miR-214-5p targets KLF5 and suppresses proliferation of human hepatocellular carcinoma cells
Jinzhong Pang and Zheng Li contributed equally to this study.
Abstract
MicroRNAs (miRNAs) are small endogenous conserved RNAs regulating genes expression through base pairing with the 3′-untranslated region (3′-UTR) of target messenger RNAs. MiR-214-5p is a newly identified miRNA with its biological role largely unknown. In this study, we explored miR-214-5p expression status in 78 paired tumor and nontumor tissues obtained from patients with hepatocellular carcinoma (HCC) by RT-qPCR. The effects of miR-214-5p expression on HCC cell proliferation, cell cycle progression, and cell migration were measured by CCK-8 assay, flow cytometry, and wound-healing assay. A dual-luciferase activity assay was performed to identify whether KLF5 was a target of miR-214-5p. Kaplan-Meier curve and log-rank test were used to investigate the effects of miR-214-5p and KLF5 on overall survival and disease-free survival of patients with HCC. We found miR-214-5p expression was sharply reduced in HCC tissues and cell lines compared with the normal tissues and cell lines. Functional assay revealed that miR-214-5p overexpression could downregulate cell proliferation, cell migration, and arrested cell cycle at G0/G1 phase. Further, we validated Krüppel-like factor 5 (KLF5) as a direct target of miR-214-5p, and was upregulated in HCC and inversely correlated with the expression of miR-214-5p. Moreover, we found the low expression of miR-214-5p and high expression of KLF5 were correlated with tumor size, tumor stage, and poorer 5-year overall survival and disease-free survival of patients with HCC. In conclusion, our results suggested miR-214-5p functions as a tumor suppressor through targeting KLF5 in HCC. Also, miR-214-5p and KLF5 were identified as potential prognostic markers and might be therapeutic targets in HCC.
1 INTRODUCTION
Hepatocellular carcinoma (HCC), with more than half a million new cases diagnosed annually, is the fifth most common type of cancer worldwide and the third leading cause of cancer-related deaths.1, 2 The treatment and diagnoses methods for HCC have improved over the past decades, but the 5-year overall survival remains quite low.3-6 Therefore, an increased understanding of HCC is very important to improve patients’ prognosis.
Krüppel-like factor 5 (KLF5) is a member of the Krüppel-like factor (KLF) family and plays crucial roles in various cellular processes including cell proliferation, apoptosis, migration, and differentiation.7-9 KLF5 consists a conserved zinc finger domain and is capable to regulate the expression of several downstream target genes, such as cyclin D1, cyclin B1, fibroblast growth factor binding protein 1, SOX4, Hypoxia-inducible factor 1 α, epithelial growth factor receptor, tumor necrosis factor-α (TNF-α)-induced protein 2, and survivin.10-18 Besides that, KLF5 expression can be regulated by miRNAs or posttranslation modification.19-22 In tumorigenesis, KLF5 has dual functions, being tumor suppressing gene and tumor promoting gene.23-26 Meanwhile, recent study revealed that KLF5 acetylation is crucial for the transition from tumor suppressor to tumor promoter.27 As for the mechanisms of how KLF5 promotes tumorigenesis, there were studies demonstrated that KLF5 overexpression increased cell cycle transition while silencing KLF5 expression decreased vascular endothelial growth factor A (VEGFA) expression in human bladder cancer.28, 29
MicroRNAs (miRNAs) are small, noncoding RNA molecular that regulate gene expression by inhibiting mRNA translation or stimulating mRNA degradation through binding to the 3′-untranslated region (3′-UTR) of its target genes.30 miR-214-5p is a product of the 110 bp miR-214 gene in the intron of the Dynamin-3 gene on human Chromosome 1-NC_000001.10.31 Very recently, Zhang et al demonstrated that miR-214-5p was downregulated and regulates cell proliferation and invasion by targeting ROCK1 in human osteosarcoma.32 However, the expression and clinical significance of miR-214-5p in HCC has not yet been investigated. Therefore, in this study, we measured the expression of miR-214-5p in HCC to investigate whether or not miR-214-5p has a role in the HCC progression process. The bioinformatic analysis and luciferase reporter assay were conducted to validate whether or not KLF5 was a direct target of miR-214-5p. Further, the clinical significance of miR-214-5p and KLF5 in HCC was investigated.
