DNA-PKcs Associates With PLK1 and Is Involved in Proper Chromosome Segregation and Cytokinesis
ABSTRACT
Accurate mitotic regulation is as important as intrinsic DNA repair for maintaining genomic stability. It is believed that these two cellular mechanisms are interconnected with DNA damage. DNA-PKcs is a critical component of the non-homologous end-joining pathway of DNA double-stranded break repair, and it was recently discovered to be involved in mitotic processing. However, the underlying mechanism of DNA-PKcs action in mitotic control is unknown. Here, we demonstrated that depletion of DNA-PKcs led to the dysregulation of mitotic progression in response to DNA damage, which eventually resulted in multiple failures, including failure to segregate sister chromatids and failure to complete cytokinesis, with daughter cells becoming fused again. The depletion of DNA-PKcs resulted in a notable failure of cytokinesis, with a high incidence of multinucleated cells. There were also cytoplasmic bridges containing DNA that continuously connected the daughter cells after DNA damage was induced. Phosphorylated DNA-PKcs (T2609) colocalizes with PLK1 throughout mitosis, including at the centrosomes from prophase to anaphase and at the kinetochores from prometaphase to metaphase, with accumulation at the midbody during cytokinesis. Importantly, DNA-PKcs was found to associate with PLK1 in the mitotic phase, and the depletion of DNA-PKcs resulted in the overexpression of PLK1 due to increased protein stability. However, deficiency in DNA-PKcs attenuated the recruitment of phosphorylated PLK1 to the midbody but not to the kinetochores and centrosomes. Our results demonstrate the functional association of DNA-PKcs with PLK1, especially in chromosomal segregation and cytokinesis control. J. Cell. Biochem. 115: 1077–1088, 2014. © 2013 Wiley Periodicals, Inc.
Cytokinesis is the final stage of mitosis. It divides a cell into two daughter cells. This process is initiated in anaphase, during which the chromosomes move towards the spindle poles. A microtubule-associated protein complex is positioned at the cell equator with the formation of a spindle midzone, followed by the assembly and constriction of a cortical actomyosin-driven contractile ring. As the cleavage furrow constricts the central spindle, the midzone microtubules become highly compacted into a transient narrow intracellular bridge at the midbody, which is the last structure connecting the two daughter cells after division and the one that localizes the abscission [Glotzer, 2005; Otegui et al., 2005; Barr and Gruneberg, 2007].
The appropriate assembly of the spindle midzone, contractile ring and midbody is critical for the accurate proceeding of cytokinesis and genetic stability maintenance. The assembly and constriction of the contractile ring is coordinated by the Rho signaling pathway in yeast [Krause et al., 2012], Drosophila [Glotzer, 2005] and mammalian cells [Vithalani et al., 1996; Piekny et al., 2005; Bastos et al., 2012]. In Drosophila, activation of the GTPase RhoA is achieved with the localization of the Rho-specific guanine nucleotide exchange factor (RhoGEF) Pebble (mammalian ortholog Ect2) to the sites of the formation of the cleavage furrow [Prokopenko et al., 1999; Somers and Saint, 2003], which is mediated by the centralspindlin complex RacGAP50C (also known as RacGAP) and the microtubule-associated motor protein Pav-KLP/MKLP1. Pebble and RacGAP are directly associated, and this association is essential for the localization of Pebble to the equator and the subsequent activation of Rho signaling [Somers and Saint, 2003]. PLK1 is a member of the polo-like kinases family. The founding member, Polo, was originally identified in a Drosophila screen for mutations affecting spindle pole behavior [Sunkel and Glover, 1988]. PLK1 has been demonstrated to be a central regulator of mitosis from the mitotic entry to exit [van de Weerdt and Medema, 2006; Randall et al., 2007]. PLK1 is necessary for the interaction between RacGAP and the RhoGEF Pebble, playing a critical role in recruiting RhoGEFs to the central spindle and leading to the activation of RhoA [Prokopenko et al., 1999; Burkard et al., 2007]. Polo kinase can directly interact with the linker protein RacGAP through its intermediate domain [Ebrahimi et al., 2010]. The absence of PLK1 or the Drosophila ortholog Polo kinase disturbs the localization of Pebble/Ect2 and Pav-KLP at the equator and subsequent ring assembly, although the centralspindlin complex still localizes correctly [Prokopenko et al., 1999; Burkard et al., 2007; Ebrahimi et al., 2010]. During midbody formation, midzone proteins on microtubules are recruited to or excluded from the stem body. PLK1 is one of the proteins with this type of changing localization pattern [Hu et al., 2012]. PLK1 colocalizes with the key stem body protein KIF4 in the midzone but is gradually separated from KIF4 during furrow ingression. After furrow ingression, PLK1 relocalizes continuously throughout the stem body late in cytokinesis. The kinase activity of PLK1 is required to form and maintain the midbody and stem body during cytokinesis [Hu et al., 2012]. PLK1 usually interacts with other mitosis-regulating proteins within these subcellular structures, and it either activates or is activated by its interacting/docking partners via colocalization. For example, the interaction of PLK1 with the protein regulator of cytokinesis 1 (PRC1) and mitotic kinesin-like protein 2 (MKLP2) promotes the timely recruitment of PLK1 to the central spindle during anaphase [Neef et al., 2007]. In turn, PLK1 recruits RhoGEFs to the central spindle, leading to the activation of RhoA and the assembly of the actomyosin contractile ring [Burkard et al., 2007]. We recently found that PLK1 forms a ternary complex with TIP60 and cyclin B1 during the mitotic phase and colocalizes in the centrosome, where PLK1 is acetylated by TIP60 [Zhang et al., 2012]. The eIF4E-binding protein 1 (4E-BP1) was recently identified as another PLK1-interacting protein in the mitotic phase, and colocalization of phosphorylated 4E-BP1 at T37/46 with PLK1 was observed at the centrosomes during mitosis. Depression of 4E-BP1 resulted in an increased incidence of polyploidy and aberrant mitosis [Shang et al., 2012].
