Expression characteristics of long noncoding RNA uc.322 and its effects on pancreatic islet function
Abstract
Increasing evidence indicates that long noncoding RNAs (lncRNAs) perform special biological functions by regulating gene expression through multiple pathways and molecular mechanisms. The aim of this study was to explore the expression characteristics of lncRNA uc.322 in pancreatic islet cells and its effects on the secretion function of islet cells. Bioinformatics analysis was used to detect the lncRNA uc.322 sequence, location, and structural features. Expression of lncRNA uc.322 in different tissues was detected by quantitative polymerase chain reaction analyses. Quantitative polymerase chain reaction, Western blot analysis, adenosine triphosphate determination, glucose-stimulated insulin secretion, and enzyme-linked immunosorbent assay were used to evaluate the effects of lncRNA uc.322 on insulin secretion. The results showed that the full-length of lncRNA uc.322 is 224 bp and that it is highly conserved in various species. Bioinformatics analysis revealed that lncRNA uc.322 is located on chr7:122893196-122893419 (GRCH37/hg19) within the SRY-related HMG-box 6 gene exon region. Compared with other tissues, lncRNA uc.322 is highly expressed in pancreatic tissue. Upregulation of lncRNA uc.322 expression increases the insulin transcription factors pancreatic and duodenal homeobox 1 and Forkhead box O1 expression, promotes insulin secretion in the extracellular fluid of Min6 cells, and increases the adenosine triphosphate concentration. On the other hand, knockdown of lncRNA uc.322 has opposite effects on Min6 cells. Overall, this study showed that upregulation of lncRNA uc.322 in islet β-cells can increase the expression of insulin transcription factors and promote insulin secretion, and it may be a new therapeutic target for diabetes.
1 INTRODUCTION
Diabetes mellitus is a disorder of glucose regulation caused by a heterogeneous collection of disorders. Type 2 diabetes mellitus develops according to the insulin resistance of multiple organs and inadequate pancreatic β-cell insulin secretion.1 Due to lifestyle modification, including increased sedentary behavior, and high-calorie foods, the prevalence of type 2 diabetes affects more than 300 million people around the world, and it is estimated that type 2 diabetes may affect more than 520 million people within 15 years.2, 3 The pathogenesis of type 2 diabetes involves multiple factors. Environmental exposure, lifestyle factors, and genetic predisposition modulate susceptibility to this disease.4
Pancreatic β-cells are a type of endocrine cells, accounting for approximately 70% of the total islet cell count. They play an important role in regulating blood glucose by secreting insulin. Impaired islet β-cell function leads to an absolute or relative lack of insulin secretion, resulting in diabetes. In recent years, it has been confirmed that long noncoding RNAs (lncRNAs) play a significant role in cell differentiation and progress of disease. They modulate gene expression at multiple levels, such as epigenetic, transcriptional, and post-transcriptional regulation.5 They also involve in the recruitment of chromatin modification, regulation of X chromosome inactivation, chromosome recombination, genomic imprinting, protein folding, and protein activity.6 Recent studies have confirmed that lncRNAs are enriched in pancreatic islets and/or are specifically expressed in pancreatic β-cells.7, 8 The role of lncRNAs in pathogenesis of diabetes has been recognized recently. A growing number of lncRNAs are involved in the regulation of blood glucose.9 For example, research has shown that knockdown of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic cells.10
We screened the differential expression lncRNAs in the pancreatic tissue of diabetic mice by lncRNA chip technology. One of the most distinct lncRNA, uc.322, attracted our attention. We further found that lncRNA uc.322 was highly expressed in pancreatic tissue. Further bioinformatics revealed that lncRNA uc.322 is highly homologous in mammals and is extremely conservative in evolution. So we speculate that lncRNA uc.322 may play an important role in the regulation of islet β-cell function. The full-length of lncRNA uc.322 is 224 bp, which is located within the exon region of the SRY-related HMG-box 6 (SOX6) gene (chr7:122, 893, 196-122, 893, 419, GRCH37/hg19).
