Developing DNA methylation-based prognostic biomarkers of acute myeloid leukemia
Chundi Gao have made a significant contribution to this work and should be considered first author.
Abstract
Acute myeloid leukemia (AML) is a heterogeneous clonal neoplasm characterized by complex genomic alterations. The incidence of AML increases with age, and most cases experience serious illness and poor prognosis. To explore the relationship between abnormal DNA methylation and the occurrence and development of AML based on the Gene Expression Database (GEO), this study extracted data related to methylation in AML and identified a methylated CpG site that was significantly different in terms of expression and distribution between the primary cells of AML patients, and hematopoietic stem/progenitor cells from normal bone marrow. To further investigate the differences caused by the dysfunction of methylation sites, bioinformatics analysis was used to screen methylation-related biomarkers, and the potential prognostic genes were selected by univariate and multivariate Cox proportional hazards regressions. Finally, five independent prognostic indicators were identified. In addition, these results provide new insight into the molecular mechanisms of methylation.
Abbreviations
-
- AML
-
- Acute myeloid leukemia
-
- PPI
-
- Protein-protein interaction network
1 INTRODUCTION
Acute myeloid leukemia (AML) is a type of malignant clonal disease derived from hematopoietic stem and progenitor cells. It has a high degree of heterogeneity in terms of cytomorphology, molecular biology, cytogenetics, immunophenotyping, and clinical manifestations, reflecting the complexities of myeloid cell differentiation. It is characterized by the accumulation of immature myeloid cells in the bone marrow that results in the dysfunction of hematopoiesis.1 On average, 4 in 100 000 individuals will develop AML, with a median age of 67 years at diagnosis, and the incidence of this disease increases with age.2, 3 Epidemiological data showed that AML has the highest mortality rate of all the different types of leukemia. The French-American-British cooperative group identified eight different subcategories of AML, M0-M7, based on the specific cell population from which AML arises.4 To further reflect the origin and nature of AML cells, MICR typing was developed based on immunology (I), cytogenetics (C), molecular biology (M), and morphology (M) to make typing more accurate and to improve the diagnostic conformance rate. Chemotherapy and hematopoietic stem cell transplantation, two standard treatments of AML, improve survival to a certain extent. However, the 5-year survival rate remains below 50%, because of chemoresistance or the toxicity of the treatments.5, 6 Most patients eventually succumb to relapsed and/or progressive disease.7 Therefore, to decrease mortality and improve the management of AML, new early diagnostic biomarkers and therapeutic targets are urgently needed to improve the detection and risk stratification of AML.
With the deepening of research in the field of genomics, the influence of epigenetic changes on diseases has gradually attracted the attention of researchers. Aberrant DNA methylation is the most common and important modification in epigenetics, and abnormal changes are considered to be the main factors leading to the development of many tumors.8 Numerous studies have suggested that the altered DNA methylation patterns in tumor tissues may silence tumor suppressor genes and activated oncogenes through hyper/hypomethylation.9, 10 In addition, DNA methylation can cause changes in chromatin structure, DNA conformation, DNA stability, and DNA and protein interactions, thus controlling gene expression. Studies have found that DNA methylation changes occur early in carcinogenesis, and therefore can be used as promising biomarkers for the early detection of cancer.11, 12 Currently, numerous DNA methylation-based biomarkers have been identified in several types of cancers, including gastric cancer and lung cancer.13, 14
The Infinium Human Methylation 450K BeadChip enables the detection of 450 000 human methylation sites in the human genome. To further explore the relationship between abnormal DNA methylation and the occurrence and development of AML based on the Gene Expression Database (GEO), this study extracted the data related to methylation of AML, the gene expression profile GSE63409,15 and identified a methylated CpG site that was significantly different in terms of expression and distribution between the primary cells of patients with AML and hematopoietic stem/progenitor cells from normal bone marrow. To further study the differences caused by methylation site dysfunction based on wANNOVAR software, which provides easy and intuitive web-based access to the most popular functionalities of the ANNOVAR software, we annotated the methylation regions that were different in AML cells, obtained the related genes, conducted a bioinformatics analysis and univariate and multivariate Cox proportional hazards regressions, and screened for biomarkers associated with methylation to help us predict the prognosis of patients with AML. Moreover, these results could provide new insight into the molecular mechanism of AML based on methylation.