The DNA-dependent protein kinase catalytic subunit (DNA-PKcs), a member of the phosphatidylinositol 3-kinase-related kinase (PIKK) family, is a well-known critical component in the non-homologous end-joining pathway of double-stranded DNA break repair [Ma et al., 2005; Zhou, 2011]. DNA-PKcs was recently discovered to be important for the stabilization of centrosome and spindle structure and the regulation of mitotic progression under the stress of DNA damage induced by irradiation [Shang et al., 2010] or the normal cell cycle [Lee et al., 2011]. The abnormal mitotic manifestations in the cells generated by inactivating DNA-PKcs [Shang et al., 2010; Lee et al., 2011] are similar to these demonstrated in cells with a defective or dysregulated PLK1 [Mundt et al., 1997; Bruinsma et al., 2012; Tandle et al., 2013], but the precise mechanism of how DNA-PKcs participates in the mitotic control is unclear. One of the most striking features of PLK1 is its dynamic localization to various subcellular structures during mitotic progression, including centrosomes, kinetochores, the central spindle and the midbody [Takaki et al., 2008]. However, it is still important and valuable to answer questions such as how PLK1 is recruited to, and functions at, such multiple subcellular structures during mitotic progression and whether there are specific coregulators at different subcellular structures. In view of the similarity between the mitotic manifestations generated by inactivation of DNA-PKcs and the dysregulation of PLK1, here we focused on such question as whether a direct association in physics as well as function exists between DNA-PKcs and PLK1 and if so, what the biological significance of this association is. In the present study, we demonstrated that the phosphorylated DNA-PKcs interacts with PLK1 through colocalization throughout the mitotic phase. Inactivation of DNA-PKcs resulted in multiple disorders of mitotic progression, especially abnormal chromosome segregation and the failure of cytokinesis. Suppression of DNA-PKcs increased the stabilization of the PLK1 protein. Although the inactivation of DNA-PKcs did not attenuate the overall level of PLK1 phosphorylation in the mitotic phase or its localizations at the kinetochores and centromsomes, it did prevent the localization of phosphorylated PLK1 at the midbody. These observations revealed the physics and functional associations of DNA-PKcs with PLK1 in mitosis.
MATERIALS AND METHODS
CELL CULTURE AND IRRADIATION
HeLa, HeLa-NC, HeLa-H1 and ATM-deficient AT5BIVA cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin in a humidified incubator at 37°C in 5% CO2. HeLa-H1 and HeLa-NC were generated from HeLa cells by stably transfecting the cells with a DNA-PKcs-specific siRNA construct targeting the catalytic motif (nucleotides 11637–11655, H1) and a control siRNA construct (NC), respectively [Shang et al., 2010]. A cobalt-60 γ-ray source (Beijing Institute of Radiation Medicine) was used to irradiate the cells at a dose rate of 1.74 Gy/min at room temperature. After irradiation, the cells were collected either immediately or after a certain time in culture and then subjected to further experiments.
ANTIBODIES AND CHEMICALS
All of the antibodies used were commercially available. The following were used in this study: anti-phospho-DNA-PKcs (pT2609) (#19716, Rockland, Gilbertsville, PA), anti-phospho-DNA-PKcs (pT2647) (#ab61045, Abcam, Cambridge, UK), anti-phospho-DNA-PKcs (pS2056) (#ab18192, Abcam), anti-DNA-PKcs total (sc-9051, H163, Santa Cruz, CA), anti-Plk1 (#ab17057, Abcam), anti-phospho-Plk1 (T210) (#ab12157, Abcam), anti-α-Tubulin (#ZM-0438, Invitrogen, Carlsbad, CA), anti-HA antibody (#AB104-02, Beijing Tiangen Company, Beijing, China), anti-β-actin (I-19-R, Santa Cruz), anti-phospho-histone H3 (pS10) (Cell Signaling Technology), HRP-conjugated anti-rabbit IgG(H + L) (ZB-2301, Zhongshan, Beijing, China), and HRP-conjugated anti-mouse IgG(H + L) (ZB-2305, Zhongshan). Nu7026, nocodazole, and cycloheximide (CHX) were purchased from Sigma (St. Louis, MO).
