MicroRNAs associated with lung squamous cell carcinoma: New prognostic biomarkers and therapeutic targets
Abstract
Background
With the development in research, the importance of microRNAs (miRNAs) in the occurrence and metastasis, and prognosis of lung squamous cell carcinoma (LUSC) have received extensive attention. This study aims to identify new biomarkers and pivotal genes associated with LUSC prognosis through bioinformatics.
Materials and methods
We downloaded miRNA and messenger RNA samples related to LUSC from The Cancer Genome Atlas (TCGA) database. Following initial data screening and preprocessing, we utilized the R platform and a range of analysis tools (miRDB, TargetScanHuman7.2, DAVID, and Cytoscape_v3.5.1) to analyze the TCGA data and identify highly specific and sensitive biomarkers.
Results
We finally identified 12 miRNAs and six central genes closely related to the overall survival of patients with LUSC. We found that high expression of 7 miRNAs (miR-301b, miR-383, miR-512, miR-515, miR-525, miR-577, and miR-5682) and low expression of five miRNAs (miR-448, miR-486, miR-4732,miR-4732, miR-516, and miR-1911) can significantly improve the overall survival rate of patients with LUSC. The six central genes DLGAP5, CENPA, CHEK1, LRRK2, CALB1, and TOP2A are directly or indirectly involved in the formation and metastasis of LUSC and could, therefore, be considered as target genes for drug therapy.
Conclusion
With the development in research, the importance of miRNAs in the occurrence and metastasis and prognosis of LUSC have received extensive attention. This study aims to identify new biomarkers and pivotal genes associated with LUSC prognosis through bioinformatics.
1 INTRODUCTION
Globally, lung cancer is not only the most common diagnostic cancer but also the leading cause of cancer death, accounting for 11.6% of total cases and 18.4% of total cancer deaths, respectively.1 Lung cancer is generally classified into two major groups; small cell lung cancer (SCLC; 15% of cases) and non–small cell lung cancer (NSCLC; 85% of cases).2 Squamous cell carcinoma of the lung (LUSC) is a common type of NSCLC with poor clinical outcomes, with only 15% of patients surviving for 5 or more years.3 Due to a lack of clear histological features for LUSC,4 although some treatments are effective there is no clear personalized treatment strategy as to whether conventional radiotherapy, chemotherapy, or targeted therapy will be most effective.5, 6 Based on the current state of onset and treatment of patients with tumor, the importance of finding relevant tumor markers has become increasingly prominent.7, 8 For example, studies have shown that phosphoinositide 3-kinases and S155R mutations can have an impact on tumor progression.9, 10 The recent development of the The Cancer Genome Atlas (TCGA) database, which adopts high-throughput genomic analysis technology, has allowed clinicians and researchers to better understand the genetics of cancer, thereby improving their ability to prevent, diagnose, and treat their malignancies of interest.
The TCGA database contains extensive and standardized clinical data, as well as a large number of samples for each cancer type, including gene expression data, microRNA (miRNA) expression data, information on copy number variation, DNA methylation, and single nucleotide polymorphisms. The introduction of technologies such as protein or DNA arrays and mass spectrometry enables the simultaneous detection of thousands of proteins and genes in a single experiment, giving us the opportunity to explore the characteristics of cancer and specific therapeutic strategies at the molecular and even atomic levels,11-13 and molecular level biological research has yielded considerable results, which is attracting widespread attention.14, 15 Some studies have shown that mutations in individual miRNAs, as well as ectopic or overexpression, can lead to serious physiological consequences by affecting cell death and immune function (for example autoimmunity, lymphatic proliferation, and cancer).16 These affects are mainly due to the regulation of messenger RNA (mRNA) by miRNA.17 In addition, various types of miRNAs display different levels of expression in specific types of malignancies.18 Therefore, the mRNA and miRNA data of a patient with LUSC samples in the TCGA database were used to find new biomarkers in our study.
