Association between the −11377 C/G and −11391 G/A polymorphisms of adiponectin gene and adiponectin levels with susceptibility to type 1 and type 2 diabetes mellitus in population from the west of Iran, correlation with lipid profile
All authors contributed equally to this study.
Abstract
Adipose tissue, an endocrine organ, secretes bioactive factors including adiponectin. Adiponectin is a protein hormone that enhances insulin sensitivity through increased fatty acid oxidation and inhibition of hepatic glucose production. We assessed the association of the adiponectin promoter region polymorphisms −11391 G/A and −11377 C/G with susceptibility to type 1 (T1DM) and type 2 (T2DM) diabetes mellitus in the population of west Iran. Also, we investigated the effect of adiponectin level and lipid profile on T1DM and T2DM development. In this case-control study, we recruited 189 patients with diabetes (100 T2DM and 89 T1DM) and 161 sex and age-matched unrelated healthy controls. Adiponectin mutations were detected by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), and the protein level was measured by the enzyme-linked immunosorbent assay. Other biochemical parameters were determined by routine laboratory methods. The G allele of adiponectin gene at −11377 position (C/G) significantly increased the risk of T1DM. With respect to genotype models, codominant (2.97 times), dominant (3.6-fold), and over-codominant (2.9-fold) patients with T1DM who carried −11377 C > G single-nucleotide polymorphisms were significantly susceptible to the development of the disease. A significantly higher level of adiponectin in T1DM was oberved compared with the control group. In contrast, patients with T2DM had lower adiponectin levels compared with healthy controls. The genotype distributions of −11391 G/A polymorphisms were the same for patients with diabetes and control groups. The presence of G allele at −11377 C/G adiponectin gene significantly increased serum adiponectin level and may be a risk factor for T1DM susceptibility among the western Iranian population.
1 INTRODUCTION
Diabetes mellitus is a systemic disease associated with lack of relative or complete insulin deficiency and function, which leads to hyperglycemia.1 The two most common types of diabetes are type 1 and type 2 diabetes mellitus (T1DM and T2DM). The global prevalence of diabetes for all age groups was estimated to be 2.8% in 2000, which increases annually and may reach as high as 4.4% in 2030.2, 3 Therefore, prevention and treatment of diabetes is considered a global health priority.4 Although insulin resistance plays a major role in the development of type 2 diabetes, other mechanisms, including a complex interaction between genes and environment factors, may be involved in pathogenesis of the disease.5, 6
Recently, the role of adipokines such as adiponectin, leptin, and interleukin (IL)-6 in the pathogenesis of diabetes has been emphasized in numerous studies.7, 8 Adiponectin, a secretory protein and the most plentiful adipokine in circulation, induces insulin sensitivity and exhibits anti-inflammation and antiatherogenic effects.9 A low plasma concentration of adiponectin is associated with an increase in body fat mass, insulin resistance, and dyslipidemia in humans.9-11 Because of its noticeable role in the pathogenesis of type 2 diabetes, a low level of adiponectin is considered as a predictor of insulin resistance and type 2 diabetes.12 The adiponectin gene is encoded by the ADIPOQ gene located on chromosome 3q27, and mutations at this gene lead to the development of metabolic syndrome and diabetes.13-15
Single-nucleotide polymorphisms (SNPs) in adiponectin gene affect the gene expression and, thus, its plasma concentration.16 The effects of several adiponectin SNPs including −11391 G > A, −11377 G > C, −10066 G > A, −7734 C > A, +3228 C > T, +45 T > G, and 276 G > T on its plasma level have been investigated.17, 18 Vasseur et al18 have shown that a high level of circulating adiponectin is associated with −11391 G > A (rs 17300539) SNP in the promoter region of the adiponectin gene, while the −11377 C > G (rs 266729) variant (cytosine is replaced by guanine) is correlated with a low level of adiponectin. In the current study, we have determined the correlation of −11391 G > A and −11377 C > G SNPs, lipid profile, and adiponectin concentration with susceptibility to T1DM and T2DM in the population from the Kermanshah province, situated in the west of Iran.