Islet transcription factors (TFs), pancreatic and duodenal homeobox 1 (PDX1) and Forkhead box O1 (FOXO1), can combine with insulin gene promoter regions, regulate insulin gene transcription, and thus affect the synthesis and secretion of insulin. PDX1 plays an important role in pancreatic development and adult β-cell function. FOXO1 is an important TF in insulin signaling downstream, and it plays an important role in insulin signaling in liver and fat during the transfer process, which can effectively regulate the proliferation and differentiation of β-cells and apoptosis in the state of insulin resistance.11 In this study, we upregulated and downregulated lncRNA uc.322 via lentivirus transfection in Min6 cells, and detected its effect on PDX1 and FOXO1 expression and cell function.
As the most important energy molecule, adenosine triphosphate (ATP) plays an important role in various physiological and pathological processes of the cells. ATP coexists with insulin in the secretory vesicles of pancreatic cells, and it releases with insulin outside of the cells, feedback regulates its own insulin secretion and the secretory activity of adjacent cells.12, 13 In rat islet cells and human islets, ATP promotes insulin secretion by acting on the P2 receptor.14 Therefore, we measured the changes in the ATP in the cells after lncRNA uc.322 overexpression and knockdown, and this could partly reflect the changes in the secretory function of islet cells.
2 MATERIALS AND METHODS
2.1 Animals and cell culture
All animal experiments were carried out in strict accordance with the rules of management used by the Experimental Animal Committee of Nanjing Medical University (Approval NO: IACUC-1781195). Five-week-old C57BL/6J male mice were purchased from the Model Animal Research Center of Nanjing Medical University. The mice were killed by the spinal dislocation method, and then the heart, liver, pancreas, spleen, kidney, muscle, and fat of the mice were removed. Total RNA of these tissues was extracted by Trizol reagent (Thermo Fisher Scientific, Shanghai, China) and the expression of lncRNA uc.322 in these tissues was detected by quantitative polymerase chain reaction (qPCR). The primers were designed by Primer 5.0 software and synthesized by Shanghai Generay Biotech Co, Ltd (Table 1).
| Primer | Sequence | Application |
|---|---|---|
| lncRNAuc.322 | F:GACATTCATTAGGACCCACACA | qPCR |
| R:GTTGTGAAGAGTTGATTGGGAAT | qPCR | |
| PDX1 | F:TACAAGCTCGCTGGGATCACT | qPCR |
| R:GCAGTACGGGTCCTCTTGTT | qPCR | |
| FOXO1 | F:CTTCAAGGATAAGGGCGACA | qPCR |
| R:GACAGATTGTGGCGAATTGA | qPCR | |
| SOX6 | F:CCACAGACAGATTGAGCAGC | qPCR |
| R:CAGGGCAGGAGAGTTGAGAC | qPCR | |
| β-Actin | F:GACGGCCAGGTCATCACTAT | qPCR |
| R:ACATCTGCTGGAAGGTGGAC | qPCR |
- FOXO1, forkhead box O1; lncRNA, long noncoding RNA; PDX1, pancreatic and duodenal homeobox 1; qPCR, quantitative PCR; SOX6, SRY-related HMG-box 6.
Min6 cells are a mouse insulinoma cell line. The line was obtained from the Institute of Endocrinology and Metabolism, Ruijin Hospital. The cells were cultured in Dulbecco modified Eagle medium high-glucose medium containing 15% fetal bovine serum, 0.5% streptomycin and penicillin, and 50 μM of β-mercaptoethanol at 37°C in 5% CO2.
2.2 Uc.322 overexpression and knockdown experiments via lentivirus transfection
For overexpression experiments, Min6 cells were infected by LV5-lncRNA uc.322, and the infection efficiency was confirmed by qPCR. For knockdown, cells at 60% confluency were transfected with uc.322-specific short hairpin RNA (shRNA) pool (LV3-uc322-shRNA-71 and LV3-uc322-shRNA-166). Control cells were treated with relative RNA negative lentivirus. The lentivirus was purchased from Shanghai GenePharma Pharmaceutical Technology Co, Ltd.