More and more evidence shows that there is a trend of differential expression of miRNA and mRNA in various malignant tumor cells, which is a good starting point for the search for tumor biomarkers. In the present study, we not only identified prognosis-related miRNA and mRNA but also constructed miRNA-mRNA interaction networks. This may help us better understand the role of miRNA-mRNA in tumors. We utilized the R package of the R platform and a range of analysis tools, namely; miRDB (http://www.mirdb.org/miRDB/), TargetScanHuman7.2 (http://www.targetscan.org/), DAVID (https://david.ncifcrf.gov/), Kaplan-Meier Plotter (www.kmplot.com), and Cytoscape_v3.5.1 software, to analyze TCGA data and identify highly specific and sensitive biomarkers associated with LUSC. In the data analysis process, we separately analyzed the difference between miRNA and mRNA and constructed the protein interaction network. Finally, we identified 12 miRNAs (miR-301b, miR-383, miR-512, miR-515, miR-525, miR-577, miR-5682, miR-448, miR-486, miR-4732, miR-516, and miR-1911) and six pivotal genes (DLGAP5, CENPA, CHEK1, LRRK2, CALB1, and TOP2A) closely related to the survival of patients with LUSC , which provides a new theoretical basis for the prognosis and targeted therapy of LUSC. However, the regulatory network between miRNA and mRNA is complex and needs further research.
2 MATERIALS AND METHODS
2.1 Data download and screening
The TCGA is currently the largest database of cancer gene information and is supervised by the National Cancer Institute and the National Human Genome Research Institute. A data set of patients with LUSC, including samples for both miRNAs and mRNAs, was downloaded from the TCGA database (https://cancergenome.nih.gov/) for analysis.
To ensure the accuracy of the TCGA database analysis, we included a preprocessing step for the data. This entailed: (1) Utilizing the R packet to separate LUSC tissue and adjacent nontumor lung tissue in downloaded samples. (2) The results of mRNA expression data were differentially analyzed using the R-pack of R (edgeR), and mRNAs with significantly different expression were selected (P cutoff < 0.05 and |logFC| > 3.0). (3) The expression profiles of miRNA were normalized by the R package and miRNAs with |logFC| > 3.0 were screened by univariate Cox regression analysis and Kaplan-Meier curves, with P < 0.05 indicating a significant correlation with survival outcomes. The false discovery rate (FDR) was applied to correct the multiple hypothesis test and FDR < 0.05 was considered to be significant.
2.2 Prediction of miRNA target genes
To ensure the integrity of the target genes, we utilized two online analysis tools, miRDB (http://www.mirdb.org/miRDB/) and TargetScanHuman7.2 (http://www.targetscan.org/) for target gene prediction. The resulting Venn diagram was applied to analyze the miRNA target genes and mRNA differential genes. This finally revealed the target genes related to the survival of LUSC described in this study.
To further understand the biological functions of the target genes, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using DAVID (https://david.ncifcrf.gov/). Jiao et al,19 P < 0.05 were considered as significant.
2.3 Construction of a protein interaction network
The official gene symbols of the predicted target genes were imported into the Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org) to assess the interactions between the genes in the protein interaction (PPI) network.20 There were 197 nodes and 396 edges in the resulting PPI network, the results of which were visualized using Cytoscape software. A medium confidence interval of 0.4 was used as the threshold.
2.4 Topology analysis and verification of central genes
CentiScaPe, an app within Cytoscape_v3.5.1 software,21 was utilized to identify central genes from the predicted target genes for subsequent topology analysis, with degree >15 as the qualification for high connectivity nodes. Subsequently, we used univariate Cox regression analysis and the online analysis tool Kaplan-Meier plotter (kmplot.com) to assess the prognostic value of the six central genes. Genes with a P < 0.05 were considered to be significantly associated with survival and identified as prognostic genes. Finally, based on the Cytoscape software, the corresponding regulatory relationship of miRNA-mRNA was observed in the miRNA-gene regulatory network.