PLASMIDS
The PLK1 coding sequence was amplified by PCR from an expressed sequence tag (EST) clone (GenBank™ accession no. BE 900300, obtained from Research Genetics) and cloned into the pCMV-HA and pGEx-4T vectors to generate pCMV-HA-Plk1 and pGEx-4T-Plk1.
CELL CYCLE ANALYSIS BY FLOW CYTOMETRY
After γ-irradiation or treatment with NU7026 (10 µM), the cells were harvested either immediately or at the indicated times post-treatment and fixed with 75% ethanol. The cells were resuspended in PBS plus 0.1% saponin and 1 µg/ml RNase A (Sigma), incubated for 20 min at 37°C, and stained with 25 µg/ml propidium iodide (PI) (Sigma). The cell cycle distribution was evaluated by flow cytometry, counting more than 10,000 cells per sample. For the detection of mitotic cells, the fixed cells were treated with 0.25% Triton X-100 in PBS for 15 min, stained with 10 mg/ml of FITC-conjugated anti-phospho-histone H3 (pS10) antibody for 1 h at room temperature in the dark, and then stained with propidium iodide (20 mg/ml). Cellular fluorescence was measured using a FACSCalibur flow cytometer (BD Pharmingen, USA). Two-dimensional dot plots were generated using ModFit LT software.
CONFOCAL IMMUNOFLUORESCENCE MICROSCOPY
Cells were grown on poly-D-lysine-coated culture slides (BD Pharmingen), washed in PBS, fixed in PBS containing 4% or 0.5% paraformaldehyde (PFA) for 15 min, and permeabilized in Triton buffer (0.5% Triton X-100 in PBS). The fixed cells were blocked in blocking solution (2% bovine serum albumin, 0.1% Tween, PBS) for 30 min at 37°C in a humidified chamber. Immunostaining was performed using an anti-phospho-DNA-PKcs (pT2609, pT2647, or pS2056) antibody (1:100), an anti-Plk1 antibody (1:200), an anti-α-tubulin antibody (1:200), or a mouse/rabbit anti-γ-tubulin antibody (1:1,000) for 2 h at room temperature in a humidified chamber and then washed three times in blocking buffer. The cells were incubated with anti-mouse/rabbit Rhodamine (Rd) (1:200) and anti-mouse/rabbit FITC (1:200) secondary antibodies. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) in mounting solution. Confocal immunofluorescence microscopy observation was performed using an LSM 510 laser-scanning confocal microscope (Zeiss, Oberkochen, Germany).
TRANSIENT TRANSFECTION OF PLASMIDS
Cells (3–5 × 105) were seeded in a 60-mm dish, and 24 h later, plasmids transfections were performed using Lipofectamine 2000, as recommend by the manufacturer (Invitrogen). Briefly, 20 µl of Lipofectamine 2000 was added to 500 µl of DMEM without antibiotics and serum and incubated at room temperature for 5 min (solution A). The plasmids (8 µg) were added to 3.5 ml of DMEM without antibiotics and serum (solution B). Solution A and solution B were mixed and incubated at room temperature for 20 min. The medium in the cell culture was removed, 4 ml of Lipofectamine 2000/plasmid DNA mixture was added to each culture well, and the cells were gently mixed. Forty-eight hours post-transfection, transfected cells were harvested for further experiments.
GST PULL-DOWN ASSAY
Glutathione Sepharose beads (GE Healthcare, CA) bound to recombinant GST-Plk1 or GST alone (as a negative control) were mixed with HeLa cell extracts and rocked for 2 h at 4°C. After washing, the proteins were eluted using 20 mM glutathione/50 mM Tris, pH 8.0. A Western blot was performed on the extracts used for the pull-down assays and the eluted fractions to identify the bound proteins.