2 METHODS
2.1 Subjects
The study protocol was approved by the Ethics Committees of the Kermanshah University of medical sciences, and an informed written consent was obtained from all subjects. One hundred eighty-nine patients with diabetes (100 T2DM, 57 females and 43 males with a mean age 56.18 ± 8.9 and body mass index [BMI] ≥ 27.4; and 89 T1DM patients, 44 females and 45 males with a mean age 19.65 ± 7.33 and BMI ≥ 21.5) and 161 healthy individuals (control subjects) matched with T1DM and T2DM patients according to the sex, age, and weight were studied. Control subjects for T2DM included 51 females and 44 males with a mean age 55.8 ± 9.1, and for T1DM, included 34 females and 32 males with a mean age 19.65 ± 7.33. Patients with T2DM and T1DM were diagnosed according to the criteria set by the American Diabetes Association 1.1
2.2 Blood sampling
After overnight fasting and obtaining an informed written consent, blood samples were collected from all subjects in tubes containing ethylenediaminetetraacetic acid for genomic DNA extraction and for determining the HbA1c level and biochemical markers.
2.3 Biochemical measurements
The serum level of adiponectin and fasting blood glucose were determined by the enzyme-linked immunosorbent assay (ELISA; Human Adiponectin ELISA from Mediagnost Catalog Number E09) and by the glucose oxidase method (Pars Azmoon kit, Iran), respectively. Total plasma cholesterol (TC) and triglyceride (TG) levels were measured by the standard enzymatic methods (Pars Azmon kit, Iran), using an automated Erba XL600 (Germany). The plasma low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol (HDL-C) were determined by using commercially available enzyme assay kits (Pars Azmoon kit, Iran).
2.4 DNA analysis
Genomic DNA was extracted from peripheral blood leukocytes using the phenol-chloroform extraction method.19 Amplification was performed using a PCR Eppendorf-mastercycler gradient (Germany). The PCR reaction mixtures of the total volume of 25 μL included genomic DNA (50 ng), 20 pmol of each primer, 200 mM of each deoxynucleotide triphosphates, and 0.5 U Taq DNA polymerase in 10X Taq DNA reaction buffer. DNA sample was denatured at 95°C for 5 minutes, and the reaction mixture was subjected to 35 cycles of 95°C for 60 seconds, 58.4°C for 30 seconds, and 72°C for 45 seconds with a final extension time of 5 minutes. The forward 5′-CATCAGAATGTGTGGCTTGC-3′ and reverse 5′-AGAAGCAGCCTGGAGAACTG-3′ PCR primers were used to detect −11377 C/G (rs 266729) polymorphisms, and the polymorphisms at position −11391G/A (rs17300539) were detected by using forward 5′-ACTCTGCTGAGATGGACGGA-3′ and reverse 5′-GGGATGAGGGTGAAGATGGG-3′ PCR primers.20, 21 The PCR products were digested with Hha1 (−11377 C/G [rs 266729]) and MspI (11391 G/A [rs17300539]) restriction endonuclease (5 U, Fermentas) overnight at 37°C, and the digested products were separated by electrophoresis on a 16% polyacrylamide gel and visualized by ethidium bromide staining. The length of digested PCR fragments for homozygote wild-type −11391 AA was 163 bp; for homozygote mutant −11391 GG, it was 137 and 26 bp; and for heterozygote mutant −11391 GA, it was 163, 137, and 26 bp as shown in Figure 1. The length of digested PCR fragments for homozygote wild-type −11377 CC was 163 bp, for heterozygote mutant −11377 CG, it was 163, 137 and 26 bp, and for homozygote mutant −11377 GG, it was 121 and 42 bp as shown in Figure 2.
