Mir-1287 suppresses the proliferation, invasion, and migration in hepatocellular carcinoma by targeting PIK3R3
Abstract
Mature microRNAs (miRNAs) are a class of small noncoding RNA molecules involved in regulation of post-translational gene expression. Although aberrant levels of miRNAs have been found in various tumor tissues, their importance in tumor development and the molecular basis of their regulatory role remain unclear. Our bioinformatic analysis on The Cancer Genome Atlas database and microarray-based comparison of miRNA in different cell lines revealed that the level of mir-1287 is suppressed in hepatocellular carcinoma (HCC) cells. When upregulated, mir-1287 can reduce the tumorigenesis phenotypes of HCC cells in several in vitro models. We further found that mir-1287 directly targets messenger RNA encoding PIK3R3, which is a tumor-promoting factor acting in several pathways linked to tumorigenesis. Our study suggests that aberrant suppression of mir-1287 is potentially responsible for the development of HCC, and miRNA-based strategies may be developed for efficient detection and treatment of HCC.
Abbreviations
-
- HCC
-
- hepatocellular carcinoma
-
- miRNA
-
- microRNA
-
- NGS
-
- next generation sequencing
-
- TCGA
-
- The Cancer Genome Atlas
1 INTRODUCTION
Hepatocellular carcinoma (HCC) is now one of the leading causes of cancer-related death worldwide.1, 2 Because of the lack of effective therapy and early detection, HCC patients endure a poor 5-year-survival rate. Hepatoma cells are aberrant in proliferation, migration, and invasion, which cause severe metastasis and recurrence. Research aimed at addressing the molecular basis of liver tumorigenesis would help develop new tools for early-stage diagnosis and effective therapy.3-5
MicroRNAs (MiRNAs) are a class of ~22-nucleotide small noncoding RNAs. They play important roles in posttranscriptional regulation of gene expression by specifically targeting messenger RNAs (mRNAs).6 MiRNA precursors are transcript in nuclear, transported into cytoplasm, and form mature fragments after cleavage by Dicer. In mammal cells, mature miRNAs are loaded into RISC, either blocking the translation, or degrading the mRNAs of target genes. The seed sequences (the 2 to 7 nucleotides) are essential for sequence-specific recognizing on the target sites in mRNA 3′-UTR regions. The functions of miRNAs are critical for many biological processes and aberrant expressions of these regulatory molecules are associated with various diseases including HCC. Accumulating evidence suggests that miRNA-based tools may be utilized for diagnosis and treatment of HCC.4
Analyses on the miRNA levels in tumor tissues, through microarray or next-generation sequencing (NGS), have found many cancer-related miRNA functions. Mir-1287 is found by NGS but not clear of its functions. Previous microarray-based studies analyzing miRNA levels in tumor tissue samples detected aberrant levels of mir-1287 in retinoblastoma, cervical cancer, malignant glioma, and larynx carcinoma, suggesting that its misregulation is associated with the development of cancers.7-12
We found that mir-1287 is downregulated in HCC cells and overexpression of this miRNA could suppress the HCC phenotypes in several in vitro model systems. The follow-up analysis suggested that mir-1287 specifically targets mRNA encoding PIK3R3, which is a tumor-promoting factor linked to enhancement of several tumorigenesis phenotypes including cell proliferation and migration. The results will stimulate efforts to further investigate the molecular details of HCC development in the absence of mir-1287 mediated silencing of PIK3R3, facilitating the optimization of strategies for HCC detection and treatment.
2 MATERIALS AND METHODS
2.1 TCGA data analysis
The miRNA expression data was retrieved from The Cancer Genome Atlas (TCGA) Research Network (https://cancergenome.nih.gov). The miRNA-seq data of 375 HCC patient samples and 50 normal tissue samples were normalized and transformed in log2 form. The TCGA IDs of the samples were listed in the supplementary materials Table S1.