2 MATERIALS AND METHODS
2.1 HCC tissue collection
This study was approved by the Ethics Review Committee of The Affiliated Hospital of Qingdao University (West Coast District) (Qingdao, China), and written consent form was obtained from all the patients. A total of 78 patients who underwent surgery at Qingdao West Coast New Area Central Hospital between March 2010 and January 2012 were recruited. All patients enrolled did not receive any anticancer treatment before surgery. The tumor tissues and matched normal tissues were immediately frozen in liquid nitrogen after surgical resection and preserved at −80°C until further use. Clinical stage for HCC was evaluated based on the seventh edition of the UICC/AJCC TNM Staging System. The clinicopathological features were collected and summarized in Table 1.
| miR-214-5p expression | KLF5 expression | ||||||
|---|---|---|---|---|---|---|---|
| Features | Total | Low | High | P value | Low | High | P value |
| Age (y) | |||||||
| ≥60 | 38 | 22 | 16 | 0.513 | 20 | 18 | 0.114 |
| <60 | 40 | 25 | 15 | 16 | 24 | ||
| Gender | |||||||
| Male | 41 | 24 | 17 | 0.407 | 19 | 22 | 0.509 |
| Female | 37 | 23 | 14 | 17 | 20 | ||
| Tumor size (cm) | |||||||
| ≥5 | 35 | 18 | 17 | 0.028 | 13 | 22 | 0.033 |
| <5 | 43 | 29 | 14 | 23 | 20 | ||
| Tumor stage | |||||||
| I-II | 36 | 18 | 18 | 0.014 | 13 | 23 | 0.023 |
| III | 42 | 29 | 13 | 23 | 19 | ||
- Abbreviations: HCC, Hepatocellular carcinoma; KLF5, Krüppel-like factor 5; miR-214-5p, microRNA-214-5p.
2.2 Follow-up survey
After surgery, all patients were followed up and the overall survival time (OS) and disease-free survival time were calculated. OS was defined as the time between the date of surgery and the patient's death from HCC. Disease-free survival was defined as the time between the date of surgery and recurrence.
2.3 Cell culture and transfection
The HCC cell lines (Hep3B and Huh7) and the normal liver cell line (HL-7702) were obtained from Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cell lines were maintained in Dulbecco modified Eagle medium (DMEM, Thermo Fisher Scientific, Inc., Waltham, MA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) in a humidified incubator at 37°C containing 5% of CO2.
Synthetic mimic (5′-UGCCUGUCUACACUUGCUGUGC-3′) or inhibitor (5′-GCACAGCAAGUGUAGACAGGCA-3′) of miR-214-5p, and negative control (NC) miRNA (5′-GUGAGAAGUACCACCGAGACAG-3′) were purchased from Ribobio (Guangzhou, China). Synthetic small interfering RNA (siRNA) targeting KLF5 (5′-GCUCCAGAGGUGAACAAUA-3′) and NC siRNA (5′-AGCGACUCAAGAUUACGAG-3′) were also purchased from Ribobio (Guangzhou). For transfection, the cells were cultured in a 6-well plate at a density of 3 × 105 cells/well, and growth to 70% to 80% confluence. Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) was used for synthetic siRNAs or miRNAs transfection according to the manufacture's recommendations. Briefly, the miRNAs or siRNAs and Lipofectamine 2000 were added into Eppendorf tubes containing 50 µL DMEM. Following, the mixture was added to well after incubating for 20 minutes at room temperature. Following transfection for 48 hours, the cells were collected to detect the expressions of miR-214-5p or KLF5. Transfection efficiency was confirmed by quantitative real-time polymerase chain reaction (RT-qPCR) or Western blot.