2.3 qPCR and Western blot analysis
The Trizol reagent (Thermo Fisher Scientific) was used to extract total RNA from Min6 cells, and the total RNA of the cells was reverse-transcribed into complementary DNA by a Revertra ace-m (TOYOBO Shanghai Biotech Co, Ltd) high-efficiency reverse transcription Kit in the PCR amplification instrument. The reaction condition was 37°C for 15 minutes, 85°C for 5 seconds, and preservation at 4°C. Then, complementary DNA was analyzed by qPCR on the ABI 7500 real-time qPCR to detect the expression of lncRNA uc.322, insulin TFs (PDX1 and FOXO1), and SOX6, which are the possible targets of lncRNA uc.322 in Min6 cells. β-Actin was used as the internal control, and the 2−∆∆Ct method was used to calculate the relative value of gene expression. PCR primers were designed with Primer5.0 software and were synthesized by Shanghai Generay Biotech Co, Ltd (Table 1).
For Western blot analysis, protein was extracted from whole-cell lysates using RIPA buffer and quantified by the bicinchonininc acid (BCA) Protein Assay Kit. A 50 µg sample of the extracted protein was separated via 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to 0.22 µm nitrocellulose membranes (Sigma), and incubated with specific antibodies. The autoradiograms were quantified using densitometry. The rabbit primary antibodies were used at a dilution of 1:1000. The secondary antibody used in these assays was horseradish peroxidase conjugated goat anti-rabbit lgG (dilution of 1:1000). The β-actin antibody was used as the control. Antibodies used in this study are PDX1 (SAB2104577-50UG, Sigma), SOX6 (SAB2102269-50UG, Sigma), FOXO1 (2880P, Cell Signaling), β-actin (4970S, Cell Signaling). All of the experiments were repeated at least 3 times.
2.4 Detecting glucose-, arginine-, and KCl-stimulated insulin secretion by enzyme-linked immunosorbent assay
Min6 cells were digested and passaged in 96-well plates until the cells were 80% confluent, and 6 accessory holes were established in each group. Then, the culture medium was replaced by 200 μL of 0.25% BSA, 2.8 mM of glucose (Glu) KRB buffer to pre-incubate for 30 minutes. After that, the 0.25% BSA, 2.8 mM of Glu KRB buffer was replaced by new 200 μl of 0.25% BSA, 2.8 mM of Glu KRB buffer to incubate at 37°C for 1 hour. Then, 100 μL of culture solution was used to detect insulin secretion under low glucose stimulation. Afterward, we added new 100 μL of 0.25% bovine serum albumin (BSA), 2.8 mM of Glu kerbs-ringerbicarbonate (KRB) buffer, and 30 mM l-arginine into the surplus 100 μL of 0.25% BSA, 2.8 mM of Glu KRB buffer to incubate at 37°C for 1 hour. Next, 100 μL of culture solution was used to detect insulin secretion under l-arginine stimulation. Then, the 0.25% BSA, 2.8 mM of Glu KRB, and 30 mM l-arginine buffer was replaced by 200 μL of 0.25% BSA, 16.7 mM of Glu KRB buffer to incubate at 37°C for 1 hour. Next, 100 μL of culture solution was used to detect insulin secretion under high glucose stimulation. Then, the 0.25% BSA, 16.7 mM of Glu KRB buffer was replaced by 200 μL of 0.25% BSA, 30 mM of KCL KRB buffer to incubate at 37°C for 1 hour. Finally, 100 μL of culture solution was used to detect insulin secretion under KCL stimulation. The 4 types of culture solutions were centrifuged (200g, 20 minutes, 4°C) to remove some of the isolated cell debris or impurities. Enzyme-linked immunosorbent assay was used to detect insulin concentrations of different standard products, and we used 3 enzyme-linked immunosorbent assay wells per sample.
2.5 Detection of mitochondrial ATP production
The ATP bioluminescence assay kit and multifunctional enzyme marker were used to detect mitochondrial ATP concentration (Shanghai Beyotime Biotechnology Co, Ltd). Min6 cells were cultured to the logarithmic phase, and then the cells were passaged in 6-well plates until 80% confluency. After that, the cell culture fluid in the 6-well plate was removed, and 200 μL of lysate was added into each pore. The sample was continuously pipetted to ensure cell lysis and then centrifuged to separate the cell mass and supernatant. The centrifugal frequency was 12000 rpm for 5 minutes at 4°C. After centrifugation, the supernatant was collected. Then, 20 μL of the supernatant and different concentrations of the standard were added to the wells prepared with an ATP detection working fluid and rapidly mixed. We used 3 ATP detection wells per sample when measuring the ATP, and the numerical values of luminous intensity were measured by the liquid scintillation analyzer. Standard curves were plotted according to the luminous intensity of standard products. The amount of ATP in cells was calculated according to the standard curve.