3 RESULTS
3.1 Identification of differentially expressed miRNAs in LUSC
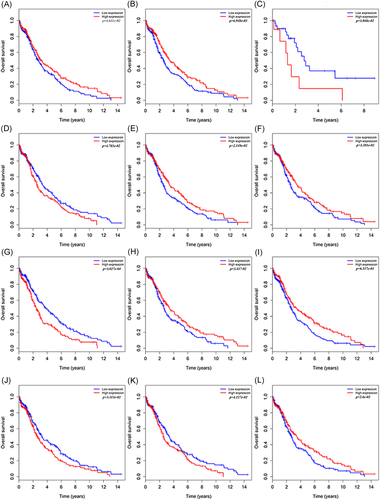
We downloaded 523 miRNA LUSC-related samples (45 normal and 478 tumors) from the TCGA online database. First, we removed information about adjacent (nontumor) tissues in the sample, then set thresholds for the relevant parameters (P cutoff < 0.05 and |logFC| > 3.0) to identify the differentially expressed miRNAs (DEMs) using the R package of the R software. A total of 99 DEMs were obtained, of which 90 were upregulated and nine were downregulated (Figure 1A). Based on these, we used Cox regression analysis, Kaplan-Meier curve, and log-rank analysis to further verify the characteristics of the DEMs and the intensity of their correlation with survival. A total of 12 miRNAs were ultimately considered to have significant differences in the survival status of patients with LUSC (Figure 2), comprising seven upregulated miRNAs (miR-301b, miR-383, miR-512, miR-515, miR-525, miR-577, and miR-5682) and five downregulated miRNAs (miR-448, miR-486, miR-4732, miR-516, and miR-1911).
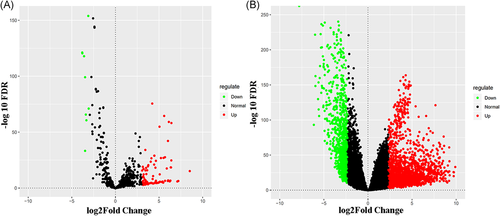
A, Volcano plot of differentially expressed miRNAs. The red dot represents upregulated miRNAs and the green dot represents downregulated miRNAs. B, Volcano plot of differentially expressed mRNAs. The red dot represents upregulated mRNAs and green dot represents downregulated mRNAs. miRNAs, microRNAs; mRNAs, messenger RNAs
Twelve differentially expressed miRNAs were associated with survival in lung squamous cell carcinoma patients by using the Kaplan-Meier curve. A, miR-301b; B, miR-383; C, miR-448; D, miR-486; E, miR-512; F, miR-515; G, miR-516; H, miR-525; I, miR-577; J, miR-1911; K, miR-4732; L, miR-5682. miRNAs, microRNAs
3.2 Identification of differentially expressed miRNAs in LUSC
Similarly, 551 mRNA LUSC-related samples (49 normal and 502 tumor) were downloaded from the TCGA database. The data set was identified and analyzed using the limma package and the edgeR package of the R platform. On the premise of P < 0.05 and |logFC| > 3.0, 2573 differentially expressed mRNAs were identified, of which 1734 were upregulated and 839 downregulated (Figure 1 B).
3.3 Prediction of target genes and functional analysis
We utilized the online target gene prediction tools, miRDB and TargetScanHuman7.2 in our target gene prediction process, which revealed a total of 3255 genes. Considering the accuracy of the biological analysis, we used a Venn diagram to identify genes closely related to miRNA as well as mRNA, resulting in 344 selected target genes. To investigate the biological effects of these genes, we performed GO and KEGG functional enrichment analyses, which showed significant enrichment in various functional characteristics. The results of the GO analysis revealed an unequal enrichment of genes across the three biological levels (biological process, cellular component and molecular function; Figure 3). In addition, GO analysis revealed a high enrichment of genes functioning on the plasma membrane. The KEGG functional enrichment analysis, on the other hand, identified an enrichment in several tumor-related pathways, such as pathways in cancer, proteoglycans in cancer, cell cycle, and cell adhesion molecules (CAMs; Table 1).