2.4 Cell proliferation assay
The cell proliferation rate was assessed by a Cell Counting Kit 8 assay (CCK-8, Beyotime, Haimen, Jiangsu, China) following the supplier's instructions. Briefly, the cells were plated in a 96-well plate at a density of 2 × 103 cells/well. Following, 10 μL CCK-8 solution was added to each well at indicated time points (0 hours, 48 hours, and 72 hours) and incubated for another 4 hours at 37°C. The optical absorbance of each well was measured at 450 nm using ELISA plate reader (Bio-tek, Winooski, VT). The experiments were repeated in triplicate.
2.5 Cell cycle analysis
For cell cycle analysis, the cells were treated with trypsin and fixed using 80% ethanol at 4°C for overnight. The cells were then washed with cold phosphate-buffered saline (PBS) and stained with propidium iodide/RNase A mixture (Beyotime) at darkness for 30 minutes. Following, cell cycle analysis was conducted using FACSCalibur™ flow cytometer (BD Biosciences, Franklin Lakes, NJ). A total of 20 000 cells were acquired and analyzed for DNA content. The results were analyzed by ModFit 3.2 software (Verity Software House Company, Topsham, ME). Histograms represent the percentage of cells in each phase of the cell cycle (G0/G1, S, and G2/M). The experiments were repeated in triplicate.
2.6 Cell migration analysis
Wound-healing assay was used to evaluate the migratory ability of the cultured cells. Scratch was created using a sterile micropipette tip in the cell surface. The detached cells were removed by washing the cell monolayer using PBS. The initial (0 hours) and the residual gap length of 24 hours after wounding were calculated from photomicrographs. The experiments were repeated in triplicate.
2.7 RNA extraction and RT-qPCR
Total RNA was extracted from all the fresh tissues with an RNAeasy tissue MiniKit (Qiagen Inc., Nordrhein-Westfalen, Germany) according to the manufacture's recommendations. The concentration of extracted RNA was measured using Nanodrop-ND-1000 (Thermo Fisher Scientific Inc.). For miRNA expression, reverse transcription was performed using a One Step PrimeScript RT Reagent Kit (Takara Bio., Inc., Dalian, China), followed by RT-qPCR with Takara SYBR Master Mix (Takara Bio., Inc.) at an ABI 7200 Real-Time PCR System (Applied Biosystems, Life Technologies GmbH, Darmstadt, Germany). PCR conditions included an initial holding period at 95°C for 10 minutes and for 1 cycle, then 95°C for 15 seconds and 60°C for 30 seconds for 40 cycles. The primer sequences used in this study were as follows: miR-214-5p forward, 5′-ACACTCCAGCTGGGTGCCTGTCTACACTTG-3′, and miR-214-5p reverse, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCACAGCA-3′; U6 snRNA forward, 5′-GCGCGTCGTGAAGCGTTC-3′, and U6 snRNA reverse, 5′-GTGCAGGGTCCGAGGT-3′. Differences in gene expression were calculated using the 2−ΔΔCq method18 and expressed as a fold-change. Each experiment was repeated in triplicate.
2.8 Protein extraction and Western blot
All the fresh tissues or cell lines were lysed with lysis buffer (150 mm NaCl, 50 mm Tris-HCl, 1% NP-40, pH 7.5; Beyotime Biotechnology). Protein samples were quantified using a Bradford Protein Concentration Determination Kit (Beyotime Biotechnology) according to the supplier's instructions. Protein samples were separated at a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and subsequently transfected onto nitrocellulose membranes (CWBiotech, Beijing, China). Following blocked with 10% non-fat milk in PBS, membranes were incubated with primary antibodies at 4°C overnight (anti-KLF5: ab24331, 1:2000, Abcam, Cambridge, MA or anti-GAPDH: ab181602, 1:1000, Abcam) independently. Subsequently, the membranes were washed with TBST for three times and then incubated with secondary antibody (sc-2491, 1:1500, Santa Cruz Biotechnology, Inc., Dallas, TX) for 3 hours at 25°C. The signals were detected using a BeyoECL Plus Kit (Beyotime). Protein expression levels were normalized to total GAPDH expression.