2 MATERIALS AND METHODS
2.1 Patient datasets
Gene expression profiles (GSE63409) associated with methylation in patients with AML were obtained from the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/). The profiles were obtained from 15 patients with AML and normal hematopoietic stem/progenitor cells from five normal bone marrow donors. R software and related R packages were used to normalize and analyze the downloaded data to obtain differentially methylated sites and regions. The GEO is a public functional genomics data repository, and tools are provided to help users query and download experiments, and curated gene expression profiles.16 The relevant data that provide are publicly available and open-ended; therefore, approval by a local ethics committee was not needed.
2.2 Gene annotation and functional enrichment analysis
The wANNOVAR database is a rapid and efficient tool that provides easy and intuitive web-based access to the most popular functionalities of the ANNOVAR software to annotate the functional consequences of genetic variation in high-throughput sequencing data detected from diverse genomes.17, 18 Using wANNOVAR software, we annotated the methylation regions that were different in AML and obtained the genes corresponding to the differentially methylated regions. To better understand the functional abnormalities caused by these differentially methylated regions, the Database for Annotation, Visualization and Integrated Discovery, DAVID (https://david.ncifcrf.gov/)19 was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of those genes. GO terms and KEGG pathways with p < 0.05 were selected.
2.3 PPI network construction and central gene screening
STRING (http://string-db.org) was used as a search tool for the retrieval of interacting genes to assess the protein-protein interaction (PPI) information.20 To further examine the potential association between these differentially methylated genes, we used STRING to characterize the PPI network of the genes, and the confidence score > 0.4 was set as the cutoff criterion. Then, we used Cytoscape software to visualize the resulting PPI network.21 Among the 345 nodes, 31 central node genes were selected based on connectivity of ≥ 10. The application Molecular Complex Detection (MCODE) was used to select for further biological analysis significant modules from the PPI network in Cytoscape_v3.5.1 with the MCODE number of nodes >10 as the standard.
2.4 Survival analysis and Cox regression, ROC curve
To verify and screen whether 31 key genes have different expression in patients with AML, the expression of genes and the corresponding clinical information of 185 AML samples and 2 normal samples were obtained from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/), which was imputed on an Illumina HiSeq RNA-Seq platform. Difference analysis found that 31 key genes were differentially expressed in AML samples. The differential expression data of those genes and sample-related survival time were obtained. A univariate Cox model was used to calculate the association between the expression level of each center of nodes and patient overall survival (OS). When the p values were <0.05, those nodes were considered to be statistically significant in the univariate Cox analysis. Next, multivariate Cox analysis was used to evaluate the contribution of these statistically significant node genes as independent prognosis factors of patient survival. The central node genes-based prognosis risk score was established based on a linear combination of the expression level multiple regression model (β) with the following formula. The Prognosis Index = (β × expression level of ESR1) +(β × expression level of CD44) + (β × expression level of TLR9) + (β × expression level of DRD4) + (β × expression level of MPO).
We constructed a prognostic signature by integrating the expression profiles of five genes and the corresponding estimated regression coefficient. Then, we calculated a risk score for each patient and ranked them in increasing order. According to the median risk score, 187 samples were classified into a high-risk group (n = 93) and a low-risk group (n = 94) to construct five biomarker prognosis models. Survival analysis was performed using the Kaplan-Meier method with a log-rank statistical test. The accuracy of prognostic performance was evaluated using time-dependent receiver operating characteristic (ROC) curves within 5 years by comparing the sensitivity and specificity of the survival prediction based on the risk score. All reported p values were two sided. All analyses were performed using R/BioConductor (version 3.4.2).
3 RESULTS
3.1 Identification of differentially methylated regions and region annotations in AML
The gene chip GSE63409 was used with the related methylation data of 15 primary patients with AML and hematopoietic stem/progenitor cells from five normal bone marrow samples. Differentially methylated data were analyzed in R with related R packages. In total, 1015 differentially methylated regions were obtained with < 0.001 as the cutoff condition. To further analyze the differences in functional enrichment due to differentially methylated sites and regions, a differentially methylated region of AML was annotated based on the web access provided by wANNOVAR software, and 1004 related genes are corresponding to differentially methylated regions were obtained. These genes were used for further functional and pathway enrichment analyses to explore functional abnormalities caused by differential methylation in normal and diseased patients.