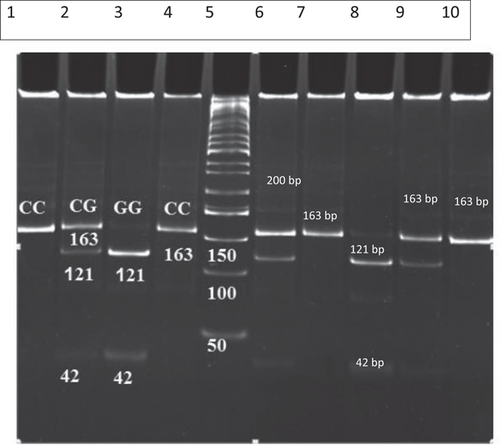
Poly acryl amid gel electrophoresis (PAGE) (12.5%) patterns for − 11377 C/G adiponectin alleles analyzed by PCR-RFLP. Lane 5, 50 bp DNA ladder; Lane 1, is an undigested PCR product; Lanes 2, 6, and 9, heterozygous mutant (C/G), Lanes 4, 7, and 10 homozygous wild-type (C/C); Lanes 3 and 8, homozygous mutant (GG). PCR, polymerase chain reaction
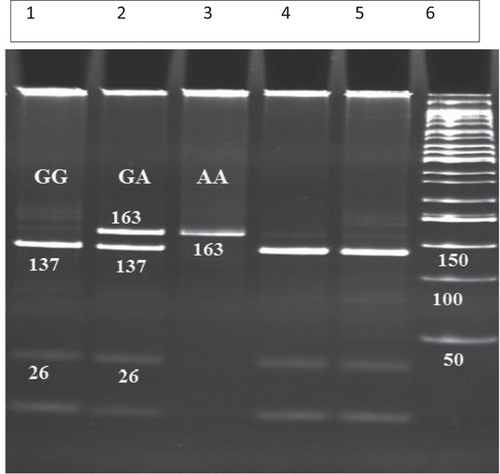
Polyacrylamid gel electrophoresis (PAGE) (12.5%) patterns for −11391 G/A adiponectin alleles analyzed by PCR-RFLP. Lane 6, 50 bp DNA ladder; Lane 2 , is an heterozygous mutant (G/A), Lanes 4 and 5 homozygous wild-type (G/G); Lanes 3 homozygous mutant (A/A). PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism
2.5 Statistical analysis
The statistical software package version SPSS 16 was used to analyze individual parameters. The allelic frequencies were calculated by the gene counting method. The adiponectin −11391G/A genotypes and allele frequencies in diabetic (T1DM and T2DM) patients were compared with controls using the chi-square (χ2) test. The χ2 test was used to verify the agreement of the observed genotype frequencies with those expected according to the Hardy-Weinberg equilibrium. The odds ratios (ORs) were calculated as estimates of relative risk for disease and 95% confidence intervals. The correlation of the serum levels of HbA1c, creatinine, HDL-C, LDL-C, TC, TG, and BMI with adiponectin −11391G/A polymorphism among the studied groups were calculated using linear regression and unpaired t test (Pearson). A two-tailed Student’s t test, analysis of variance (ANOVA), and nonparametric independent-sample Mann-Whitney analyses were used to compare the quantitative data. A P value less than 0.05 was assumed statistically significant.
3 RESULTS
The demographic and laboratory features of T1DM and T2DM patients and their control groups are presented in Table 1. Significantly reduced serum concentrations of HDL-C (42.5 [38.8-48] vs 49 [43-54], P ≤ 0.001) and higher adiponectin (19.4 [14-30.3] vs 16.2 [9.2-21.7] μg/L, P = 0.001) levels in T1DM patients compared with the control group were observed. The serum levels of cholesterol (191 [168-210] vs 171 [140-208], P ≤ 0.001), triglyceride (140 [111-192] vs 111 [77.8-160], P = 0.019), and LDL-C (109 [97-119] vs 101 [87.3-118], P ≤ 0.001) were significantly higher in T2DM patients compared with the control group. In contrast, T2DM patients had significantly lower levels of adiponectin (9.1 [6.3-12.4] vs 14.5 [8.2-21.5], P ≤ 0.001) and HDL-C 40 [34.3-45] vs 44 [37-50], P ≤ 0.001) compared with its healthy controls. In addition, there was a significantly increased BMI (P < 0.001) in T2DM patients compared with control subjects.