CO-IMMUNOPRECIPITATION AND IMMUNOBLOTTING ASSAY
For general cell lysis and co-immunoprecipitation (Co-IP) of DNA-PKcs, PLK1 and HA, one experiment used an anti-Plk1 antibody or an anti-DNA-PKcs antibody to co-immunoprecipitate DNA-PKcs or PLK1 from HeLa cell extracts. Another experiment used an anti-HA antibody to co-immunoprecipitate DNA-PKcs from the extracts of HEK293T cells transfected with HA-Plk1 vectors or the empty vector. The co-immunoprecipitations were performed using the Immunoprecipitation Kit (Protein A/G, Roche Molecular Biochemicals) according to the manufacturer's instructions, as described previously [Shang et al., 2010]. For immunoblotting, the above co-IP products were denatured, resolved by SDS–PAGE, and subjected to Western blot analysis. Otherwise, the cells were harvested and washed twice in ice-cold PBS. Cell pellets were treated with lysis buffer as indicated above, with one protease inhibitor cocktail tablet in a 50 ml solution, and the total protein was isolated. The protein (50 µg) was resolved using SDS–PAGE (8%) and then transferred onto a polyvinylidene fluoride (PVDF) membrane for Western blot analysis.
FLUORESCENCE TIME-LAPSE IMAGING
To analyze the dynamics of mitosis in DNA-PKcs-deficient cells following 4 Gy irradiation, the culture dish was incubated in a chamber with 5% CO2 for the indicated period of time and placed on the stage of an inverted Andor Revolution xD Live cell confocal fluorescence microscope equipped with a Yokogawa CSU-X1 spinning disk confocal scanner, iXON EM CCD camera (512 × 512 pixels), and 488 and 561 nm laser lines. To facilitate the in vivo time-lapse analysis, DNA-PKcs-deficient cells were co-transfected with H2B-GFP and HSPC300-RGP expression plasmids to visualize the nuclear and cell morphology, respectively. Twenty-four hours post-transfection, the DNA-PKcs-deficient cells were irradiated as described above, and imaged 24 h post-irradiation. Five different fields of view were recorded at 60× magnification every 2 min for 20 h. The images from the z-stacks were combined in a maximum projection, and the time series of image projections was exported to a time-lapse video with 8 frames/s. The videos and still images taken from the videos were processed using Andor IQ confocal software.
ANALYSIS OF PLK1 STABILITY
HeLa-H1, HeLa-NC, and NU7026-pretreated HeLa-NC cells were treated with 40 µg/ml cycloheximide (CHX) (Sigma) at 37°C to block novel protein synthesis. The cells were harvested at the indicated times after CHX treatment, and subjected to the immunoblotting analysis with the anti-PLK1 antibody. To test the effect of inactivating DNA-PKcs on the stability of PLK1, HeLa-NC cells were pretreated with 10 µM NU7026 for 2 h to inhibit the DNA-PKcs activity and then subjected to the CHX treatment concomitant with NU7026 treatment.
RESULTS AND DISCUSSION
DNA-PKcs IS REQUIRED FOR APPROPRIATE MITOTIC PROGRESSION IN RESPONSE TO DNA DAMAGE
We have recently shown that shRNA-mediated suppression of DNA-PKcs resulted in a prolonged G2-M blockage of HeLa cells following ionizing radiation (IR)-induced DNA damage, along with a significantly increased occurrence of multipolar spindles and polyploidy cells [Shang et al., 2010]. Even without DNA damage stress, the suppression of DNA-PKcs also led to a significantly delayed transition of prometaphase-to-anaphase transition under normal growing conditions [Lee et al., 2011]. To verify the role of DNA-PKcs on the IR-induced G2/M boundary arrest and mitotic arrest, the cells were analyzed by flow cytometry after immunostaining using an antibody against phospho-histone H3/Ser10 (pH3), a known molecular marker for mitotic cells [Juan et al., 1998]. As shown in Figure 1A,B, the frequency of pH3-positive cells in both DNA-PKcs-deficient HeLa-H1 cells and control HeLa-NC cells was dramatically reduced 2 h after 4 Gy irradiation, suggesting that an early G2/M boundary arrest was induced independent of the presence of DNA-PKcs. However, the percentage of pH3-positive cells was higher in HeLa H1 cells than in HeLa-NC cells 4 h post-irradiation, suggesting that the G2/M arrest in DNA-PKcs deficient cells was released earlier in HeLa H1 cells than that in control cells. At 8–12 h post-irradiation, G2/M arrest in control HeLa-NC cells was also released, as demonstrated by the increased frequency of pH3-positive cells. However, the frequency of H3-positive cells in HeLa-NC cells was significantly higher than in HeLa-H1 cells, which might be due to efficient transient spindle-checkpoint arrest in normal cells. However, 24 h post-irradiation, the frequency of mitotic phase cells returned to the normal level in HeLa-NC cells, while it remained at a high level in HeLa-H1 cells, which indicated prolonged mitotic arrest in DNA-PKcs deficient cells.