3.2 GO function and KEGG pathway enrichment analyses
To further understand the functional role of differentially methylated sites and regions in AML, GO functions and KEGG pathway enrichment analyses were conducted. The results of the GO analysis showed that genes corresponding to differentially methylated regions were particularly rich in the classifications of molecular functions, biological processes, and cell components (Table 1). As shown in Table 1, in the biological processes group, these genes were mainly enriched in cell adhesion, negative regulation of transcription by the RNA polymerase II promoter, and cAMP-mediated signaling. The molecular function group was mainly enriched in a variety of combinations, such as protein kinase binding, sequence-specific DNA binding, and regulatory region DNA binding. In addition, the cellular component terms were mainly involved in the plasma and postsynaptic membranes. The results of the enrichment of functions and signaling pathways (Table 2) showed that these genes are involved in 20 pathways, with a p < 0.05 cutoff. The KEGG pathways were significantly enriched in neuroactive ligand-receptor interactions, chemokine signaling pathways, proteoglycans in cancer, the calcium signaling pathway, and the ErbB signaling pathway. These most significantly enriched GO terms and KEGG pathways showed the interactions of genes at the functional level.
| Term | Count | p value | |
|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0007156~homophilic cell adhesion via plasma membrane adhesion molecules | 35 | 1.91E−13 |
| GO:0007399~nervous system development | 35 | 1.79E−06 | |
| GO:0007155~cell adhesion | 44 | 3.63E−05 | |
| GO:0050707~regulation of cytokine secretion | 6 | 1.01E−04 | |
| GO:0042552~myelination | 10 | 3.35E−04 | |
| GOTERM_CC_DIRECT | GO:0005886~plasma membrane | 282 | 1.30E−11 |
| GO:0014069~postsynaptic density | 26 | 2.39E−06 | |
| GO:0005887~integral component of plasma membrane | 104 | 1.03E−05 | |
| GO:0008076~voltage-gated potassium channel complex | 13 | 9.53E−04 | |
| GO:0005578~proteinaceous extracellular matrix | 24 | 0.004941 | |
| GOTERM_MF_DIRECT | GO:0005509~calcium ion binding | 64 | 2.09E−06 |
| GO:0019901~protein kinase binding | 36 | 1.29E−04 | |
| GO:0030295~protein kinase activator activity | 7 | 0.001177 | |
| GO:0015179~L-amino acid transmembrane transporter activity | 5 | 0.002609 | |
| GO:0001077~transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | 22 | 0.004601 |
- Abbreviation: AML, acute myeloid leukemia.
| Term | Count | p value | Genes |
|---|---|---|---|
| hsa04921:Oxytocin signaling pathway | 18 | 0.001001 | CACNA2D1, CACNG8, OXT, CAMK2G, ADCY6, PRKAG2, NPR1, PRKCG, CACNA2D3, KCNJ12, TRPM2, CACNA2D4, CCND1, ADCY9, RYR1, GUCY1A2, PIK3R5, and PIK3R1 |
| hsa04080:Neuroactive ligand-receptor interaction | 24 | 0.004644 | GPR83, GABRG3, PRLHR, PTH2R, DRD3, TACR3, GRIK2, ADCYAP1R1, DRD5, GABRA5, DRD4, F2RL1, PLG, LEP, GRM5, HTR1B, GPR35, GRM2, HRH2, GIPR, GRM6, P2RX2, MC3R, and TSHR |
| hsa04911:Insulin secretion | 11 | 0.005743 | KCNMA1, FXYD2, KCNN4, ADCY9, CAMK2G, ADCYAP1R1, ADCY6, PRKCG, ATP1A1, PCLO, and KCNJ11 |
| hsa04670:Leukocyte transendothelial migration | 13 | 0.008405 | CYBB, CLDN6, BCAR1, PRKCG, AFDN, PIK3R5, ACTN3, JAM3, PIK3R1, CLDN23, CLDN14, CTNNA3, and CLDN15 |
| hsa04960:Aldosterone-regulated sodium reabsorption | 7 | 0.00855 | FXYD2, SGK1, PRKCG, ATP1A1, PIK3R5, NEDD4L, and PIK3R1 |
- Abbreviation: AML, acute myeloid leukemia.