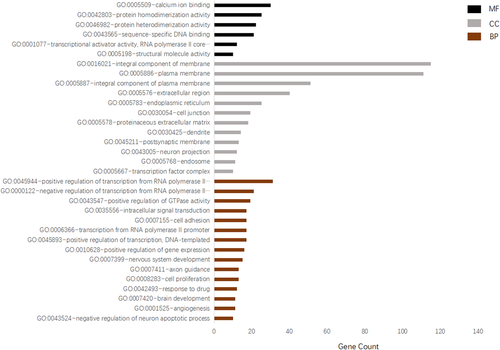
The significant enriched GO biological processes of target genes. GO, Gene Ontology
| Term | Count | P value | Genes |
|---|---|---|---|
| Pathways in cancer | 15 | .013596468 | FZD9, F2RL3, E2F2, WNT10A, WNT16, EPAS1, TGFBR2, SKP2, EGLN3, DAPK2, CCNE2, CCNE1, RASGRP4, HHIP, WNT6 |
| Neuroactive ligand-receptor interaction | 12 | .013400886 | CRHR1, F2RL3, MCHR2, AGTR2, S1PR1, CCKBR, SSTR1, GRIK2, ADRA1A, CALCRL, GRM1, GABRP |
| Proteoglycans in cancer | 10 | .011676387 | FZD9, WNT10A, WNT16, CAV1, ERBB4, CAMK2B, WNT6, GPC1, HOXD10, TWIST1 |
| Axon guidance | 9 | .002362515 | EPHA7, SEMA3G, EFNA3, NTNG1, DPYSL5, DPYSL2, EPHB1, SLIT2, SLIT3 |
| Cell adhesion molecules (CAMs) | 9 | .00469574 | CLDN18, PTPRM, CADM1, ITGB8, CLDN1, NTNG1, NRXN1, NEGR1, CDH5 |
| Adrenergic signaling in cardiomyocytes | 7 | .053406473 | AGTR2, ADRA1A, SCN4B, CAMK2B, ATP1A2, PPP2R2C, SCN5A |
| Protein digestion and absorption | 6 | .023264405 | COL9A1, COL6A6, COL7A1, COL22A1, COL2A1, ATP1A2 |
| Cell cycle | 6 | .079728036 | CCNE2, CCNE1, E2F2, SKP2, CHEK1, ORC6 |
| Basal cell carcinoma | 5 | .01850775 | FZD9, WNT10A, WNT16, HHIP, WNT6 |
| Proximal tubule bicarbonate reclamation | 3 | .066880427 | CA4, ATP1A2, SLC4A4 |
- Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.4 PPI network and verification of central genes
We utilized the STRING online tool to further observe the interactions between our target genes and visualized the results as a PPI network using Cytoscape software (Figure 4). The PPI network contained 197 nodes and 396 edges. According to the view that highly connected genes can have a major impact on disease, we calculated the connectivity between the nodes using topological analysis through CentiScaPe software. This identified six nodes (with degree >15) as high connectivity genes in our study. Additionally, in the Kaplan-Meier diagrams, the P values of the central six genes were far less than 0.05. This shows that the 10-year survival rate of patients with LUSC is significantly increased when one (LRRK2) gene is expressed to a high level and five (DLGAP5, CENPA, CHEK1, CALB1, and TOP2A) genes are expressed to a high level (Figure 5). By retrospective examination of the miRNA-mRNA regulatory network, we intuitively identified that the six central genes are regulated predominantly by three miRNAs. miR-515 regulates the expression of DLGAP5 and CENPA, miR-577 regulates the expression of GALB1 and TOP2A, and miR-383 regulates the expression of CHEK1.