2.2 Gene chip for miRNA expression
The miRNA microarray used was miRCURY LNA microRNA Array, sixth generation—hsa, mmu, & rno (EXIQON). The gene chip was checked and analyzed by UCLA Department of Pathology and Laboratory Medicine David Geffen School of Medicine. The cell samples were QGY-7703 which we chose as representative hepatoma cells, and a subpopulation hepatoma cells with suppressed malignant phenotypes which we treated as normal cells for control.13
2.3 Cell culture
Human HCC cell lines, QGY-7703 and SMMC-7721 are cultured with RMPI-1640, supplemented with 10% FBS and 100 U/mL of penicillin-streptomycin, at 37°C in a humidified chamber supplemented with 5% CO2.
2.4 Xenograft assay on nude mice
The nude mice used were the male BALB/c athymic nude mice. The cells stably expressing mir-1287 or empty vector were suspended with phosphate-buffered saline and subcutaneously injected nude mice at a total cell number of approximately 2 × 106 each mouse. Sixteen days after injection, when tumors were palpable, the tumor volumes in mice were measured with a slide caliper every 4 days until the scarification. And all the tumors were excised and weighed in the end.
2.5 Soft agar colony formation assay
The cells stably expressing mir-1287 or empty vector were seed (300 cells each) onto the plate with 0.7% low melting point agarose in growth media. After incubated at 37°C for 2 weeks, the cells were stained with 1% neutral red.
2.6 Quantitative real-time PCR
Total RNA samples were extracted from cancer cells with TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Then total RNA was reverse transcripted with M-MuLV reverse transcriptase kit (Shanghai Pufei Biotech, Shanghai, China). Quantitative real-time polymerase chain reaction (PCR) was operated on SYBR Green PCR kit (Bio-Rad, Hercules, CA). The primers was purchased from Genepharm (Shanghai, China).
2.7 Cell proliferation assay
QGY-7703 and SMMC-7721 was seeded on 96-well plates 24 hours before transfection of miRNA mimics or inhibitors together with negative control. At 0, 24, 48, and 72 hours, the culture medium was removed and then replaced by fresh medium with 10% concentration of CCK-8 (Dojindo, Kumamoto, Japan) for each well. And the cells were incubated for 2 hours at 37℃, before the absorbance of suspension was measured the at 450 nm by microplate reader (Thermo Scientific Multiskan MK3, Waltham, MA).
2.8 Cell cycle analysis with flow cytometer
Cell cycle was assessed with Flow Cytometer (BD FACS Calibur, San Jose, CA). All the cells were cultured in serum-free medium for 48 hours to synchronize the cell cycle before transfection of miRNA mimics, inhibitors or negative control. The cells was collected, washed with phosphate-buffered saline and fixed with 70% ethanol at 4℃ overnight. Then the cells were stained with propidium iodide at room temperature for 30 minutes before analysis on flow cytometry.
2.9 In vitro cell invasion and migration assay
Cell invasion and migration was evaluated by transwell assay respectively with matrigel and without matrigel in each well. After transfection of miRNA mimics, inhibitors or negative control, cells were cultured in serum-free medium for 24 hours. Then the cells were resuspended and seeded 1 × 105 cells each in the upper chamber of the transwell chamber (Corning Costar, Corning, NY), and culture medium with 10% FBS was added in the lower chamber. The incubation time was 24 hours for migration and 48 hours for invasion. And the migratory or invasive cells were stained with crystal violet and manually counted at ×200 magnification.
2.10 Luciferase report assay
The 3′-UTR sequence of PIK3R3 containing the predicted target site of mir-1287 was cloned onto psiCHECK-2 vector (C8021; Promega, Madison, WI). And a mutant plasmid on the target site was constructed with the Site-directed Gene Mutagenesis kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. The luciferase constructs and the mir-1287 mimics or negative control were cotransfected into QGY-7703 cell with lipofectamine 2000. Forty-eight hours after transfection the cells were collected and examined the luciferase activity using Dual-Luciferase Reporter Assay System (E1910; Promega).