3.3 PPI network construction and module selection
The STRING online database and Cytoscape software were used to visualize the acquired genes and to obtain a PPI network with 345 nodes and 693 edges (Figure 1). Based on the connectivity of ≥ 10, from the 345 nodes, 31 highly differentially expressed node genes were selected. And the 31 central genes were CCND1, TNF, ERBB2, ESR1, ADCY9, ADCY6, CD44, PIK3R1, LEP, SST, CAMK2G, LYN, BCR, CCL5, ERBB4, TAC1, JAK1, PNPLA7, HELZ2, TLR9, GDNF, HTR1B, IRF8, TRAF6, ARRB2, BCAR1, DRD4, MPO, OXT, PRKAR1B, and TSHR (Figure 2). To further explore the functional role of the corresponding genes in the differentially methylated region, we selected two important modules from the PPI network complex based on the Cytotype MCODE plugin. The enrichment analysis showed that module 1 consisted of 15 nodes and 64 edges (Figure 3A), which were mainly enriched in protein phosphorylation, positive regulation of heterotypic cell-cell adhesion, receptor binding, and the regulation of calcium ion transport. In addition, module 2 consisted of 24 nodes and 56 edges (Figure 3B), which were involved in the cell surface receptor signaling pathway, the adenylate cyclase-inhibiting G-protein-coupled receptor signaling pathway, G-protein-coupled receptor internalization, and cAMP-mediated signaling. Moreover, seven genes based on 31 nodes belonged to module 1, and nine genes belonged to module 2. These results suggested that these central genes are closely related and interact to promote the development of disease, which may suggest new therapeutic approaches for AML.
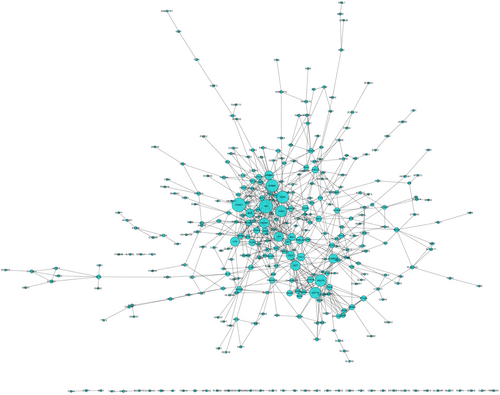
The PPI network of the genes corresponding to the differential methylation regions in AML. The greater the degree, the larger the diameter. AML, acute myeloid leukemia; PPI, protein-protein interaction
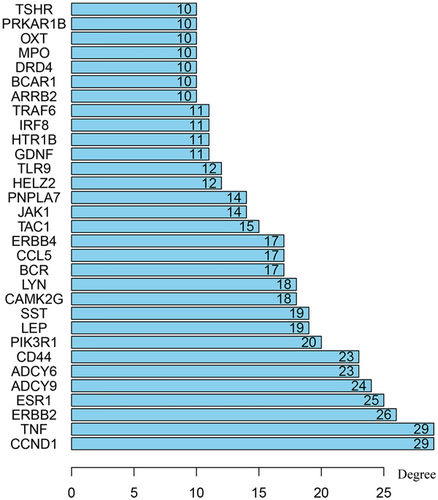
Top 31 hub genes with higher degree of connectivity
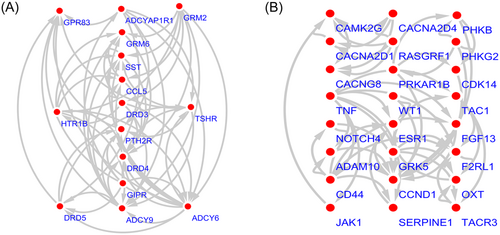
Top 2 modules from the protein-protein interaction network. A, module 1; B, module 2
3.4 Prognostic assessment of differentially methylated regions corresponds to genes and clinical features
We conducted a univariate Cox regression between 31 central genes in patients with AML, and the results showed that a total of seven genes (MPO, CD44, ESR1, TLR9, DRD4, ARRB2 and TSHR) were significantly associated with OS with p values > 0.05 (Table 3). Multivariate Cox proportional regression was applied to confirm the above results, and we found that five genes (ESR1, CD44, TLR9, DRD4 and MPO) were independent prognostic indicators for AML (Figure 4). The prognostic index was imputed as follows: (0.1057 × expression level of ESR1) + (− 0.1488 ×expression level of CD44) + (0.1373 × expression level of TLR9) + (− 0.2402 × expression level of DRD4) +(− 0.0616 × expression level of MPO). We calculated the prognostic index for each sample and divided the samples into high-risk