2.6 Statistical analysis
SPASS l9.0 statistical software was used for statistical analyses. The data were presented as mean ± standard deviation and the P values of the experiment were calculated using 1-way analysis of variance or unpaired 2-tailed Student t test with a significance of P < .05. The experiment was repeated more than 3 times.
3 RESULTS
3.1 lncRNA uc.322 is an extremely conservative lncRNA and it is highly expressed in pancreas tissues of mice
Conservation analysis of the lncRNA uc.322 locus found it to be highly conserved in rat, human, dog, orangutan, horse, and opossum (Figure 1A). Sequence alignment by DNAMAN (http://www.lynnon.com) of lncRNA uc.322 between human and mouse genomes showed 100% identity. qPCR was performed to detect the expression of lncRNA uc.322 in different tissues of adult C57BL/6J male mice, including pancreas, fat, muscle, spleen, kidney, heart, and liver. As shown in Figure 1B, lncRNA uc.322 appeared to be highly expressed in pancreas tissue, followed by adipose tissue.
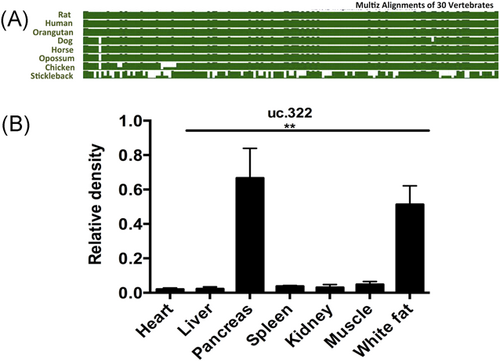
Tissue quantification of lncRNA uc.322. (A) Molecular evolutionary analyses of lncRNA uc.322. (B) The expression of lncRNA uc.322 in different tissues of mice. The experiment was repeated more than 3 times. Data are presented as mean ± SD. **P < .01 (1-way ANOVA). lncRNA, long noncoding RNA; SD, standard deviation
3.2 lncRNA uc.322 overexpression increases the expression of insulin TFs and insulin secretion
To estimate whether lncRNA uc.322 directly affected the expression of insulin TFs (PDX-1 and FOXO1) and insulin secretion, Min6 cells were transduced with a lentivirus expressing lncRNA uc.322. The expression of lncRNA uc.322 in Min6 cells was significantly increased after transfection (Figure 2A). qPCR and Western blot analysis revealed that lncRNA uc.322 overexpression resulted in elevated expression of the insulin TFs, PDX1 and FOXOl (Figure 2B,C).