2.11 Western blot
Cell lysis were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred on to poly(vinylidene difluoride) membranes (Millipore, Billerica, MA). The membranes were blocked with nonfat milk at room temperature for 2 hours, followed by incubation with primary antibodies for 1.5 hours. After washing with 2‰ TBST 3 times, the membranes were incubated with secondary antibodies for 45 minutes and washed another 3 times before exposure with ECL (Thermo Fisher Scientific, Waltham, MA). The intensity of the protein bands was analyzed by Image J system (NIH, Bethesda, MD). The primary antibodies used were as follows: PIK3R3 (Abgent, Wuxi, China) Akt, p-Akt, NF-κB pathway sampler kit (Cell Signaling Technology, Danvers, MA).
2.12 Statistical analysis
All data were evaluated with the software R version 3.3.2 and Origin 9.1 (OriginLab, Northampton, MA). A 2-tailed Student t test was performed to compare the data. Difference was considered significant at P < .05.
3 RESULTS
3.1 The expression of mir-1287 is suppressed in HCC cells
To understand miRNA homeostasis in HCC cells, we analyzed miRNA levels derived from microarray profiles and found that a number of miRNAs are aberrantly expressed in human HCC cells (Figure 1A). Among these, mir-190, mir-10a, and mir-34a are reported to be involved in tumorigenesis.14-16 While mir-1287 is also largely suppressed in tumor cells, indicating that the normal expression level of this microRNA is potentially important for tumor suppression.
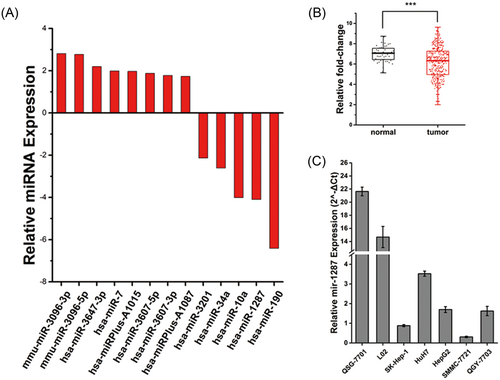
The expression of mir-1287 is suppressed in hepatocellular carcinoma (HCC) cells. (A) Gene chip analysis of significantly aberrant expression microRNAs (miRNAs) in hepatoma cells in contrast with normal hepatocytes. (B) The fold-change of mir-1287 expression in normal tissue and HCC tumor tissue (7.00 ± 0.79 and 6.14 ± 1.61, respectively). ***P < .001. Data is downloaded from TCGA miRNA-seq database. (C) Relative expression of mir-1287 in normal liver cell lines QSG-7701 and L02, together with HCC cell lines SK-Hep-1, Huh7, HepG2, SMMC-7721, and QGY-7703 by real-time qPCR. qPCR, quantitative polymerase chain reaction; TCGA, The Cancer Genome Atlas
To determine whether the suppression of mir-1287 is a universal phenomenon in HCC, we further compared the miRNA expression levels in HCC tumor and adjacent normal tissues using data downloaded from TCGA database (Figure 1B). The normalized data was log 2 transformed to demonstrate the fold-change of the mir-1287 level between tumor tissues and normal tissues. The results indicated that the expression of mir-1287 was downregulated in HCC tumor tissues than in normal tissues (6.14 ± 1.61 and 7.00 ± 0.79, respectively; P < .001).
To confirm the aberrant level of mir-1287 in a cell model, we further used real-time PCR to validate the expression level of mir-1287 in 7 types of human liver cell lines (Figure 1C). Among them, the normal liver cells QSG-7701 and L02 showed a remarkably high expression of mir-1287, while in the rest 5 types of HCC cell lines, the expression levels were relatively low, indicating that mir-1287 was downregulated in HCC cells.
3.2 Mir-1287 suppresses the tumor growth and the colony formation
We next sought to determine whether the overexpression of mir-1287 would suppress the tumor growth. We exogenously expressed mir-1287 to increase the levels of this miRNA in the QGY-7703 and SMMC-7721 tumor cells. These cells formed fewer colonies in soft agar assays (Figure 2A,B). Furthermore, they formed much smaller tumors after being xenografted into nude mice (Figure 2C,D).