or low-risk groups according to the median cutoff point. ESR1 and TLR9 were highly expressed in both groups, while CD44, DRD4, and MPO had low-expression levels in the AML group. Survival analysis was performed using the Kaplan-Meier method with a log-rank statistical test. The results showed that samples in the high-risk group have significantly worse OS than samples in the low-risk group (p < 0.001; Figure 5). Finally, we used time-dependent ROC curves to evaluate the accuracy of prognostic of the five biomarkers. The AUC for the five biomarkers prognostic model was 0.717 at 5 years of OS (Figure 6).
| Genes | HR | p value |
|---|---|---|
| MPO | 0.9048 | 0.000911 |
| CD44 | 0.765222 | 0.001128 |
| ESR1 | 1.178452 | 0.003116 |
| TLR9 | 1.227279 | 0.00468 |
| DRD4 | 0.751644 | 0.016727 |
| ARRB2 | 0.80717 | 0.028522 |
| TSHR | 1.133077 | 0.033071 |
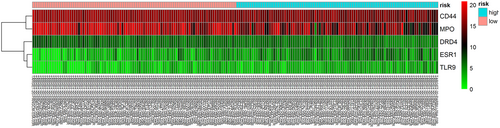
The heatmap of five independent AML-related prognostic genes. 2 genes were upregulated genes and 3 genes were downregulated genes. The color from green to red shows a trend from low-expression to high expression. AML, acute myeloid leukemia
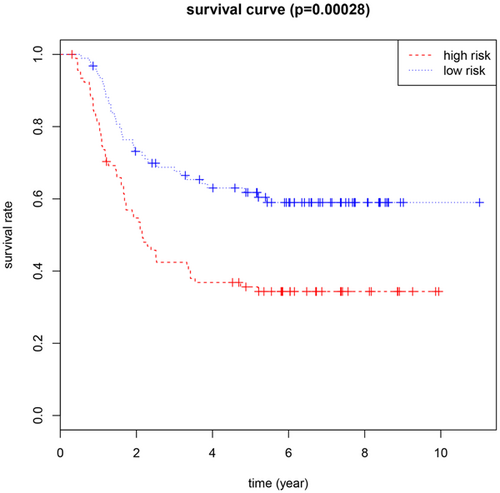
Kaplan-Meier survival curves for overall survival outcomes according to the risk cutoff point
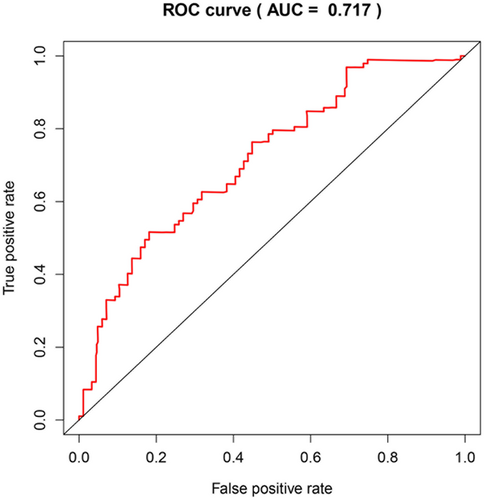
Time-dependent ROC curves analysis for 5-year survival prediction by the five key genes. ROC, receiver operating characteristic
4 DISCUSSION
AML is an aggressive malignancy characterized by the bone marrow infiltration of immature leukemia cells,22 and the acquisition of chromosomal abnormalities, somatic mutations, and epigenetic changes that result in a considerable degree of biological and clinical heterogeneity. The incidence of AML increases with age, and most cases experience serious illness and have poor prognoses. The 5-year survival rate is approximately 20%-25%, and AML can often be life-threatening if not treated in time. With the continuous development of molecular biology, molecular immunology, epigenetics, and other disciplines, more in-depth the understanding of the pathogenesis of AML and its related treatment has become increasingly possible. Epigenetics has led to the discovery that in addition to cytogenetic changes, epigenetic abnormalities are involved in the pathogenesis of AML, involving DNA methylation, histone modification, chromatin remodeling noncoding RNA regulation, and other mechanisms. Advances in genomic technologies have increasingly shown the complexity and heterogeneity of genetic, and epigenetic alterations in AML. Among the genetic alterations occurring in AML, alterations frequently occur in genes involved in the epigenetic control of the DNA methylome and histone methylome. Therefore, it is critical to explore the molecular mechanism of AML progression and the associated epigenetic changes to determine new biomarkers for improving AML early diagnosis, treatment, and prognosis.
