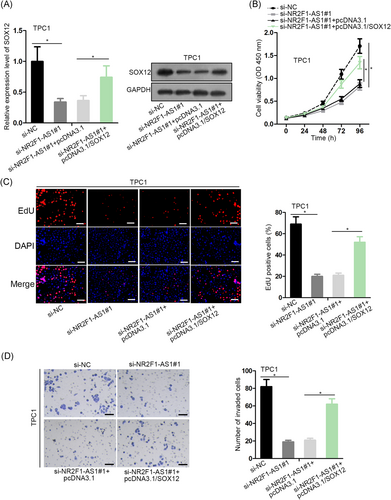
Finally, we interrogated whether NR2F1-AS1 could regulate PTC proliferation and invasion through SOX12. RT-qPCR and Western blot analysis depicted that SOX12 expression was decreased by si-NR2F1-AS1#1, and recovered by the cotransfection of pcDNA3.1/SOX12 in TPC1 cells (Figure 4A). CCK-8 and EdU assays delineated that silence of NR2F1-AS1 reduced proliferation of TPC1 cells, and such reduction was reversed by the overexpression of SOX12 (Figure 4B,C). Also, the invasion of TPC1 cells was hindered by the depletion of NR2F1-AS1, and reversed by the forced expression of SOX12 (Figure 4D). In a word, it was implied that NR2F1-AS1 accelerated proliferation and invasion of PTC cells through a SOX12-dependent way.
4 DISCUSSION
During past decades, rising attention has been paid toward the regulatory roles of lncRNAs in tumorigenesis and the development of PTC. Association of dysregulated lncRNAs with PTC progression has been established by a number of studies.7-9 However, the function of a large proportion of lncRNAs in PTC remains unknown. Formerly, Huang et al10 stated that NR2F1-AS1 contributed to oxaliplatin resistance of HCC, and silence of NR2F1-AS1 suppressed the migration, invasion, and tumor growth of HCC cells, indicating the role of NR2F1-AS1 as an oncogene. The current study firstly discovered through bioinformatics analysis that the NR2F1-AS1 level was increased in THCA samples. PTC is known to account for almost 90% of all thyroid cancer cases.1 Hence, NR2F1-AS1 might participate in PTC. Expectedly, we confirmed that NR2F1-AS1 was highly expressed in PTC cell lines. Functionally, loss-of-function assay suggested that inhibiting NR2F1-AS1 could retard proliferation and invasion of PTC cells, indicating that NR2F1-AS1 positively regulated PTC progression.
Mechanistically, lncRNAs are widely demonstrated to serve as miRNAs sponge to regulate cancers,12-14 including PTC.15 miRNAs are renowned regulator of gene expression because they recognize and target the 3′-UTR of mRNAs to trigger the degradation of mRNA or suppress the translation of functional proteins.11 Our study first found that miR-423-5p was a target for NR2F1-AS1. Previously, the downregulation of miR-423-5p and its tumor-suppressive role have been reported in a diversity of cancers, such as osteosarcoma,16 brain metastasis of lung adenocarcinoma,17 nasopharyngeal carcinoma,18 and colon cancer.19 Herein, our findings showed the downregulation of miR-423-5p in PTC cell line and confirmed the interplay between miR-423-5p and NR2F1-AS1. In addition, we revealed the reciprocal repression between NR2F1-AS1 and miR-423-5p expressions in PTC.
Thereafter, we found that SOX12 was one of the three candidate targets for miR-423-5p through bioinformatics prediction. SOX12 was identified to present an upregulated level in PTC cell lines, indicating its involvement of SOX12 in PTC. It is known that SOX12 belongs to the SOX transcription factors, which are responsible for the activation of target genes.20-22 Multiple previous studies unveiled that SOX12 functioned as an oncogene in cancers by promoting diverse cellular activities such as proliferation, invasion, and migration.23-25 The present study was the first to uncover the participation of SOX12 in PTC. Moreover, we validated the interaction of miR-423-5p with SOX12 and demonstrated that NR2F1-AS1 positively modulated NR2F1-AS1 expression through sponging miR-423-5p. Finally, the abrogation of the inhibitory effect of NR2F1-AS1 silence on PTC cell proliferation and invasion by SOX12 overexpression suggested that NR2F1-AS1 accelerated PTC progression through SOX12.
In conclusion, the current study first revealed that NR2F1-AS1 was upregulated in PTC, and demonstrated the function of NR2F1-AS1 in promoting proliferation and invasion of PTC cells. Mechanistically, we revealed a novel competitive endogenous RNA network NR2F1-AS/miR-423-5p/SOX12 in PTC cells. These findings suggested NR2F1-AS1 as a biomarker in PTC progression and provided new clues to push forward the development of PTC treatment.
ACKNOWLEDGMENT
The authors would like to thank Shengjing Hospital of China Medical University for their technical support.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
CY wrote the manuscript. CY, ZL, XC, and WX contributed to the experiments. JG, FC, DC conducted the data and analyzed it. All authors participated in identifying the final version of the manuscript.