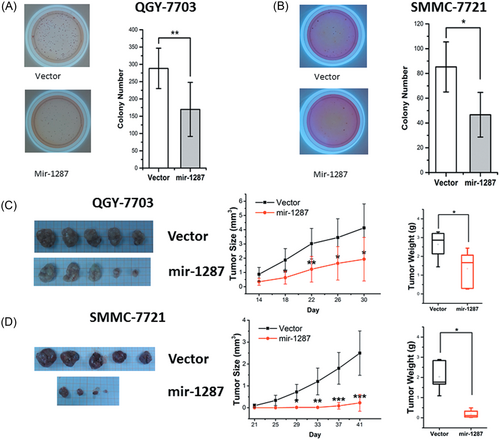
The colony formation and tumor growth of HCC cell lines was suppressed by overexpression of mir-1287. The soft agar assays of QGY-7703 (A) and SMMC-7721 (B) stable clones, with empty vector or mir-1287. Representative images of colony assay were aside with the bar graphs. The xenograft assays of the stable clones of QGY-7703 (C) and SMMC-7721 (D). Representative tumor images were aside with the graphs. The tumor size and tumor weight data represented the mean ± SD of 5 nude mice. *P < .05; **P < .01; ***P < .001. HCC, hepatocellular carcinoma; SD, standard deviation
3.3 Mir-1287 suppresses the cell proliferation in QGY-7703 and SMMC-7721 and arrest the cell cycle at G1/S phase
Given the robust phenomenon that mir-1287 suppressed the tumor growth, we next identified the cellular mechanism underlying this miRNA. We further analyzed the proliferation of the QGY-7703 and SMMC-7721 tumor cells by CCK-8 assays (Figure 3A,B). The cells transfected with miR-1287 mimics showed apparently reduced proliferation rates. This was associated with increased numbers of cells at the G1 phase (Figure 3E,F), indicating a possible impact of miR-1287 on cell cycle arrest. In contrast, the cells transfected with miR-1287 inhibitors (antisense RNA of miR-1287) proliferated faster and fewer cells were arrested at the G1 phase (Figure 3C,D,G,H).
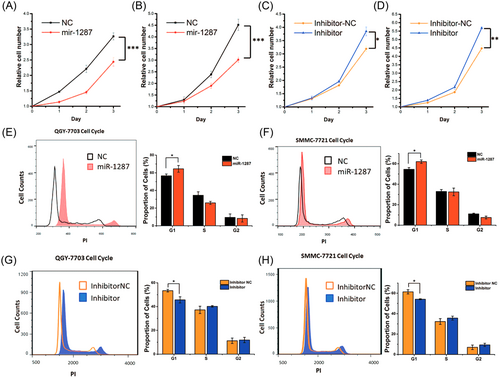
The overexpression of mir-1287 lead to the cell cycle arrest and resulted in the decay of proliferation. (A-D) The CCK-8 analyses of cell proliferation. (A, B) QGY-7703 and SMMC-7721 transfected with miR-1287 mimics and mimics negative control (NC). (C, D) QGY-7703 and SMMC-7721 transfected with miR-1287 inhibitor and inhibitor NC. The results were represented by mean ± SD (n = 3). *P < .05; **P < .01; ***P < .001. (E-H) The flow cytometer analyses of cell cycle of the QGY-7703 (E, G) and SMMC-7721 (F, H). Cells were transfected with miR-1287 mimics or inhibitors, 24 hours after cell cycle synchronization. The percentages of cell counts in different cell cycle stages were represented by mean ± SD (n = 3). *P < .05, in the histograms on the right columns. SD, standard deviation
3.4 Mir-1287 inhibits the migration and invasion of HCC cell lines
To determine the function of mir-1287 in tumor metastasis, we used transwell assays to test the migration and invasion of the QGY-7703 (Figure 4A,B) and SMMC-7721 (Figure 4C,D) tumor cells. The cells transfected with the miR-1287 mimics showed reduced migration and invasion. In contrast, transfection of the miR-1287 inhibitors led to enhanced cell migration and invasion.