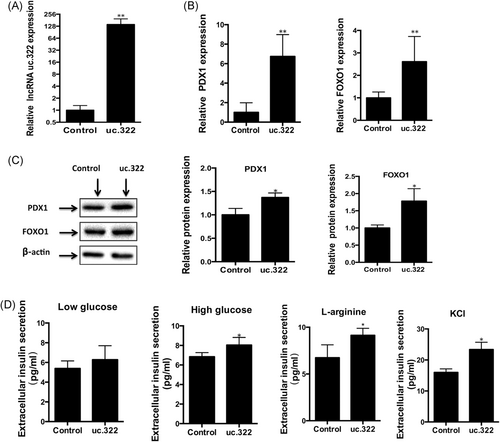
lncRNA uc.322 overexpression increases the expression of insulin transcription factors and insulin secretion. (A) Expression of lncRNA uc.322 in Min6 cells was significantly increased after lentivirus expressing lncRNA uc.322 transfection. (B) Real-time PCR revealed that lncRNA uc.322 overexpression resulted in elevated expression of the insulin transcription factors, PDX1 and FOXOl. (C) Western blot analysis showed increased expression of insulin transcription factors, PDX1 and FOXO1, induced by overexpression of lncRNA uc.322. (D) Overexpression of lncRNA uc.322 results in increased insulin secretion stimulated by high glucose in Min6 extracellular fluid. The experiment was repeated more than 3 times. Data are presented as mean ± SD. *P < .05, **P < .01 (Student t test). FOXO1, forkhead box O1; lncRNA, long noncoding RNA; PDX1, pancreatic and duodenal homeobox 1; PCR, polymerase chain reaction; SD, standard deviation
The effects of lncRNA uc.322 overexpression on insulin secretion were detected by glucose, l-arginine, and KCL. As shown in Figure 2D, Insulin secretion in the lncRNA uc.322 overexpression group was significantly higher than that of the control group under high glucose, l-arginine, and KCL stimulation (P < .05). However, there was no significant difference in insulin secretion under low-glucose conditions (P > .05). It has been reported that Min6 cells have impaired insulin secretion after a high number of passages, and the overall response in insulin release in the Min6 cells under high glucose and low glucose was lower than expected. Considering that l-arginine and KCL are potentiators of glucose-stimulated insulin secretion, we used l-arginine and KCL to evaluate the insulin release response, and found that l-arginine- and KCL-stimulated insulin secretion in Min6 cells was significantly greater than with glucose (Figure 2D).
3.3 lncRNA uc.322 knockdown decreases the expression of insulin TFs and insulin secretion
To estimate the impact of lncRNA uc.322 loss of function on the expression of insulin TFs (PDX-1 and FOXO1) and insulin secretion, we performed an RNAi-knockdown experiment using a pool of shRNAs to transfect Min6 cells. As expected, lncRNA uc.322 shRNA significantly decreased the expression of lncRNA uc.322 in Min6 cells (Figure 3A). qPCR and Western blot analysis revealed that lncRNA uc.322 knockdown resulted in decreased expression of the insulin TFs, PDX1, and FOXO1 (Figures 3B and 2C).
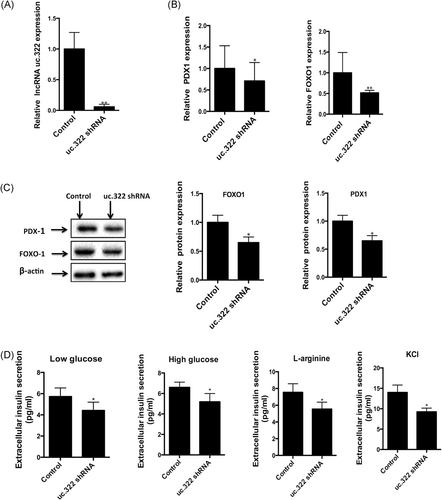
Knockdown of lncRNA uc.322 decreases the expression of insulin transcription factors and insulin secretion. (A) Expression of lncRNA uc.322 in Min6 cells was significantly decreased after uc.322-specific short hairpin RNA (shRNA) pool transfection. (B) Real-time PCR revealed that silencing of lncRNA uc.322 leads to decreased expression of insulin transcription factors, PDX1 and FOXOl. (C) Western blot analysis showed decreased expression of insulin transcription factors, PDX1 and FOXO1, induced by knockdown of lncRNA uc.322. (D) Silencing of lncRNA uc.322 results in decreased insulin secretion stimulated by both high glucose and low glucose in Min6 extracellular fluid. The experiment was repeated more than 3 times. Data are presented as mean ± SD. *P < .05, **P < .01 (Student t test). FOXO1, forkhead box O1; lncRNA, long noncoding RNA; PDX1, pancreatic and duodenal homeobox 1; PCR, polymerase chain reaction; SD, standard deviation
The effects of lncRNA uc.322 knockdown on insulin secretion were also detected by glucose, l-arginine, and KCL. As shown in Figure 3D, insulin secretion in the lncRNA uc.322 knockdown group was significantly lower than that of the control group under high-glucose, low-glucose, l-arginine, and KCL stimulation (P < .05). Consistent with the above research, l-arginine and KCL-stimulated insulin secretion in Min6 cells was significantly greater than with glucose (Figure 3D).