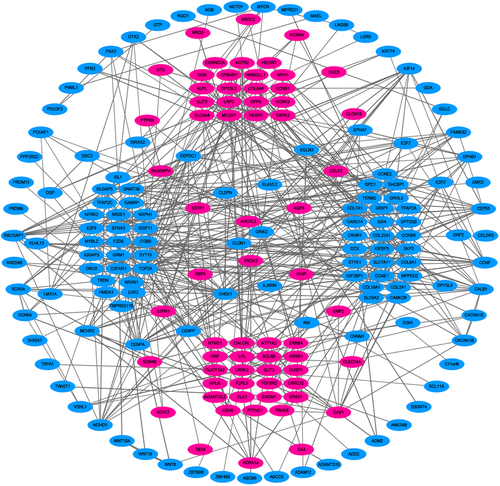
PPI network of lung squamous cell carcinoma related target genes and related genes. The blue dot represents upregulated genes and the pink dot represents downregulated genes. PPI, protein interaction
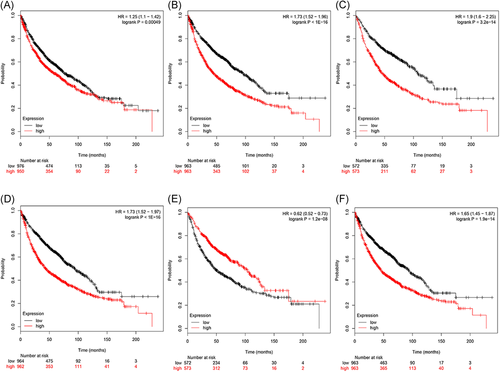
Six central mRNAs were associated with survival in lung squamous cell carcinoma patients by using the Kaplan-Meier curve. A, CALB1; B, CENPA; C, CHEK1; D, DLGAP5; E, LRRK2; F, TOP2A. mRNAs, messenger RNAs
4 DISCUSSION
Approximately 85% of patients with lung cancer worldwide are diagnosed with NSCLC, and most are difficult to treat due to the presence of complex advanced disease. For these patients, drug-based chemotherapy can moderately extend survival time. However, the effect of therapy currently seems to be in a stagnant phase, with no significant improvement in response rate (20%-35%) or median survival (8-12 months).22-24 The treatment status for LUSC, the main type of LUSC, is also not optimistic. Previous studies have revealed that there are still viable unexplored genetic changes in squamous cell lung cancer.25, 26 Therefore, new ideas for the diagnosis and treatment of LUSC could be provided by analyzing the tumor-related data in TCGA to obtain the LUSC–related miRNA-mRNA regulatory network and predict new tumor markers.
An increasing number of studies have recently attracted researchers’ attention to the importance of miRNAs. By univariate Cox regression analysis and Kaplan-Meier curves, we identified 12 miRNAs as significant in terms of survival status of patients with LUSC, including seven upregulated miRNAs (miR-301b, miR-383, miR-512, miR-515, miR-525, miR-577, and miR-5682) and five downregulated miRNAs (miR-448, miR-486, miR-4732, miR-516, and miR-1911). Several of these miRNAs have been previously associated with the molecular mechanisms of tumors. In lung cancer, high expression of miR-301b is known to modulate FOXF2, and ectopic expression of its 3′-untranslated region (3′-UTR) reduced the sensitivity of cells to chemotherapy.27, 28 miR-383 is an inhibitor of lung cancer, which can target the 3′UTR of endothelial PAS domain protein 1 mRNA, which is significantly associated with poor prognosis in patients.29, 30 Expression of miR-448 has been linked to the molecular mechanisms of various tumors, such as LUSC,31 osteosarcoma,32 pancreatic ductal adenocarcinoma,33 colorectal cancer,34 breast cancer,35, 36 and others. Among these many studies, one report has shown that miR-448 expression is associated with TNM staging of LUSC.37 There have also been multiple reports on the role of miR-486 in tumorigenesis, some concluding that it acts as a tumor suppressor of LUSC.38-40 miR-515 has also been reported as a tumor suppressor,41, 42 and similarly, studies have shown that miR-512 not only inhibits glycolysis but also accelerates apoptosis.43, 44 So far, there are few reports of a role for miR-516 or miR-525 in LUSC, however, there is evidence that they can regulate target genes in other tumors to affect protein expression.45, 46 miR-577 affects cell expression by regulating the Wnt signaling pathway, which subsequently affects cell growth and cell viability. Wang et al,47, 48 to our knowledge, there are currently no reports relating miR-1911, miR-4732, or miR-5682 to lung squamous cell malignancies. Therefore, more research is needed to investigate the function of these miRNAs in LUSC, and with the exponential increase of important findings in this area, we believe this could give exciting new insights.