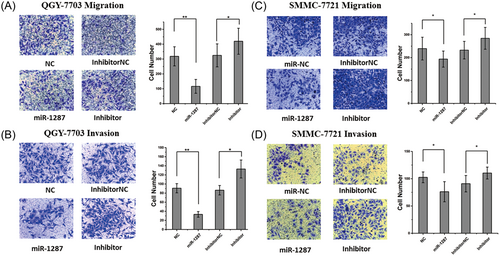
Upregulated mir-1287 induced suppression on migration and invasion in HCC cell lines. (A) The migration and (B) invasion analysis of QGY-7703 cells by transwell assay. The right column showed the histogram of the migratory and invasive cells. And the similar assay was conducted for the migration (C) and invasion (D) of SMMC-7721 cells. All the results were represented as mean ± SD (n = 3). *P < .05; **P < .01. HCC, hepatocellular carcinoma; SD, standard deviation
3.5 PIK3R3 is the specific target of miR-1287
To identify the direct target of mir-1287, a number of mRNAs, the expression levels of which are linked to HCC, have been predicted as targets of miR-1287 mediated regulation. We picked out 11 candidates, which were most relative with HCC, from 140 multiple predicted mRNAs. We further checked their levels and binding sites (see Supplementary Material Figure S2), and found one direct target PIK3R3, a regulatory subunit of PI3K complex, as well as a novel oncogene in many cancers including HCC.17-19
To validate the targeting site of mir-1287 on PIK3R3, we cloned the predicted miR-1287’s binding site on PIK3R3 onto the dual-luciferase report assay, and checked the luciferase level after transfection of miR-1287 mimics. A mutant on binding site was also designed as contrast. The luciferase decreased in the wild-type group, while the mutant group showed no significant difference (Figure 5A).
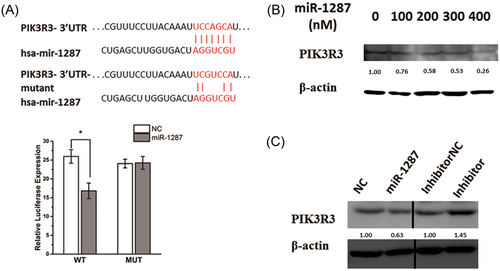
PIK3R3 is a direct target of miR-1287 in HCC. (A) Diagram of the predicted binding site, and the corresponding mutant site, for miR-1287 on the 3′-UTR of PIK3R3 gene. The dual-luciferase report assay was used for validating the target site, and the luciferase activity was showed in the histogram. The data were normalized and represented as mean ± SD (n = 3). *P < .05. (B, C) Western blot analysis of the PIK3R3 expression. SMMC-7721 cells were transfected with the concentration gradient of miR-1287 mimics (B), and either mimics or inhibitors (C), in contrast with their NCs, respectively
We further confirmed the protein level of PIK3R3 by Western blot. With the increased volume of miR-1287 transfected, the expression of PIK3R3 protein showed a decrease (Figure 5B). The expression of PIK3R3 was upregulated by the transfection of miR-1287 inhibitors (Figure 5C). These results confirmed that PIK3R3 is the direct target gene of miR-1287.
3.6 Overexpression of miR-1287 leads to the inhibition of PIK3R3 involving cell signaling pathways
We next aimed to determine whether PIK3R3 is the biological effectors of mir-1287, and the molecular mechanism underlying the mir-1287 regulation. PIK3R3 can directly bind with the Rb protein, an important regulator in cell cycle progression.20, 21 We defined the phosphorylation level of Rb protein is decreased by the increased amount of transfected miR-1287 mimics through Western blot (Figure 6A).