2.9 Immunohistochemistry (IHC)
HCC tumor tissues and matched normal tissues were previously formalin-fixed and paraffin-embedded were sliced into 4 μm sections and underwent deparaffination and then rehydration. Antigen retrieval, suppression of endogenous peroxidase activity, and 10% skim milk blocking were performed before primary antibody incubation. An anti-KLF5 (ab24331, Abcam) antibody was used as a primary antibody and incubated with the sections overnight at 4°C. The section was subsequently incubated with peroxidase conjugated secondary antibody (ab6721, Abcam) for 90 minutes, and DAB solution was used for signal development for 5 minutes. The sections were counterstained with hematoxylin followed by dehydrating and mounting.
2.10 Luciferase reporter assay
Both the wild-type (WT) and mutant (Mut) 3′-UTR of KLF5 containing the putative binding sequence of miR-214-5p were inserted into the pmiRGLO dual-luciferase expression vector (Promega, Madison, WI). The renilla luciferase expression vector was used as an internal control. The cells were transfected with miR-214-5p mimic or NC miRNA and luciferase reporter constructs (WT or Mut) using lipofectamine 2000 (Thermo Fisher Scientific, Inc.). 48 hours post transfection, the luciferase activity was measured using the dual-luciferase reporter assay system (Promega).
2.11 Statistical analysis
Data were presented as mean ± SD. Graphpad Prism version 5.0 software (GraphPad, San Diego, CA) was used for data analysis. The corrections between miR-214-5p or KLF5 expression and clinicopathological features were assessed by χ2 test. Student t test was conducted to analyze the difference between two groups. One-way analysis of variance and Tukey test were performed to compare the difference among three or above groups. Kaplan-Meier curve and log-rank test were used for survival analysis. Cox proportional hazards regression model was used for multivariate survival analysis to assess prognostic factors that were significant in the univariate analysis. P-value <0.05 was considered to be statistically significant.
3 RESULTS
3.1 The general characteristics of patients in this study
The clinical characteristics of the 78 patients with HCC were summarized in Table 1. The average age of patients with HCC was 58.6 ± 9.9 years (range, 33-80 years). The number of male patients was 41, while that of female was 37. The clinicopathological features used in this study includes age, gender, tumor size, and tumor stage.
3.2 Expression of miR-214-5p in HCC tissues and cell lines
The expression of miR-214-5p in 78 pairs HCC tissues and HCC cell lines was examined by RT-qPCR. We found miR-214-5p downregulation was occurred in 60.3% (47/78) of HCC tissues when compared with matched normal tissues (>2 folds decrease, Figure 1A). Therefore, these patients were classified into low miR-214-5p expression group. The rests of the enrolled patients were classified into high miR-214-5p expression group accordingly. Moreover, we found the expression level of miR-214-5p was also significantly downregulated in HCC cell lines than in normal liver cell line (P < 0.05, Figure 1B).