To understand the biological functions of miRNAs more deeply, we constructed a PPI network and carried out target gene prediction, followed by functional analysis on the 344 resulting target genes. This revealed that the target genes form part of a variety of tumor-related functional categories, such as CAMs, proteoglycans in cancer, pathways in cancer, basal cell carcinoma, and cell cycle (Table 1). A large body of evidence supports the role of adhesion molecules in the communication between vascular endothelial cells and tumor cells, which may lead to tumor cell invasion and ultimately metastasis of malignant tumors.49 For example, cadherin-11 affects the activity levels of crac1 and STAT 3, which in turn regulates the P53 signaling pathway.50 Proteoglycans have an indispensable role in the tumor microenvironment, and as a result, many studies have described their potential use as target genes, biomarkers, and active agents.51, 52 Moreover, it has been found that mutations in cell cycle genes can affect the differentiation of squamous cells in the course of modern anticancer treatment research.53, 54 Pathways in cancer, which describes important signaling pathways such as Wnt signaling and T-cell receptor signaling, is also an important factor in the progression of NSCLC and development of squamous cell carcinoma of the lung.55, 56
To further validate and screen the target genes, we performed a topological analysis in the PPI network. The six genes with a connectivity score >15 were selected as central genes, on which survival analysis was performed again. The results showed that the central genes; DLGAP5, CENPA, CHEK1, CALB1, LRRK2, and TOP2A, were significantly associated with 10-year survival rates of a patient with LUSC. The noncoding small RNA molecules known as miRNAs have the function of inhibiting transcription and translation and can cleave target mRNA promoting its degradation. From examining the miRNA-mRNA regulatory networks, the results of our research are consistent with this. miR-515, miR-577, and miR-383 all silenced these six pivotal genes separately when they were highly expressed. DLGAP5 is associated with the development of a variety of tumors, such as ovarian cancer, breast cancer, and NSCLC, due to its ability to inhibit mitosis and block cell cycle progression.57, 58 Calcium is one of the key factors in stabilizing the cell cycle, and CALB1 can interfere with the resorption and homeostatic processes of calcium, ultimately regulating cell apoptosis.59 In addition, studies have shown that CALB1 can play a role in early tumor diagnosis and recurrence prediction.60, 61 TOP2A, or topoisomerase IIa, has been shown to be involved in proliferation, apoptosis, and tumor resistance mechanisms in various studies.62-64 CHEK1 is also a regulator of cell proliferation; in a study of microtubule-targeting agents, it was shown that knockdown of CHEK1 had a synergistic effect in inhibiting cell growth.65 In a similar vein, CENPA is a centromere protein and is therefore essential for maintaining chromosome stability.66, 67 Finally, LRRK2 is considered to be a common mutant gene in Parkinson's disease.68, 69 Although few studies have reported a direct role for CENPA and LRRK2 in malignancy, future studies targeted to certain processes or cancer types should be designed to discover more about their biological functions.
5 CONCLUSION
In conclusion, we adopted bioinformatics methods to systematically analyze LUSC-related mRNAs and miRNAs in the TCGA database. Twelve miRNAs (seven upregulated miRNAs: miR-301b, miR-383, miR-512, miR-515, miR-525, miR-577, and miR-5682 and five downregulated miRNAs: miR-448, miR-486, miR-4732, miR-516, and miR-1911) and six pivotal genes (DLGAP5, CENPA, CHEK1, CALB1, LRRK2, and TOP2A) were identified as potential biomarkers for the prognosis of patients with LUSC. Our research has deepened our understanding of the treatment and prognosis of LUSC at the molecular level, which could improve the understanding of the pathogenesis and potential molecular events of LUSC, contribute to the timely diagnosis and prognosis of patients with LUSC, and lay a foundation for future clinical research. However, it is worth emphasizing that the mechanism of action of miRNAs and the regulatory network of miRNA-mRNA interactions are particularly complex. We have provided an analysis of theoretical knowledge and clinical outcomes, but more scientific research is needed to confirm our findings, and investigate their potential for clinical application in improving the outlook for patients with LUSC.
CONFLICT OF INTERESTS
The authors declare that there is no conflict of interests.
AUTHOR CONTRIBUTIONS
LQ and CS contributed to the design of the study protocol. LQ and CG contributed to the writing of the study protocol. FF, TZ, and YY have downloaded data. XW, CL performed statistical analysis. JL and JLdrawed the pictures. All authors approved the final version of the manuscript.