3.4 lncRNA uc.322 is located in the exon region of the SOX6 gene, and it influences mitochondrial ATP production in Min6 cells
The full-length lncRNA uc.322 is 224 bp, which is located in the exon region of the SOX6 gene (chr7: 122, 893, 196-122, 893, 419, GRCH37/hg19; Figure 4A). qPCR and Western blot analysis revealed that uc.322 overexpression resulted in elevated expression of SOX6. However, lncRNA uc.322 knockdown resulted in decreased expression of SOX6 (Figure 4B,C). To estimate whether lncRNA uc.322 directly affected the expression of mitochondrial ATP production, luciferase assay was used to detect the changes in mitochondrial ATP production in Min6 cells. The results revealed that lncRNA uc.322 overexpression resulted in elevated content of ATP in the mitochondria of Min6 cells (P < .05). However, lncRNA uc.322 knockdown resulted in decrease of content of ATP in the mitochondria of Min6 cells (P < .05; Figure 4D).
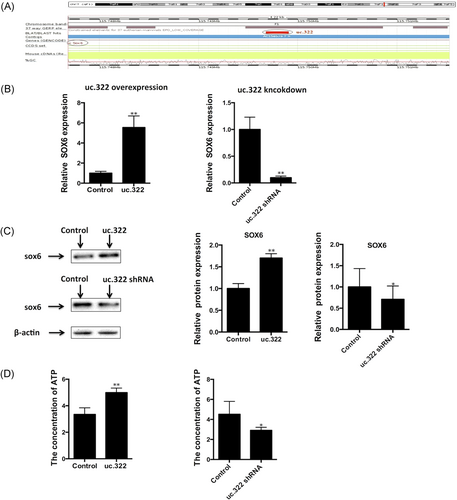
Genetic loci of lncRNA uc.322 and SOX6 expression after lncRNA uc.322 overexpression and knockdown. (A) lncRNA uc.322 is located in the exon region of the SOX6 gene (chr7:122,893,196-122,893,419, GRCH37/ hg19). (B,C) Real-time PCR and Western blot analysis revealed that uc.322 overexpression resulted in elevated expression of SOX6, and lncRNA uc.322 knockdown resulted in decreased expression of SOX6. (D) lncRNA uc.322 overexpression resulted in elevated content of ATP, and lncRNA uc.322 knockdown resulted in decreased content of ATP in the mitochondria of Min6 cells. The experiment was repeated more than 3 times. Data are presented as mean ± SD. *P < .05, **P < .01 (Student t test). ATP, adenosine triphosphate; lncRNA, long noncoding RNA; PCR, polymerase chain reaction; SD, standard deviation; SOX6, SRY-related HMG-box 6
4 DISCUSSION
Although most of the human genome is transcribed, only a small number of transcribed sequences translate to protein products.15, 16 The remainder of the human genome is dominated by noncoding RNAs (ncRNA). The great majority of ncRNAs are constitutively expressed and have verified roles in splicing and translation.17 However, the functional exploration of new ncRNAs is complex; these molecules have contributed to the regulation of gene expression and cell biology over the past several years.
lncRNAs belong to a heterogeneous class of regulatory ncRNAs with transcript lengths >200 nucleotides.18 Recent studies have shown that lncRNAs play an important role in many biological processes, such as X chromosome inactivation, dosage compensation, genomic imprinting, chromatin modification, and remodeling. In general, lncRNAs can regulate the expression of genes in 3 ways: epigenetic regulation, transcriptional regulation, and post-transcriptional regulation.6, 19 The whole genome transcriptional profile of the human pancreas was found to be more than 1100 lncRNAs. However, in the susceptible genes of type 2 diabetes, at least 9 major single nucleotide polymorphisms (150 kb) contain lncRNAs.7 Islet-specific lncRNAs are inactivated in embryonic pancreatic progenitor cells and are activated in the differentiation of pancreatic islet cells.
lncRNA uc.322 was obtained using the lncRNA chip technology, which is highly conserved in mammals. In this study, we found that lncRNA uc.322 was highly expressed in mouse pancreatic tissue. This result suggests that lncRNA uc.322 may play an important role in pancreatic tissue. To confirm this, we upregulated and downregulated the expression of lncRNA uc.322 in Min6 cells. The intracellular insulin TFs (PDX1 and FOXO1) and insulin secretion levels were then measured after the intervention. The results showed that upregulation of the lncRNA uc.322 expression increased the expression of insulin TFs (PDX1 and FOXO1) in Min6 cells. However, knockdown of lncRNA uc.322 decreased the expression of insulin TFs (PDX1 and FOXO1).