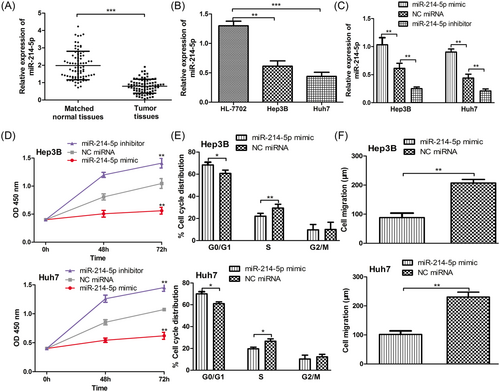
miR-214-5p was frequently downregulated in HCC and its downregulation promotes cell proliferation, cell cycle progression, and cell migration. A, RT-qPCR to measure the expression of miR-214-5p in HCC tumor tissues and matched normal tissues; B, RT-qPCR to measure the expression of miR-214-5p in HCC cell lines (Hep3B and Huh7) and the normal liver cell line (HL-7702); C, RT-qPCR to measure the expression of miR-214-5p in HCC cell lines (Hep3B and Huh7) post miR-214-5p mimic, inhibitor, and NC miRNA transfection; D, CCK-8 assay to measure cell proliferation rate in HCC cell lines (Hep3B and Huh7) post miR-214-5p mimic, inhibitor, and NC miRNA transfection; E, Flow cytometry to measure cell cycle distribution in HCC cell lines (Hep3B and Huh7) post miR-214-5p mimic and NC miRNA transfection; F, Wound-healing assay to measure cell migration in HCC cell lines (Hep3B and Huh7) post miR-214-5p mimic and NC miRNA transfection. (***P < 0.001; **P < 0.01; *P < 0.05). HCC, hepatocellular carcinoma; NC, negative control; miR-214-5p, microRNA-214-5p; RT-qPCR, quantitative real-time PCR
3.3 Effect of miR-214-5p expression on cell proliferation, cell cycle, and cell migration
Following, we investigated the biological role of miR-214-5p since it was downregulated in HCC. The miR-214-5p mimic, inhibitor, and NC miRNA were transfected into HCC cell lines (Hep3B and Huh7) to manipulate miR-214-5p expression. RT-qPCR analysis showed that miR-214-5p expression was increased by miR-214-5p mimic but decreased by miR-214-5p inhibitor transfection (P < 0.05, Figure 1C). CCK-8 analysis demonstrated that miR-214-5p overexpression inhibited the cell proliferation of HCC cell lines, whereas miR-214-5p downregulation promoted cell proliferation (Figure 1D). Furthermore, we measured the cell cycle distribution by flow cytometry. The results indicated miR-214-5p mimic transfection increased the HCC cells at G0/G1 phase but decreased the cells at the S phase compared with NC miRNA transfected counterpart (Figure 1E). Importantly, we examined the cell migration using the wound-healing assay and we revealed that miR-214-5p mimic reduced the cell migration of HCC cell lines compared with the NC miRNA transfected counterpart (Figure 1F). These results indicate that miR-214-5p may inhibit cell proliferation, arrest cell cycle at G0/G1 phase, and inhibit cell migration.
3.4 KLF5 was a direct target of miR-214-5p in HCC
To understand the underlying mechanism of miR-214-5p in HCC, we searched for the potential targets of miR-214-5p using TargetScan. The results implied that KLF5 might be a possible target of miR-214-5p since it contains a putative binding sequence for miR-214-5p in its 3′-UTR (Figure 2A). To confirm this prediction, the dual-luciferase assay was conducted. As presented in Figure 2B, miR-214-5p mimic suppressed the relative luciferase activity of cells containing WT 3′-UTR of KLF5 construct but did not alter that of cells containing Mut 3′-UTR of KLF5 construct. To further validate KLF5 was a direct target of miR-214-5p, we measured KLF5 expression in miR-214-5p mimic or NC miRNA transfected cells. As expected, the protein expression of KLF5 was decreased by miR-214-5p overexpression (Figure 2C), which implied KLF5 was a direct target of miR-214-5p in HCC. Moreover, we investigated the expression KLF5 in HCC tissues and we found KLF5 expression was upregulated in 53.8% (42/78) of HCC tissues when compared with matched normal tissues (Figure 2D). Also, we examined the expression of KLF5 and miR-214-5p in HCC tissues and found they were inversely correlated (Figure 2E). Furthermore, the hyperexpression status of KLF5 in HCC tumor tissues was confirmed by IHC (Figure 2F).