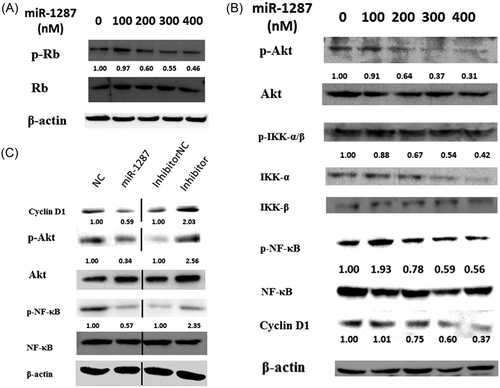
The signaling pathways downregulated by mir-1287. Western blot were used to analyze the phosphorylation or expression of (A) Rb and (B) AKT, IKK-α/β, NF-κB, and cyclin D1. SMMC-7721 cells were transfected with a concentration gradient of mir-1287 mimics. (C) The cells were transfected, with mir-1287 mimics or inhibitors, in contrast with their NCs, respectively. The density of each band was evaluated with Image J, and the data were showed beneath the brands
Furthermore, PIK3R3 is a regulatory subunit of PI3K complex, which activates PI3K/Akt pathway and the downstreaming NF-κB pathway.22 IKK-α/β links the activation of Akt and NF-κB. The NF-κB, when phosphorylated, transcriptionally regulates cyclin D1.
Western blot is used to check the phosphorylation levels of these proteins targeted by PIK3R3, including AKT, IKK-α/β, NF-κB, and cyclin D1. With the increased amount of transfected miR-1287 mimics, all the proteins presented loss of phosphorylation (Figure 6B), indicating that a cascade reaction occurred through downregulation of PIK3R3 in the PI3K/Akt and NF-κB pathway.
The transfection of miR-1287 inhibitors increased the amount of cyclin D1, and phosphorylation of Akt and NF-κB (Figure 6C).
3.7 Overexpression of PIK3R3 rescues the phenotypes aroused by miR-1287 in HCC cells
We further examine whether ectopic expression of PIK3R3 would rescue the tumor suppressive effects of miR-1287. We exogenously expressed the CDS sequences of PIK3R3 gene in both QGY-7703 and SMMC-7721 cell lines. After transfecting with miR-1287 mimics, both cells showed upregulated proliferation rates compared to those expressing empty vectors (Figure 7A,B). The transwell assay also showed increased invasion and migration in contrast to the cells without PIK3R3 ectopic expression (Figure 7C–F).
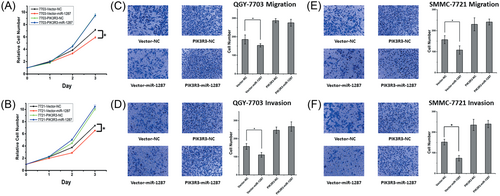
Overexpression of PIK3R3 rescues the proliferation, invasion and migration in HCC cells after miR-1287 transfection. (A, B) The CCK-8 analysis of the cell proliferation. The QGY-7703 and SMMC-7721 cells, which stably expressed the empty Vector or PIK3R3, were transfected with NC or miR-1287 mimics. (C-F) The transwell migration (C, E) and invasion (D, F) assays of PIK3R3 high expressing clones of QGY-7703 and SMMC-7721 cells. Cells were transfected with NC or miR-1287 mimics. Representative images were showed above the bar graphs. The results were represented by mean ± SD (n = 3). *P < .05; **P < .01; ***P < .001. SD, standard deviation
4 DISCUSSION
In this study, we found the tumor-suppressing function of mir-1287 in HCC cells. The overexpression of miR-1287 could lead to the cell cycle arrest and inhibition of cell proliferation. The migration and invasion were suppressed likewise. PIK3R3 gene was the novel target of miR-1287. Decreased level of PIK3R3, induced by miR-1287 overexpression, would suppress the activation of Rb protein, the PI3K/Akt pathway, and the phosphorylation of NF-κB. Thus, we demonstrate the mechanism of mir-1287 to suppress the HCC cells in Figure 8.