The PDX1 gene is an important gene for the development and maintenance of the normal secretion of pancreatic islets. It plays an important role in the formation and development of pancreatic islets in the early development of individuals, and it can regulate the growth and development of the pancreas by activating the genes associated with the formation of the pancreas. If the PDX1 gene is not expressed in the early stages of embryonic development, it can lead to pancreatic hypoplasia in both mice and humans.20 Another study found that PDX1 is an important TF in the regulation of insulin gene transcription, maturation, and secretion of insulin granules and activates the transcription process by binding to the insulin gene transcription regulation region. It plays an important role in regulating the expression of insulin and the homeostasis of blood glucose.21
FOXO1 is an important TF in insulin signaling downstream, and it plays an important role in insulin signaling in liver and fat during the transfer process, which can effectively regulate the proliferation and differentiation of β-cells and apoptosis in the state of insulin resistance.11 It also regulates glucose and fat metabolism, as well as vascular endothelial cells, muscle cells, and adipocyte differentiation. Studies have shown that deacetylation of FOXO1 can reduce mitochondrial lipid utilization, protect islet function, and promote insulin secretion.22
The plasma FOXO1 of high-glucose–fed mice showed obvious dephosphorylation, which led to the decrease of the TF, PDX1, and the apoptosis of β-cells.23 Ildem et al have uncovered a regulatory network in which lineage-specific lncRNAs and TFs control common genes. Furthermore, they have shown that lncRNAs frequently regulate genes associated with clusters of islet enhancers, which have been previously shown to be the primary functional targets of islet-specific TFs.24 Thus, we can only guess that lncRNA uc.322 and the TFs (PDX1 and FOXO1) may control common genes.
We further detected the levels of insulin secretion and intracellular ATP concentration after lncRNA uc.322 overexpression and knockdown, respectively. The results showed that both the extracellular insulin secretion and the content of ATP in Min6 cells were increased after lncRNA uc.322 overexpression. However, when lncRNA uc.322 was downregulated, both the extracellular insulin secretion and the content of ATP in Min6 cells were decreased. ATP is stored as an energy molecule in insulin-containing vesicles. When the vesicles were fused with the cell membranes, they were released to the extracellular space with insulin, suggesting that the secretion function of pancreatic β-cells and insulin secretion were increased after lncRNA uc.322 overexpression. It has been reported that Min6 cells have impaired insulin secretion after high number of passages. Considering this aspect, we used l-arginine and KCL to further evaluate the insulin release response, and we found that Min6 cells responded well to l-arginine and KCl. KCl-stimulated insulin secretion in Min6 cells was significantly greater than glucose and l-arginine. This result is similar to the study of Cheng et al.25
The results also showed that the expression level of SOX6 was significantly increased after lncRNA uc.322 overexpression and decreased significantly after lncRNA uc.322 knockdown. SOX6 is a reverse complement of lncRNA uc.322, and it is a nuclear TF of the group D SOX family of proteins, with essential roles in cell differentiation.26 It has been found that the SOX gene can promote the proliferation and differentiation of pancreatic islet cells.27 Prior studies have suggested that SOX4 is necessary for adult β-cell replication and can cooperate with neurogenin 3 to regulate endocrine pancreas formation.28 Studies have shown that SOX6 attenuates glucose-stimulated insulin secretion, and knockdown of SOX6 can induce the insulin-producing cell differentiation.29, 30
In summary, we explored that upregulation of lncRNA uc.322 can increase the insulin TFs (PDX1 and FOXO1) and SOX6 expression that contributes to ATP generation and insulin secretion in pancreatic islet β-cells. In conclusion, this study provides lncRNA uc.322 as a new target for improving in vitro β-cell function.
CONFLICTS OF INTEREST
All authors read and approved the final manuscript. The authors have no conflict of interests to report.