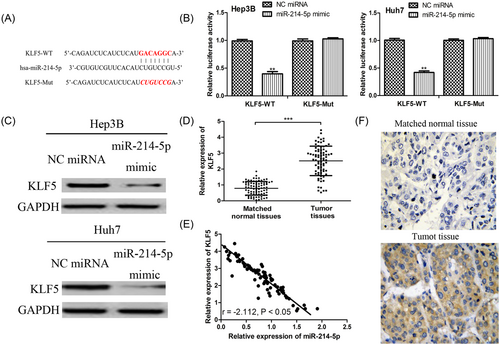
KLF5 was a direct target of miR-214-5p. A, The putative binding sequence for miR-214-5p in the 3′-UTR of KLF5; B, Luciferase activity assay revealed miR-214-5p mimic suppressed KLF5 3′-UTR WT luciferase activity, while it had no effect on KLF5 3′-UTR Mut luciferase activity in HCC cell lines (Hep3B and Huh7); C, Western blot to measure the protein expression of KLF5 in HCC cell lines (Hep3B and Huh7) post miR-214-5p mimic and NC miRNA transfection; D, Western blot to measure the expression of KLF5 in HCC tumor tissues and matched normal tissues; E, The expression of KLF5 was inversely correlated with the expression of miR-214-5p in HCC; F, IHC analysis of KLF5 expression in HCC tumor tissue and matched normal tissue. Magnification 200×. (***P < 0.001; **P < 0.01). HCC: hepatocellular carcinoma; IHC, immunohistochemistry; KLF5, Krüppel-like factor 5; miR-214-5p, microRNA-214-5p; Mut, mutant; NC, negative control; UTR, untranslated region; WT, wild-type
3.5 Involvement of KLF5 in HCC cell proliferation, cell cycle progression, and cell migration processes mediated by miR-214-5p
To further investigate whether KLF5 was a mediator for the tumor suppressive role of miR-214-5p in HCC, we performed loss-of-function studies. As shown in Figure 3A, the transfection of siRNA targeting KLF5 decreased the protein expression of KLF5. Meanwhile, we found knockdown of KLF5 resulted in cell proliferation inhibition and cell cycle arrested in G0/G1 phase (Figure 3B,C). Moreover, we found the knockdown of KLF5 could abolish the effect of miR-214-5p downregulation on cell proliferation and cell cycle progression (Figure 3B,C). Notably, we also found the si-KLF5 inhibited cell migration and partially reversed the effect of miR-214-5p inhibitor on cell migration (Figure 3D).
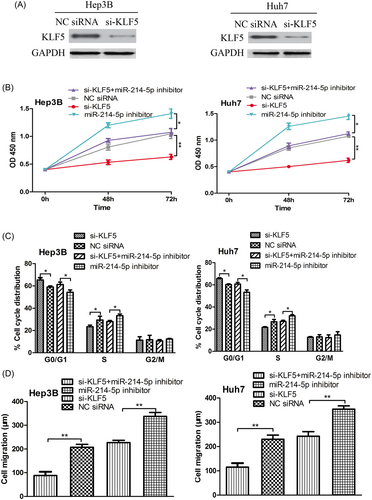
Knockdown expression of KLF5 inhibits cell proliferation, cell cycle progression, and cell migration. A, Western blot to measure the protein expression of KLF5 in HCC cell lines (Hep3B and Huh7) post si-KLF5 and NC siRNA transfection; B, CCK-8 assay to measure cell proliferation rate in HCC cell lines (Hep3B and Huh7) post si-KLF5 and NC siRNA transfection; C, Flow cytometry to measure cell cycle distribution in HCC cell lines (Hep3B and Huh7) post si-KLF5 and NC siRNA transfection; D, The wound-healing assay to measure cell migration in HCC cell lines (Hep3B and Huh7) post miR-214-5p inhibitor, si-KLF5, NC siRNA transfection or miR-214-5p inhibitor, and si-KLF5 cotransfection. (**P < 0.01; *P < 0.05). HCC, hepatocellular carcinoma; KLF5, Krüppel-like factor 5; miR-214-5p, microRNA-214-5p; NC, negative control; si-KLF5, siRNA targeting KLF5; siRNA, small interfering RNA
3.6 Clinical significance of miR-214-5p and KLF5 expression in HCC
To further investigate the clinical significance of miR-214-5p and KLF5 expression in HCC, we investigated the correlations between miR-214-5p or KLF5 expression with the clinicopathological variables. As summarized in Table 1, the results indicated low miR-214-5p expression was correlated with tumor size (P = 0.028) and tumor stage (P = 0.014). Meanwhile, we also found the high KLF5 expression was correlated with the tumor size (P = 0.033) and tumor stage (P = 0.023). However, the correlations of miR-214-5p or KLF5 expression with gender and age were not statistically significant (all P > 0.05, Table 1).