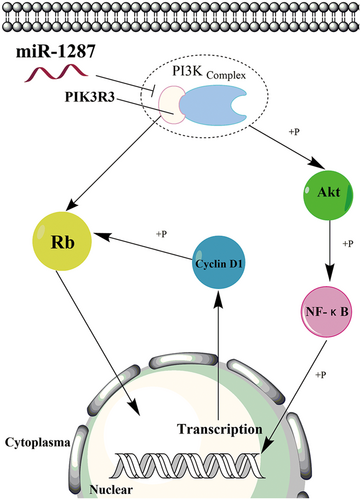
Model of miR-1287 molecular mechanism in HCC cells. The mature miR-1287 silences the PIK3R3 expression. Decrease of PIK3R3 directly downregulates the phosphorylation of Rb protein. Alternatively, decrease of PIK3R3 will interfere with the PI3K complex, downregulating the phosphorylation of Akt protein. Thus, the activation of NF-κB is also suppressed. Loss of NF-κB activation will also decrease the cyclin D1 level, suppressing the phosphorylation of Rb protein and resulting in the cell cycle arrest. HCC, hepatocellular carcinoma; NF-κB, nuclear factor κB
As is mentioned before, PIK3R3 is an oncogene, which promotes tumor growth or metastasis in many kinds of cancers. The interaction between PIK3R3 and Rb protein has been proved by Xia.21 The unique NH2 terminus of PIK3R3 can directly bind to some key proteins in cell cycle regulation, especially Rb protein.17, 23 PIK3R3 is also associated with the activation of class IA PI3K complex, as one of its regulatory subunits. Downregulation of PIK3R3 has been proved to suppress the Akt phosphorylation in many kinds of cancers.24-27 The NF-κB pathway is also suppressed due to PIK3R3 according to Wang’s report, where Akt phosphorylation is involved in this regulation.9, 10 NF-κB can regulate the transcription of cyclin D1 through binding to the site on its promoter.28 Cyclin D1 when combined with CDK4 or CDK6, can phosphorylate Rb protein to regulate the cell cycle at phase G1.29
These reports explain the functions of PIK3R3, and support our theory on the mechanism of mir-1287 in HCC cells. The suppression on HCC cells’ proliferation can be explained by the loss of Rb phosphorylation, induced by miR-1287 mediated PIK3R3 decrease. PIK3R3 directly binds to Rb protein or regulates its phosphorylation through PI3K/Akt, and NF-κB pathway might also be involved in this regulatory mechanism. The invasion and migration can also be suppressed by miR-1287 through the downregulation on PI3K/Akt pathway. Besides, upregulation of PIK3R3 in HCC cells could rescue the tumor suppressive functions caused by miR-1287. Thus, mir-1287 suppresses the proliferation, migration, and invasion of HCC cells, through directly targeting its biological effector PIK3R3.
PIK3R3 can also be suppressed by other miRNAs including miR-148b, miR-193a-3p, and miR-193a-5p, miR-181a2/181b2, miR-511, and miR-132.24, 26, 27, 30-32 Overexpression of these miRNAs can also decrease the level of PIK3R3, inactivate the PI3K/Akt pathway and suppress the tumor cells. Most of these research focus on the mTOR pathway to explain the downstreaming mechanism. In our study, we were trying to find a different way of regulation, other than the mTOR pathway, which was possibly mediated by the loss of phosphorylation of NF-κB aroused by the inactivation of PI3K/Akt pathway. The decrease of Rb phosphorylation is an alternative mechanism. These results imply a unique way of regulation for miR-1287 in HCC cells, which would broaden the view of miRNA regulation network in HCC cells. Furthermore, a combination of miR-1287 and other PIK3R3 targeting miRNAs could become a more effective and universal treatment of HCC.
Our study revealed a new mechanism of miRNA mediated suppression on HCC cells through targeting PIK3R3. However, miR-1287, like other miRNAs, is believed to target hundreds of other genes than PIK3R3. These genes apparently would have associated functions with PIK3R3 during tumorigenesis. We assume the further research on the targets of miR-1287 would help to understand the tumor promoting mechanism of PIK3R3 in HCC cells, and find new oncogenes as diagnostic and therapeutic targets.
ACKNOWLEDGMENTS
This study is granted by National Major Scientific and Technological Special Project of China, 2013ZX09102057.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHORS’ CONTRIBUTIONS
JL designed the study and wrote most of the paper. LT performed and analyzed the experiments in Figure 3. YX designed and performed the data analysis in Figure 1. KG and JH provided technical assistance and contributed to the preparation of the figures. MG revised and wrote part of the paper. JZ convinced the conception and revised the paper. QH convinced the conception and contributed all of the materials in this paper. All authors reviewed the results and approved the final version of the manuscript.