Although AML is a malignant clonal disease of the hematopoietic system, the exact pathogenesis of AML is not clear. However, cytogenetic abnormalities lead to changes in the signal transduction pathway and the loss of normal cell metabolism, which are important mechanisms of AML development and progression. DNA methylation as one of the core, most widely studied epigenetic modifications have been found to be altered in many cancers and is often associated with clinically relevant information (ie, subtype, prognosis, and drug response).23 Recent studies on genome-wide DNA methylation have emphasized the importance of methylation abnormalities in AML both from biological and clinical perspectives.24, 25 Aberrant DNA methylation has also been found to be suitable as a prognostic biomarker.26, 27 The recent study used whole-genome MCC-Seq to detect prognostic DNA methylation markers in patients with AML.28 Another study showed that in patients with acute lymphoblastic leukemia, AML, and multiple myeloma, there is hypermethylation of the pro-apoptotic BNIP3 gene locus, leading to BNIP3 gene silencing.29 In addition, in acute leukemia, plasma cell disease, nonHodgkin’s lymphoma, and myelodysplastic syndrome, the p15 gene is hypermethylated; early patients with normal p15 genes undergo methylation as the disease progresses.30 The independence and stability of DNA methylation analysis render it suitable as a prognostic biomarker in AML. The epigenetics of DNA methylation and histone methylation control changes at the gene level. Therefore, exploring the functional enrichment of genes in differentially methylated regions may provide clinicians with new tools that can be used to treat AML and predict its prognosis.
To gain a deeper understanding of the molecular functions of differentially methylated regions, DAVID bioinformatics tools for enrichment and functional analysis revealed that the corresponding genes in the differentially methylated regions were involved in neuroactive ligand-receptor interaction, leukocyte transendothelial migration, the chemokine signaling pathway, proteoglycans in cancer, the calcium signaling pathway, the ErbB signaling pathway, and focal adhesion. Studies have shown that chemokines play an important role in tumor growth and angiogenesis. Some chemokines, such as CXCL8, play an autocrine role in a variety of tumors such as malignant melanoma, liver cancer, pancreatic cancer, and colon cancer,31 and it is closely related to the epidermal growth factor receptor, indicating a close relationship between growth factors and the chemokine pathway, the overexpression of which can promote tumor cell growth.32 The calcium signaling pathway as one of the most abundantly enriched regions in our study. Calcium plays a crucial role in neuronal transmission, muscle contraction, cell motility, cell growth, and proliferation.33, 34 Studies have shown that calcium signaling pathways are associated with the development and progression of various cancers, such as lung adenocarcinoma,35 breast cancer,36 colorectal cancer,37 Burkitt’s lymphoma,38 and others. The calcium signaling may play a key role in the development and progression of AML. A recent study showed that aberrant activation of the target gene of the ErbB signaling pathway can enhance the proliferation and invasiveness of gallbladder carcinoma cells and is a key driver in gallbladder carcinomas. Targeted treatment with drug intervention in ErbB signaling molecules can improve the disease control rate and prolong the survival of advanced gallbladder cancer.39 As a multifunctional molecule, proteoglycan participates in a variety of cellular functions during morphogenesis, wound healing, inflammation, and tumorigenesis, and it regulates the phenotypes and properties of cancer cells, the development of drug resistance, and the generation of tumor stroma blood vessels.40 In addition, as shown in Table 1, the GO analysis revealed that these enriched genes are mainly involved in cell adhesion, the negative regulation of transcription from the RNA polymerase II promoter and cAMP-mediated signaling, protein kinase binding, sequence-specific DNA binding, and regulatory region DNA binding. These significantly enriched GO terms and KEGG pathways showed the interactions of genes at the functional level.