| Variable | T1DM | Control type 1 | T2DM | Control type 2 | *P value |
|---|---|---|---|---|---|
| Sex (female/male) | 44 (49.4%)/45 (50.8%); P = 0.994 | 34 (51.5%)/32 (48.5%) | 57 (57%)/43 (43%) | 51 (53.7%)/44 (46.3%) | 0.29 |
| Age, y | 19.7 7.3; P = 0.23 | 18.1 8.3 | 56.2 8.9 | 55.8 9.1 | 0.21 |
| BMI, kg/m2 | 21.9 4.1; P = 0.45 | 22 3.1 | 27.5 3.9 | 26.8 2.2 | <0.001 |
| Waist circumstance, cm | 76 11.8; P = 0.72 | 76.7 11.3 | 101.45 11.35 | 102.16 8.30 | <0.001 |
| Cholesterol, mg/dL | 166.4 29.6; P = 0.11 | 174.2 30.5 | 191 (168-210) | 171 (140-208) | <0.001 |
| Triglyceride, mg/dL | 112 51.6; P = 0.07 | 127.7 55.6 | 140 (111-192) | 111 (77.8-160) | 0.019 |
| LDL-C, mg/dL | 86 (78-94.5); *P < 0.001 | 100 (90-115) | 109 (97-119) | 101 (87.3-118) | <0.001 |
| HDL-C, mg/dL | 49 (43-54); *P < 0.001 | 42.5 (38.8-48) | 40 (34.3-45) | 44 (37-50) | <0.001 |
| Adiponectin, µg/L | 19.4 (14-30.3); *P = 0.001 | 16.2 (9.2-21.7) | 9.1 (6.3-12.4) | 14.5 (8.2-21.5) | <0.001 |
| Duration of diabetes, y | 3 (2-6) | - | 4 (6-12) | - | <0.001 |
| HbA1C, % | 8.5 1. | - | 8.2 1.41 | - | 0.09 |
| Fasting blood sugar, mg/dL | 180.2 79 | - | 192.4 66.6 | - | 0.24 |
- Abbreviations: BMI, body mass index; FBS, fasting blood sugar; HbA1c, hemoglobin glycosylates; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; T1DM, type 1 diabetes mellitus patients; T2DM, type 2 diabetes mellitus patients.
- Statistical analyses were done using t-test and by *nonparametric two independent samples Mann-Whitney test.
- * P compared clinical parameters between T1DM with T2DM.
The correlations between adiponectin concentrations with BMI, duration of diabetes, age of onset of diabetes, waist circumstance, HbA1C, FBS, cholesterol, LDL-C, HDL-C, and TG in T1DM and T2DM patients are shown in Table 2. In T1DM patients, adiponectin concentration was positively corrected with HbA1C and FBS (r = 0.255, P = 0.016, and r = 0.21, P = 0.045), but it was negatively correlated with the duration of diabetes (r = −0.22, P = 0.038) and BMI (r = −0.2, P = 0.046). In T2DM patients, adiponectin concentration was positively correlated with HDL-C level (r = 0.31, P = 0.001) and the age of onset of diabetes (r = 0.21, P = 0.033).
| Variable | Adiponectin level in T1DM patients | Adiponectin level in T2DM patients |
|---|---|---|
| Onset age of diabetes | r = −0.074, p = 0.49 | r = 0.21, p = 0.033 |
| Age, y | r = −0.196, p = 0.065 | r = 0.194, p = 0.053 |
| BMI, kg/m2 | r = −0.2, p = 0.046 | r = −0.02, p = 0.8 |
| Waist circumstance, cm | r = −0.179, p = 0.093 | r = 0.12, p = 0.23 |
| Cholesterol, mg/dL | r = −0.141, p = 0.19 | r = 0.12, p = 0.22 |
| Triglyceride, mg/dL)- | r = −0.02, p = 0.85 | r = 0.01, p = 0.9 |
| LDL-C, mg/dL | r = −0.05, p = 0.64 | r = 0.085, p = 0.39 |
| HDL-C, mg/dL | r = −0.096, p = 0.38 | r = 0.31, p = 0.001 |
| Duration of diabetes | r = −0.22, p = 0.038 | r = −0.05, p = 0.6 |
| HbA1C, % | r = 0.255, p = 0.016 | r = 0.03, p = 0.8 |
| Fasting blood sugar, mg/dL | r = 0.21, p = 0.045 | r = 0.1, p = 0.32 |
- Abbreviation: BMI, body mass index; HbA1c, hemoglobin glycosylates HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; T1DM, type 1 diabetes mellitus patients; T2DM, type 2 diabetes mellitus patients.
The odds ratio and distribution of adiponectin −11377 C/G genotypes and alleles in T1DM, T2DM patients, and their corresponding control groups are presented in Table 3. A significant difference was found between the genotype frequencies of adiponectin −11377 C > G SNP in T1DM patients and their controls (χ2 = 9.05, df = 2, P = 0.011). The −11377 codominant (C/G vs C/C, 2.97, (1.4-6.4), P = 0.005), dominant (C/G + G/G vs C/C, 3.6, (1.5-6.5), P = 0.004), and over-codominant (C/G vs C/C + G/G, 2.9, (1.37-6.2), P = 0.006) genotypes considerably increased the risk of T1DM. In addition, the frequency of adiponectin −11377 G allele was significantly higher in T1DM patients compared with their control group, and it significantly increased the risk of T1DM development by 2.6 times (P = 0.007).