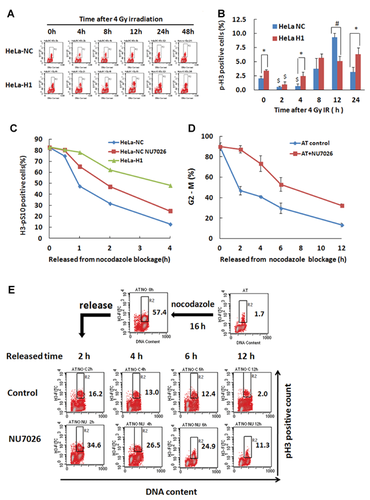
To further characterize the phenotype of the prolonged mitotic arrest, the cells were blocked at metaphase using nocodazole and then released for the detection of mitotic progression. As shown in Figure 1C, the control HeLa-NC cells progressed more quickly than HeLa-H1 cells and HeLa cells treated with the DNA-PKcs specific inhibitor NU7026.
ATM is another member of the PIKK family, sharing many common targets with DNA-PKcs. It was reported that a deficiency of DNA-PKcs led to the downregulation of ATM [Peng et al., 2005]. To confirm the dependency of the mitotic progression on DNA-PKcs, we further investigated the mitotic progression of ATM-deficient AT5BIVA cells after release from a nocodazole-induced metaphase blockage. As shown in Figure 1D, the release of the cells from mitotic arrest was largely attenuated by inactivating DNA-PKcs. Furthermore, the p-H3 antibody-labeled mitotic cell population also displayed a decreased release of mitotic arrest caused by the inactivation of DNA-PKcs (Fig. 1E). Taken together, these data indicate that DNA-PKcs deficiency could lead to premature release of the G2/M arrest and persistent mitotic arrest in response to DNA damage.
INACTIVATION OF DNA-PKcs LEADS TO ABNORMAL CHROMOSOME SEGREGATION AND CYTOKINESIS FAILURE
A delay in the prometaphase-to-anaphase transition was detected following the inactivation of DNA-PKcs [Lee et al., 2011], which partially contributed to the accumulation of mitotic cells. To obtain further information regarding the delay in mitotic progression, we used time-lapse video microscopy to monitor the mitotic progression of cells following 4 Gy irradiation. A representative cell with normal mitotic entry and daughter cell division, which finished within 120 min, is shown in Figure 2A-I. In contrast, we found that there were multiple cell phenotypes underlying abnormal mitotic progression associated with defective chromosome segregation in DNA-PKcs-deficient HeLa-H1 cells following irradiation. Some cells entered mitosis but underwent inadequate chromosome condensation during metaphase, which resulted in an anaphase bridge, lagging chromosomal material, and cell death (Fig. 2A-II). Alternatively, cells initiated mitosis and chromosome condensation, but were unable to segregate sister chromatids to drive the chromosomes towards the spindle poles (Fig. 2A-III). Finally, some cells entered mitosis, promoted chromosome condensation, segregated the sister chromatids, and initiated defective cytokinesis, which eventually resulted in two daughter cells remaining connected (bridged) and fusing to form multinucleated cells (Fig. 2A-IV).
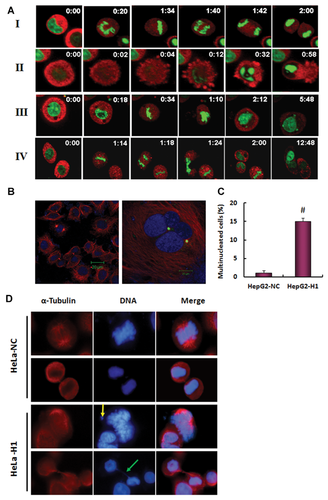
Multinucleation is a typical phenotype of failure of cytokinesis, which was also displayed in DNA-PKcs-inactivated HeLa cells post-irradiation [Shang et al., 2010]. A DNA-PKcs-depleted HepG2 cell line was generated by shRNA-targeting of DNA-PKcs, and a significantly increased occurrence of multinucleated cells was observed even under normal growing conditions (Fig. 2B,C). The defect in abscission of the cleavage furrow was considered to be attributable to various effects, one of which is continued cell division of the resultant daughter cells connected by the cytoplasmic bridge, with consequent cell death or fusion leading to formation of multinucleated cells [Huang et al., 2008]. In addition to the fusion of daughter cells (Fig. 2A-VI) and multinucleation phenotypes (Fig. 2B,C) [Shang et al., 2010], we also detected free unattached DNA in metaphase cells (Fig. 2D, yellow arrow) and the cytoplasmic DNA-containing bridges connecting the daughter cells, which were a major manifestation of abnormal chromosome segregation (Fig. 2D, green arrow). Thus, the cytoplasmic DNA bridges linking the adjacent nuclei could be a dominant cause of the failure of cytokinesis as a consequence of DNA-PKcs inactivation.