Furthermore, the patients’ 5-year OS time were collected and analyzed by Kaplan-Meier curve and log-rank test. We found patients with low miR-214-5p expression (P < 0.05, Figure 4A) or high KLF5 expression (P < 0.05, Figure 4B) tend to had a poorer 5-year OS time and disease-free survival time. Univariate analysis showed that variables including miR-214-5p expression (P = 0.033), KLF5 expression (P = 0.021), tumor size (P = 0.032), and tumor stage (P = 0.031) had significantly prognostic influences on OS and disease-free survival time (Tables 2 and 3). However, variables including gender and age did not have significantly influence on OS and disease-free survival time (all P > 0.05, Tables 2 and 3). Moreover, multivariate analysis using Cox proportional hazard model to analyze the variables that were significant in the univariate analyses revealed that miR-214-5p expression (P = 0.026) and KLF5 expression (P = 0.019) were independent prognostic factors for OS, along with tumor size (P = 0.036) and tumor stage (P = 0.016, Table 2). Furthermore, we found miR-214-5p expression (P = 0.021) and KLF5 expression (P = 0.029) were also independent prognostic factors for disease-free survival, along with tumor size (P = 0.024) and tumor stage (P = 0.022, Table 3).
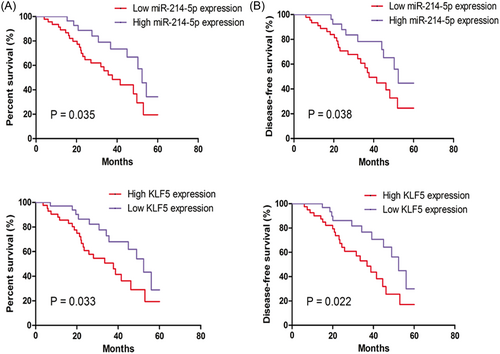
Correlations between miR-214-5p or KLF5 expression with the 5-year overall survival and disease-free survival of patients with HCC. A, Low expression of miR-214-5p; or B, High expression of KLF5 predicts poorer 5-year overall survival and disease-free survival of patients with HCC. HCC, hepatocellular carcinoma; KLF5, Krüppel-like factor 5; miR-214-5p, microRNA-214-5p
| Variables | Univariate analysis | P value | Multivariate analysis | P value | ||
|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |||
| miR-214-5p | 2.075 | 1.062-4.053 | 0.033 | 2.275 | 1.102-4.698 | 0.026 |
| KLF5 | 2.265 | 1.135-4.522 | 0.021 | 2.262 | 1.146-4.464 | 0.019 |
| Age | 1.910 | 0.875-4.167 | 0.104 | – | – | – |
| Gender | 1.822 | 0.822-4.038 | 0.140 | – | – | – |
| Tumor size (cm) | 2.079 | 1.064-4.063 | 0.032 | 2.065 | 1.045-4.079 | 0.036 |
| Tumor stage | 2.161 | 1.071-4.361 | 0.031 | 2.178 | 1.156-4.105 | 0.016 |
- Abbreviations: CI, confidence index; HR, hazard ratio; miR-214-5p, microRNA-214-5p.