| Adiponectin genotypes | T1DM (n = 89) | Control T1DM (n = 66) | T2DM (n = 100) | Control T2DM (n = 95) |
|---|---|---|---|---|
| C/C | 53 (59.6%) | 54 (81.8%) | 61 (62.9%) | 66 (70.2%) |
| C/G | 35 (39.3%) | 12 (18.2%) | 36 (37.1%) | 28 (29.8%) |
| G/G | 1 (1.1%) | 0 (0%) | 3 (3%) | 1 (1.1%) |
| (χ2 = 9.05, df = 2, P = 0.011) | (χ2 = 2.07, df = 1, P = 0.35) | |||
T1DM vs control T1MDOR (95% confidential interval) |
Control T1DM | T2DM vs Control T2MDOR(95%confidential interval) |
Control T2DM | |
| Codominant | ||||
| C/G vs C/C | 2.97 (1.4-6.4, P = 0.005) | (n = 12 vs n = 54) | 1.39 (0.76–2.5, P = 0.28) | (n = 28 vs n = 66) |
| (n = 35 vs n = 53) | (n = 36 vs n = 61) | |||
| Dominant | ||||
| C/G + G/G vs C/C | 3.6 (1.5–6.5, P = 0.004) | (n = 12 vs n = 54) | 1.46 (0.8–2.6, P = 0.21) | (n = 28 vs n = 66) |
| (n = 36 vs n = 53) | (n = 39 vs n = 61) | |||
| Over-codominant | ||||
| C/G vs C/C + G/G | 2.9 (1.37–6.2, P = 0.006) | (n = 12 vs n = 54) | 1.35 (0.74–2.5, P = 0.33) | (n = 28 vs n = 66) |
| (n = 35 vs n = 54) | (n = 36 vs n = 64) | |||
| 11377 C/G alleles | T1DM vs control T1MD OR (95%confidential interval) | Control T1DM | T2DM vs Control T2MD OR (95%confidential interval) | Control T2DM |
| C | n = 141 | n = 120 | n = 158 | n = 152 |
| G | 2.63 (1.3- 5.3, P = 0.007, n = 37) | n = 12 | 1.35 (0.8–2.26, P = 2.26, n = 42) | n = 30 |
| (χ2 = 7.8, df = 1, P = 0.005) | (χ2 = 1.3, df = 1, P = 0.26) | |||
- Abbreviation: T1DM, type 1 diabetes mellitus patients; T2DM, type 2 diabetes mellitus patients.
Adiponectin −11377 C/G genotype and allele were not associated with the risk of developing T2DM in our studied population. In addition, the genotypes and allele frequencies of adiponectin −11377 C > G SNP were similar between T2DM patients and their control group.
The genotype and allele frequencies of adiponectin −11391 G > A SNP in T1DM and T2DM patients and their corresponding control groups are demonstrated in Table 4. The distribution of the adiponectin − 11391G/A genotypes in T1DM (χ2 = 0.95, df = 1, P = 0.33) and in T2DM (χ2 = 0.45, df = 1, P = 0.5) patients were not considerably different from that of their corresponding control groups.
| Adiponectin genotypes | T1DM (n = 89) | Control T1DM (n = 66) | T2DM (n = 100) | Control T2DM (n = 95) |
|---|---|---|---|---|
| G/G | 79 (88.8%) | 55 (83.3%) | 84 (84%) | 83 (87.4%) |
| G/A | 10 (11.2%) | 11 (16.7%) | 16 (16%) | 12 (12.6%) |
| 0.64 (0.25–1.6, P = 0.33, n = 10) | (χ2 = 0.95, df = 1, P = 0.33) | 1.32 (0.6–2.9, P = 0.5, n = 16) | (χ2 = 0.45, df = 1, P = 0.5) | |
| 113391 G/A alleles | ||||
| G | n = 168 | n = 121 | n = 184 | n = 178 |
| A | 0.65 (0.27–1.6,p = 0.35, n = 10) | n = 11 | 1.29 (0.6–2.8, P = 0.52, n = 16) | n = 12 |
| (χ2 = 0.9, df = 1, P = 0.34) | (χ2 = 1.75, df = 1, P = 0.18) | |||
- Abbreviations: T1DM, type 1 diabetes mellitus patients; T2DM, type 2 diabetes mellitus patients.