PHOSPHORYLATED DNA-PKcs COLOCALIZES WITH PLK1 THROUGH MITOTIC PHASE
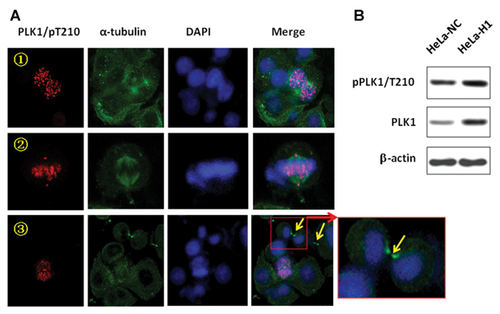
It is clear that DNA-PKcs deficiency leads to the dysregulation of mitotic progression in response to DNA damage, which eventually results in a series of phenotypes of mitotic failure. This finding suggests that DNA-PKcs is an important regulator in the overall progression of mitosis, especially following DNA damage. The phosphorylated DNA-PKcs/Thr2609 was shown to localize to both centrosomes and kinetochores during prometaphase and completely overlapped PLK1 staining [Lee et al., 2011] and phosphorylated CHK2/Thr68 [Shang et al., 2010]. We further determined the subcellular colocalization of DNA-PKcs with PLK1 throughout mitotic progression. PLK1 is a well-known critical regulator of cell division, and the most striking feature of PLK1 is its dynamic localization to various subcellular structures during mitosis [Takaki et al., 2008]. PLK1 initially associates with centrosomes during prophase, accumulates at the kinetochores in prometaphase and metaphase, is recruited to the central spindle in anaphase, and finally becomes enriched in the midbody during cytokinesis. DNA-PKcs was previously found to localize to the centrosome [Zhang et al., 2007; Shang et al., 2010; Lee et al., 2011]. Colocalization of phosphorylated DNA-PKcs (T2609) with PLK1 was clearly observed when an antibody specific for phosphorylated DNA-PKcs (T2609) was used for immunostaining HeLa cells. As shown in Figure 3A, the phosphorylated DNA-PKcs (T2609) protein displayed specific colocalization with PLK1 at various subcellular structures through mitotic progression, including at centrosomes from prophase to anaphase and at kinetochores in prometaphase and metaphase, with accumulation at the midbody when the cell was approaching the end of cytokinesis. We detected similar colocalization of DNA-PKcs/pT2609 and PLK1 in A549 cells (data not shown). The midbody localization of DNA-PKcs/pT2609 was further demonstrated by co-immunostaining with antibodies against phospho-DNA-PKcs (T2609) and α-tubulin (Fig. 3B). However, as the cells approached the end of cytokinesis, the phospho-DNA-PKcs (T2609) at the midbody did not colocalize with α-tubulin. Instead, phospho-DNA-PKcs (T2609) was localized adjacent to and between the α-tubulin proteins; in other words, phospho-DNA-PKcs was flanked by α-tubulin in the central part of the midbody structure (Fig. 3B, red arrow).
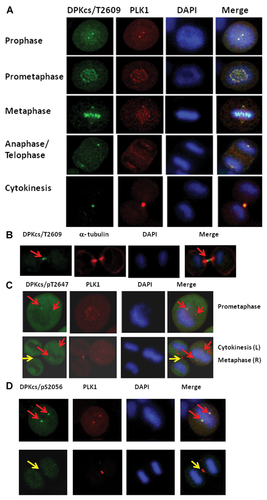
Although DNA-PKcs phosphorylated at T2647 (Fig. 3C, red arrows) and S2056 [Lee et al., 2011] (Fig. 3D, red arrows) was found at the centrosomes, there was no obvious localization at the midbody (Fig. 3C,D, yellow arrows). This result suggests that different phosphorylated forms/sites of DNA-PKcs may have different localization features and biochemical functions. To exclude the potential cross-reaction of the phospho-specific antibodies at the mitotic structures, the antibodies against phospho-DNA-PKcs/T2609/T2647/S2056 were tested in the glioma-derived DNA-PKcs deficient cell line M059-J. There was no specific immunostaining signal at the mitotic structures for the phosphorylated DNA-PKcs antibodies used in this study (Supplementary Fig. S1).