In this study, we aimed to explore the differentially expressed methylation sites and regions between patients with AML and normal samples to identify in TCGA some of the prognostic biomarkers associated with differentially methylated regions. The differentially expressed methylated regions were screened, and the corresponding genes in the differentially methylated region were annotated. Univariate and multivariate Cox analyses of the data from TCGA were performed to identify the potential of these genes to predict the prognosis of AML, based on the visualization software Cytoscape and a plugin. Finally, we identified five genes (ESR1, CD44, TLR9, DRD4, and MPO) associated with differentially methylated regions that are independent predictors of prognosis in patients with AML. ESR1, as an estrogen receptor, is closely related to BMD. It plays important roles in bone growth, bone mass maintenance, and bone loss prevention.41 Its role in cancer progression is poorly reported. In our study, ESR1 correlated with the prognosis of AML, and further experiments are needed to verify this conclusion. Several studies have reported that CD44 is associated with AML survival. In elderly patients with refractory AML, the overall survival of CD44-negative patients is longer than that of CD44-positive patients.42 Knockdown of CD44 enhances the chemo-sensitivity of AML cells to adriamycin and cytosine arabinoside.43 TLR9, a member of the Toll-like receptor family, has high expression levels in B-cell chronic lymphocytic leukemia cells, breast cancer cells, and lung cancer cells.44, 45 Dopamine receptor D4 (DRD4) is a G protein-coupled receptor that is widely expressed in the central nervous system (CNS)46 and is aberrantly methylated and repressed in pediatric CNS tumors.47 The expression of myeloperoxidase (MPO), a microbicidal protein, is a definitive marker for the diagnosis of AML.22 Studies have shown that MPO expression is also positive in lung cancer and gastric cancer.48, 49 The results showed that the AUC of the ROC curve for the prediction of 5-year survival by the five-gene signature was 0.717. The area under the ROC curve (AUROC) > 0.700 is defined as the clinically useful prognostic score, and it can be seen that the prognostic model has certain accuracy and sensitivity in assessing the prognosis of patients. These five gene tags perform well in survival prediction, although additional studies are needed to validate these findings.
Since gene expression may change due to abnormalities in the methylated region, we can use the expression of the corresponding gene to indirectly reflect the functional regulation of the differentially methylated region. Finally, screen out prognostic biomarkers related to differentially methylated regions, which may provide clinicians with new tools that can be used to treat AML and predict its prognosis. In this study, we obtained regions that are differentially methylated in patients with AML and the corresponding genes by data mining. Through the bioinformatics analysis of genes, the related functions of these differentially methylated regions were identified, and 31 highly differentially expressed node genes identified from the PPI networks. Univariate and multivariate Cox regressions showed that 5 genes (ESR1, CD44, TLR9, DRD4, and MPO) proved to be independent prognostic markers of AML. Although confirmatory studies are needed, our study reveals a novel molecular mechanism that may drive tumorigenesis and provides a bioinformatics foundation for predicting the prognosis of the disease.
ACKNOWLEDGMENTS
This study is supported by the grants from National Natural Science Foundation of China (81673799) and National Natural Science Foundation of China (81703915).
CONFLICTS of INTEREST
The authors declared no conflict of interest.
AUTHOR CONTRIBUTIONS
Changgang Sun and Chundi Gao conceived and designed the study; Jing Zhuang, Chao Zhou and Lijuan Liu performed data analysis; Cun Liu, Huayao Li, Gongxi Liu contributed analysis tools; Chundi Gao wrote the paper.