To investigate the effect of interaction between adiponectin SNP at positions −11377 G allele and −11391 A allele on the development of T1DM and T2DM, we analyzed the haplotypes in T1DM, T2DM, and their corresponding control group by software haplotype analysis http://bioinfo.iconcologia.net/snpstats/start.htm, and the results are presented in Tables 5 and 6. We noticed that lack of A allele at −11391 position and the presence of G allele at the −11377 position of adiponectin gene increases the chances of T1DM development (OR, 3.9 [1.7-9]; P = 0.002). There is no obvious interaction between 11391 A and 11377 G alleles in T2DM and its control group.
| 11377 G | 11391 A | Group.Ca n(%) | Group.Co n(%) | OR (95% CI) | P value |
|---|---|---|---|---|---|
| C | G | 45 (50.6%) | 46 (69.7%) | - | - |
| G | G | 34 (38.2%) | 9 (13.8%) | 3.9 (1.7-9) | 0.002 |
| C | A | 8 (9%) | 8 (12.1%) | 1.02 (0.36-3) | 0.98 |
| G | A | 2 (2.2%) | 3 (4.5%) | 0.68 (0.1-4.3) | 0.68 |
- Abbreviations: CI, confidence interval; OR, odds ratio; T1DM, type 1 diabetes mellitus patient.
- Haplotype frequencies association with response (n = 155, crude analysis). Global haplotype association P value: (χ2 = 11.6, df = 3, P = 0.009).
| 11377 G | 11391 A | T2DM n(%) | Control T2DM n (%) | OR (95% CI) | P value |
|---|---|---|---|---|---|
| C | G | 72 (72%) | 73 (77.1%) | - | - |
| G | G | 20 (20%) | 15 (16.15%) | 1.35 (0.7- 2.3) | 0.34 |
| C | A | 7 (7%) | 7 (6.7%) | 1.01 (0.3- 2.9) | 0.5 |
| G | A | 1 (2.2%) | 0 (0%) | - | - |
- Abbreviations: T1DM, type 1 diabetes mellitus patients; T2DM, type 2 diabetes mellitus patients.
- Global haplotype association P value: (P = 0.51).
According to Table 7, a high level of LDL-C (110.2 ± 14.4 vs 85.7 ± 15.5, P = < 0.001) and a low level of HDL-C (42.9 ± 6.2 vs 48.9 ± 8.2, P = < 0.001) in T1DM patients were associated with −11377 CC genotype of adiponectin, whereas the elevated level of adiponectin and low LDL-C and TC serum concentrations in T1DM were associated with −11377 G/G + C/G genotypes.
| T1DM patients | Control T1DM | P values | |
|---|---|---|---|
| C/C | |||
| Adiponectin, μg/L | 23.9 ± 13 | 18.6 ± 11.2 | 0.26 |
| LDL-C, mg/dL | 85.7 ± 15.5 | 110.2 ± 14.4 | <0.001 |
| HDL-C, mg/dL | 48.9 ± 8.2 | 42.9 ± 6.2 | <0.001 |
| TC, mg/dL | 170 ± 26.8 | 174 ± 31 | 0.46 |
| TG, mg/dL | 113.5 ± 58 | 129 ± 55 | 0.16 |
| Waist circumstance, cm | 74.9 ± 11.3 | 79.2 ± 14.9 | 0.1 |
| G/G + C/G | |||
| Adiponectin, μg/L | 25 ± 18.2 | 13.2 ± 6.3 | 0.035 |
| LDL-C, mg/dL | 85.5 ± 18.6 | 107.7 ± 13.4 | <0.001 |
| HDL-C, mg/dL | 48.1 ± 9.6 | 44.8 ± 11.1 | 0.33 |
| TC, mg/dL | 161 ± 33 | 189 ± 31.3 | 0.014 |
| TG, mg/dL | 110 ± 41.5 | 131 ± 49.3 | 0.14 |
| Waist circumstance, cm | 77.7 ± 12.7 | 85.3 ± .18.8 | 0.11 |
- Abbreviations: LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; TC, total plasma cholesterol; TG, triglyceride
4 DISCUSSION
Various genetic polymorphisms in the adiponectin gene have been detected and their association with T2DM have been studied although these results are debatable.22-24 In this study, we assessed the association of adiponectin gene polymorphisms at −11391 G/A and −11377 C/G position, adiponectin level and serum lipid profile concentrations with the risk of T1DM and T2DM in the population from the Kermanshah province in the west of Iran. The results of the current study showed that the overall distribution of the adiponectin genotypes in T1DM patients was different from that of the control group. According to codominant, dominant, and over-codominant models of adiponectin gene at −11377 position, the presence of C/G vs C/C, C/G + G/G vs C/C, and C/G vs C/C + G/G genotypes significantly increases the risk of type I diabetes by 2.97, 3.6, and 2.9 folds, respectively. The G allele of adiponectin at position −11377 increased the risk of T1DM by 2.63-fold. Neither genotypes nor alleles of adiponectin −11377 C/G polymorphism were associated with T2DM.