DNA-PKcs INTERACTS WITH PLK1 IN MITOTIC PHASE
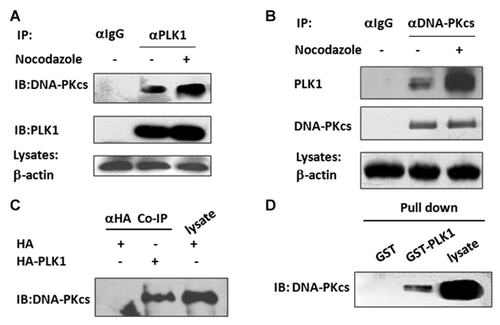
Because it is an important mitotic regulator, dysregulation of PLK1 can lead to a series of phenotypes of mitotic failure, including the failure of the bipolar spindle assembly, the formation of multicentrosomes, the development of multinucleated cells, and failure in cytokinesis [Garcia-Alvarez et al., 2007; Lenart et al., 2007; Archambault and Carmena, 2012]. The depletion or mutation of the PLK1-associated proteins, for example, Ataxin-10 [Li et al., 2011], and 4E-BP1 [Shang et al., 2012], has been shown to lead to similar fates induced by inactivation of PLK1. DNA-PKcs depletion resulted in similar phenotypes. The co-localization of DNA-PKcs with Thr2609 phosphorylation and PLK1 at centrosomes/kinetochores during mitosis and at the midbody during cytokinesis indicates that there is a functional connection between DNA-PKcs and PLK1 kinase. To test this hypothesis, mitotic cells were collected by shaking off the prometaphase HeLa cells that were synchronized using nocodazole. The asynchronous and M-phase synchronized HeLa cell lysates were then subjected to co-immunoprecipitation analysis. As shown in Figure 4A, DNA-PKcs was precipitated by anti-PLK1 antibodies. Conversely, PLK1 was precipitated by the anti-DNA-PKcs antibody (Fig. 4B). Their interaction was significantly enhanced in mitotic phase cells. These results consistently indicate a functional connection between DNA-PKcs and PKL1 during cell cycle progression through mitosis. We then transfected a HA-PLK1 expression vector or an HA empty vector into HEK293T cells and performed immunoprecipitation using an antibody against HA. DNA-PKcs co-immunoprecipitated with the recombinant HA-PLK1 expressed in cells, but not in cells transfected with the empty vector (Fig. 4C). We further performed glutathione S-transferase (GST) pull-down assays using recombinant GST-PLK1. A direct physical interaction was observed between the DNA-PKcs and GST-PLK1, but not with GST alone (Fig. 4D). The interaction of DNA-PKcs with PLK1 has also been detected by Benjamin P.C. Chen's group (Division of Molecular Radiation Biology, Department of Radiation Oncology, University of Texas Southwestern Medical Center at Dallas, USA) (communication). These results indicate that there is a direct interaction between DNA-PKcs and PLK1 both in vitro and in vivo. The results confirmed the specificity of the in vitro protein-protein interaction analysis and demonstrated the direct interaction between PLK1 and DNA-PKcs.
DNA-PKcs DEFICIENCY LEADS TO INCREASED STABILITY OF THE PLK1 PROTEIN
PLK1 is a multifunction mitotic regulator, and its protein level is highly regulated throughout the cell cycle, with its peak level in mitotic phase and a decline upon mitotic exit. However, little is known about the regulation of PLK1 protein stability except that it is degraded upon mitotic exit [Lindon and Pines, 2004]. As shown in Figure 5A, the constitutive level of PLK1 in DNA-PKcs depleted HeLa H1 cells was higher than that in the control HeLa-NC cells. There was an increased level of PLK1 in HeLa-NC cells at 8–12 h after 4 Gy irradiation. However, this IR-induced increase in PLK1 protein in HeLa-H1 cells continued until at least 24 h post-irradiation. It has also been reported that inactivation of DNA-PKcs by 6-bromine-5-hydroxy-4-methoxybenzaldehyde resulted in an increased level of phospho-PLK1 (T210) [Zhang et al., 2011].
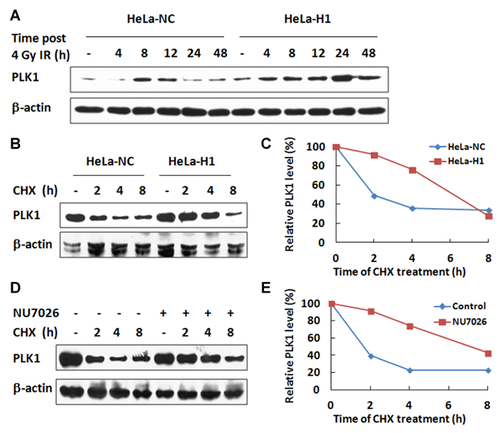
To determine how the depletion of DNA-PKcs led to increased levels of PLK1, we first quantified the mRNA expression level by quantitative real-time PCR and found that depletion of DNA-PKcs did not affect the transcription of the PLK1 gene (data not shown). We then investigated the stability of the PLK1 protein. The kinetics of PLK1 degradation was measured in HeLa cells after blocking protein synthesis with cycloheximide (CHX). We compared the PLK1 protein levels between DNA-PKcs-depleted HeLa H1 cells and the control HeLa-NC cells at 0, 2, 4, and 8 h after CHX treatment (Fig. 5B). At 2 h after CHX treatment, the PLK1 protein level decreased approximately 50% in control HeLa-NC cells but only approximately 10% in HeLa-H1 cells. PLK1 levels were reduced to approximately 40% and reached the lowest level in control HeLa-NC cells at 4 h after CHX treatment. The lowest level was not observed until 8 h after CHX treatment in the DNA-PKcs depleted HeLa-H1 cells. A similar alteration in the pattern of PLK1 degradation kinetics was also found after inactivating DNA-PKcs in HeLa cells through the specific inhibitor NU7026 (Fig. 5C). Clearly, DNA-PKs depletion resulted in increased stability of PLK1. Cell cycle analysis was performed for HeLa-H1 cells and the control HeLa-NC cells after CHX treatments. There was no difference in the cell cycle distributions (Supplementary Fig. S2), indicating that the increased stability of PLK1 caused by DNA-PKcs inactivation is not due to mitotic arrest.