| Variables | Univariate analysis | P value | Multivariate analysis | P value | ||
|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |||
| miR-214-5p | 2.025 | 1.044-3.929 | 0.037 | 2.255 | 1.130-4.501 | 0.021 |
| KLF5 | 1.950 | 1.033-3.680 | 0.039 | 2.139 | 1.081-4.235 | 0.029 |
| Age | 1.819 | 0.921-3.595 | 0.085 | – | – | – |
| Gender | 1.696 | 0.851-3.382 | 0.133 | – | – | – |
| Tumor size (cm) | 2.058 | 1.073-3.949 | 0.030 | 2.099 | 1.102-3.995 | 0.024 |
| Tumor stage | 1.960 | 1.038-3.699 | 0.038 | 2.246 | 1.126-4.482 | 0.022 |
- Abbreviations: CI, confidence index; HR, hazard ratio; miR-214-5p, microRNA-214-5p.
4 DISCUSSIONS
The development and progression of cancers require several key steps including alteration of cell proliferation, survival, metastasis, and evasion of apoptosis.13, 14 Increasing numbers of studies have demonstrated KLF5 functions as an oncogenic transcription factor in several cancer types, including bladder, breast, lung, and cervical.10, 13, 14, 17, 33 Besides that, high expression of KLF5 was reported could promote cell proliferation and migration while the knockdown of KLF5 expression could cause the opposite effects.33 Since KLF5 expression could be regulated by miRNAs10, 18, 19, 22 and the expression and role of miR-214-5p in HCC remains unknown, therefore, it is attractive for us to investigate the correlation between miR-214-5p and KLF5.
In this current study, we first examined the expression level of miR-214-5p in HCC tissues and cell lines. We found miR-214-5p expression was significantly lower in HCC tissues when compared with that in matched normal tissues, which was the same as the expression status of miR-214-5p in human osteosarcoma.32 Also, we found miR-214-5p overexpression could inhibit cell proliferation, migration, and induce cell cycle arrested in G0/G1, indicating the miR-214-5p may play a tumor suppressor role in the development and progression of HCC. Following, we identified the targeting gene of miR-214-5p in regulating cell proliferation, cell migration, and cell cycle since it is widely recognized that miRNA exerts its biological function through regulating the expression of its targets.30, 32 Therefore, by bioinformatic analysis and luciferase activity assay, we demonstrated KLF5 might be a potential target of miR-214-5p in HCC. Furthermore, we found the expression of KLF5 was inversely correlated with the expression of miR-214-5p, which provide new evidence that KLF5 was a direct target of miR-214-5p. Moreover, we revealed KLF5 is an important functional mediator for the exertion of the antitumor role of miR-214-5p.
To further elucidate the clinical significance of miR-214-5p and KLF5 in HCC, we analyzed the correlation between miR-214-5p or KLF5 expression and clinicopathological features. We demonstrated that patients with low miR-214-5p or high KLF5 expression was significantly related to tumor size and tumor stage. Furthermore, the results of Kaplan-Meier curve indicated that patients with low miR-214-5p or high KLF5 expression showed poorer OS and disease-free survival compared with those with high miR-214-5p or low KLF5 expression. The multivariate analysis using Cox hazard model suggested that miR-214-5p and KLF5 expression were independent predictors for the poor overall survival and disease-free survival of patients with HCC, along with the tumor size and tumor stage. These results suggested that low miR-214 and high KLF5 expression had an adverse outcome in HCC. Therefore, miR-214-5p and KLF5 might be used as prognostic markers for prognosis of patients with HCC.
At the molecular level, previous studies have identified several target genes of KLF5 in several tumor cell lines,10, 12-18 which helped us to better understand the role of KLF5 in tumor. Here, we found a link between miR-214-5p and KLF5 in HCC but we did not further investigate the downstream targets of KLF5 and we have to admit this is a limitation of this study. More efforts are needed to perfecting the miR-214-5p/KLF5 axis.
Taken together, our current study for the first time revealed miR-214-5p was downregulated in HCC. Also, we found low miR-214-5p expression and high KLF5 expression were valuable indicators to predict the prognosis of HCC.
CONFLICTS OF INTEREST
The authors declare that there are no competing interests associated with this manuscript.