Adiponectin can inhibit inflammatory reactions, thus an increased level of adiponectin as a result of a mutation at position −11377 of the gene leads to increased inflammation and the chance of developing type 1 diabetes. The current study suggested that the G allele of adiponectin at position −11377 is a good predictor of T1DM development. Contrary to our findings, Han et al25 showed that there is a direct association between the −11377 C/G adiponectin gene polymorphism and type 2 diabetes. It has been reported that −11377 G allele might be a risk factor for type 2 diabetes among white individuals. Choi et al reported that the −11377 G allele might be responsible for T2DM incidence among Korean population.26 In contrast, AlSaleh et al27 failed to demonstrate such an association in South Asian, Southeast Asian, Black, Middle Eastern, and Far Eastern populations.
We found that adiponectin −11391 G > A SNP was not significantly associated with susceptibility to either T2DM or T1DM. Contrary to our findings, the finding of Vasseur et al18 indicated that the adiponectin −11391 gene polymorphisms were significantly correlated with T2DM and adiponectin concentration in French Caucasian individuals. Olckers et al28 showed that the −11391 A/G heterozygote in Black South African subjects with type 2 diabetic patients had a protective effect on the T2DM. El-Shal et al29 indicated that −11391 GA or −11391 AA genotypes and A allele were significantly correlated with macroalbuminuria in T2DM patients.
In this study, we found that the presence of G allele at position −11377 and lack of A allele at positions −11391 of adiponectin gene increased the incidence of T1DM by 3.9 times (P = 0.002). In addition, a high level of adiponectin serum concentration in T1DM patients (25 ± 18.2) compared with control (13.2 ± 6.3, P = 0.035) was associated with the −11377 C/G dominant model (G/G + C/G vs C/C). In addition, the adiponectin level in T1DM patients was found to be positively correlated with HbA1C and fasting blood sugar, but it was negatively associated with BMI and duration of the diabetes. Evidence suggests that adiponectin is inversely associated with BMI, glucose, insulin and triglyceride levels, degree of insulin resistance, and visceral fat accumulation.30 We have also found a significant positive correlation between adiponectin concentration in T2DM patients with the onset age of diabetes and HDL-C level. Mohammadi et al31 suggested that improvement of biomarkers of insulin sensitivity, including adiponectin and lipid metabolism, have a crucial therapeutic role in obese patients with T2DM.
5 CONCLUSION
In summary, the frequencies of adiponectin −11377 C/G genotypes and alleles were considerably higher in T1DM patients than in healthy nondiabetic individuals and −11377 G allele significantly increased the risk of the disease by 2.6 times. Considerably increased risk of developing type I diabetes was observed in individuals with adiponectin −11377 C/G codominant, dominant and over-codominant genotypes. The adiponectin level was significantly higher in T1DM patients with −11377 G/G + C/G genotype whereas, LDL-C and TC concentrations were significantly higher in healthy individuals (control group) with the same genotype. A significant direct association was found between HbA1C and fasting blood sugar with adiponectin level in T1DM patients. A positive correlation was demonstrated between the adiponectin level and the age at the onset of diabetes and HDL-C in T2DM patients. The overall distribution of adiponectin genotypes at the −11391 position was not different between patients with T1DM and T2DM compared with their control groups.
ACKNOWLEDGMENTS
This study was performed in partial fulfillment of requirements for an MSc degree in the Clinical Biochemistry, Kermanshah University of Medical Sciences, Kermanshah, Iran (O. Hesami). This study was funded by Kermanshah University of Medical Sciences, Kermanshah, Iran (Grant #924304).
CONFLICTS OF INTEREST
There are no conflicts of interest.