Dysregulation of PLK1 expression leads to various defects in cell cycle progression, which is attributed not only to decreased expression but also to the overexpression of PLK1. Inappropriate levels (higher or lower) of PLK1 prevent the proper execution of cytokinesis. Overexpression of PLK1 has frequently been found in various cancers [Yamamoto et al., 2006; Zhao et al., 2010; Liu et al., 2012], and tumors with overexpressed PLK1 were shown to correlate more closely with the centrosome amplification, DNA aneuploidy, and chromosomal instability [Yamamoto et al., 2006]. Another study indicated that cells overexpressing wild-type PLK1 were able to enter mitosis and establish an apparently normal bipolar spindle, while progression through mitosis was delayed and cytokinesis appeared to be disturbed, reflected by a significant increase in large cells with a multinucleated phenotype [Mundt et al., 1997]. This appearance is similar to the defects induced by the inactivation of DNA-PKcs which is accompanied with overexpression of PLK1.
In addition to abnormal cell cycle progression, dysregulation of PLK1 expression also leads to other adverse effects, especially in tumorigenesis. PLK1 overexpression frequently occurs in many tumors as an early event in malignant transformation and cancer progression, which correlates with tumor progression, and prognosis [Ito et al., 2004; Feng et al., 2009; Lu and Yu, 2009], and/or plays an antiapoptotic role by regulating survivin [Feng et al., 2009]. The direct impact of PLK1 has been demonstrated by PLK1 overexpression, which has been shown to transform NIH3T3 fibroblasts into oncogenic foci in soft agar and lead to tumor formation when injected into nude mice [Smith et al., 1997]. Tumorigenesis-related genomic instability has been well documented as a phenotype of DNA-PKcs defects. Modulation of PLK1 protein stability provides new insight into and a molecular basis for the anti-oncogenic effect of DNA-PKcs beyond its maintenance of genomic stability.
LOCALIZATION OF PLK1 AT THE MIDBODY IS ATTENUATED BY DEPLETING DNA-PKcs
Finally, the effect of DNA-PKcs depletion on the subcellular localization of phospho-PLK1 (pT210) was investigated. As shown in Figure 6A, the localization of PLK1 at the kinetochores and centrosomes was clearly demonstrated in DNA-PKcs-depleted HeLa H1 cells, although these structures primarily contained supernumerary spindle poles. Normally, the localization of PLK1 at the midbody was clearly observed (Fig. 3A,C,D). However, the depletion of DNA-PKcs resulted in the attenuation of PLK1 recruitment to the midbody (Fig. 6A, yellow arrow). Even in the same microscopic display, the localization of PLK1 was very clear at the other nucleus subcellular apparatus, for example, kinetochores, but was barely seen at the midbodies. Depletion of DNA-PKcs did not attenuated the overall PLK1 phosphorylation level (Fig. 6B). The loss of PLK1 localization at midbody could be another cause for the cytokinesis failure displayed in DNA-PKcs inactivated cells.
CONCLUSION
Dysregulation of mitosis and cytokinesis defects have been suggested to result in the generation of cells with a phenotype of genetic instability, which predisposes them to oncogenic transformation. In the present study, we have further determined that phosphorylated DNA-PKcs (T2609) and PLK1 colocalize throughout mitosis, from entry to cytokinesis. The depletion of the DNA-PKcs resulted in a series of defects associated with abnormal chromosome segregation and cytokinesis failure in response to DNA damage, which were very similar to defects observed following the inactivation and even overexpression of PLK1 [Mundt et al., 1997]. Mechanistic studies demonstrate that DNA-PKcs associates with PLK1. Inactivation of DNA-PKcs increases the stability of the PLK1 protein and attenuates its localization at the midbody. DNA-PKcs is a well-known critical component of the double-stranded DNA break repair pathway, and here, we demonstrated that DNA-PKcs has another important role in the regulation of mitotic chromosome segregation and cytokinesis following DNA damage. This knowledge may contribute to our understanding of the molecular mechanisms that couple mitotic progression to the DNA damage response and repair pathway.
ACKNOWLEDGEMENTS
The authors thank Professor J.H. Hendry (Manchester University, UK) for helpful suggestions and editorial assistance with this paper. This work was supported by the Chinese National Natural Science Foundation (Grant 81071678 and 81272994), the Distinguished Youth Scientist Foundation of NFSC, China (Grant 30825011), and the Lotus Scholars Program of Hunan Provincial People's Government.




