Aldolase sequesters WASP and affects WASP/Arp2/3-stimulated actin dynamics
Abstract
In addition to its roles in sugar metabolism, fructose-1,6-bisphosphate aldolase (aldolase) has been implicated in cellular functions independent from these roles, termed “moonlighting functions.” These moonlighting functions likely involve the known aldolase–actin interaction, as many proteins with which aldolase interacts are involved in actin-dependent processes. Specifically, aldolase interacts both in vitro and in cells with Wiskott–Aldrich Syndrome Protein (WASP), a protein involved in controlling actin dynamics, yet the function of this interaction remains unknown. Here, the effect of aldolase on WASP-dependent processes in vitro and in cells is investigated. Aldolase inhibits WASP/Arp2/3-dependent actin polymerization in vitro. In cells, knockdown of aldolase results in a decreased rate of cell motility and cell spreading, two WASP-dependent processes. Expression of exogenous aldolase rescues these defects. Whether these effects of aldolase on WASP-dependent processes were due to aldolase catalysis or moonlighting functions is tested using aldolase variants defective in either catalytic or actin-binding activity. While the actin-binding deficient aldolase variant is unable to inhibit actin polymerization in vitro and is unable to rescue cell motility defects in cells, the catalytically inactive aldolase is able to perform these functions, providing evidence that aldolase moonlighting plays a role in WASP-mediated processes. J. Cell. Biochem. 114: 1928–1939, 2013. © 2013 Wiley Periodicals, Inc.
Fructose-1,6-bisphosphate (Fru 1,6-P2) aldolase (aldolase) is well characterized in its roles in glycolysis, gluconeogenesis, and fructose metabolism, catalyzing the reversible cleavage of carbon-carbon bonds in these processes. Recently, aldolase been implicated in other cellular functions that do not involve catalysis. These “moonlighting functions” of aldolase include roles in signal transduction [Koppitz et al., 1986; Orosz et al., 1988a; Baron et al., 1995; Kim et al., 2002; Singh et al., 2004; Ishida et al., 2005], vesicle trafficking [Kao et al., 1999; Lu et al., 2001, 2004, 2007; Lundmark and Carlsson, 2004; Benziane et al., 2007; Merkulova et al., 2011], and cell motility [Buscaglia et al., 2003, 2006; Jewett and Sibley, 2003; Ritterson Lew, 2012]. Furthermore, aldolase is a known F-actin binding protein, both in vitro and in vivo [Pagliaro and Taylor, 1992; Wang et al., 1996, 1997; Schindler et al., 2001], which is likely important for its moonlighting functions. Although the exact amino acid residues mediating this interaction are unknown, mutations that affect aldolase enzymatic activity do not necessarily affect its ability to bind F-actin, and mutations that diminish F-actin binding do not necessarily affect enzymatic activity [Wang et al., 1996].
The best-defined moonlighting function of aldolase is its binding to the thrombospondin-related Apicomplaxan protein (TRAP) family of transmembrane proteins found in parasites responsible for diseases such as malaria and toxoplasmosis (as reviewed in [Sibley, 2003]). Aldolase forms a bridge between the actin cytoskeleton and TRAP, which allows these parasites to move and infect their hosts [Jewett and Sibley, 2003]. The minimal sequence of the binding site for aldolase on TRAP has been defined [Buscaglia et al., 2003], and contains a span of acidic amino acids encompassing a tryptophan residue. A bioinformatics search using variations of this sequence found other proteins that may interact with aldolase [Buscaglia et al., 2006], including the Wiskott–Aldrich Syndrome protein (WASP) family (WASP and N-WASP), and the related WASP-family Verprolin-homologous (WAVE) proteins (WAVE-1, WAVE-2, and WAVE-3), which regulate actin polymerization and actin-related cell motility.
WASP is expressed in the hematopoetic system, while N-WASP is expressed ubiquitously in mammals [Millard et al., 2004]. WASP and N-WASP are structurally similar multidomain proteins. When inactive, they exist in an autoinhibited state in the cell. Upon activation by cdc42 and PIP2, the C-terminus is exposed, and WASP can interact with targets such as the Arp2/3 complex, a seven-membered protein complex responsible for stimulating branching polymerization of actin [for review, see Higgs and Pollard, 2001]. The C-terminal WA domain of WASP, consisting of the WASP Homology 2, central, and acidic regions of the protein (see Fig. S1 in Supplementary Material), is both necessary and sufficient for activation of the Arp2/3 complex [Higgs et al., 1999]. Direct binding of aldolase at the acidic portion of the WA domain has been confirmed by GST-pulldown, immunoprecipitation, and X-ray crystallography [Buscaglia et al., 2006; St-Jean et al., 2007]. Using cell lysates, it has been shown that aldolase only binds to the activated form of the WASP protein, when its C-terminus is exposed [Buscaglia et al., 2006]. Intriguingly, the C-terminal acidic domain of WASP is also responsible for binding to the Arp2/3 complex [Rohatgi et al., 1999].
Although the binding of aldolase on WASP is well defined, the function of the interaction remains unknown. In this report, the mechanism by which aldolase modulates actin-cytoskeleton dynamics through its interactions with WASP is explored. The effect of wild type aldolase and aldolase variants was tested in in vitro pyrene-actin polymerization assays stimulated with a glutathione-S-transferase (GST)-tagged WA domain (WA, see Fig. S1) and Arp2/3. It was found that aldolase inhibits WASP/Arp2/3-mediated branching polymerization in vitro by interaction with WASP. The consequence of this interaction in cells was investigated using RNAi. Knockdown of aldolase in NIH-3T3 cells caused a decrease in the rates of cell motility and cell spreading, two processes that involve WASP-mediated actin polymerization [Jimenez et al., 2000; Misra et al., 2007], both of which were rescued by expression of exogenous aldolase. Aldolase variants deficient in actin binding or enzymatic activity showed that these effects on WASP-mediated processes were dependent on moonlighting functions, not the catalytic activity, of aldolase.
MATERIALS AND METHODS
Protein Purification
Actin was purified from rabbit muscle acetone powder using previously published methods [Pardee and Spudich, 1982]. Actin was flash frozen in liquid nitrogen and stored in G-buffer (5 mM Tris pH 8.0, 0.3 mM CaCl2, 0.2 mM ATP, 2 mM dithiothreitol [DTT]) until used. Recombinant wild-type rabbit aldolase A and its variants were purified following previously published procedures [Morris and Tolan, 1993].
Pyrene-Labeling of Actin
G-actin was polymerized by addition of salts (100 mM KCl, 2 mM MgCl2), then pelleted (Beckman TLA 100.4, 150,000g, 80 min), and depolymerized by dialysis against G-buffer without DTT (three buffer changes over 48 h). After dilution to 1 mg/ml, actin was repolymerized and labeled by addition of sevenfold molar excess N-(1-pyrene) iodoacetamide (pyrene) in dimethylformamide at room temperature for 24 h. Pyrene labeled actin was centrifuged (150,000g, 80 min), redissolved in G-buffer, and then dialyzed versus G-buffer as described above to remove any excess free pyrene. Labeled G-actin was centrifuged (150,000g, 80 min) and passed over a PD-10 column equilibrated with G-buffer to remove any remaining F-actin.
In Vitro Actin Polymerization Assay
Actin (pyrene-labeled and unlabeled) was dialyzed overnight in G-buffer and centrifuged at 90,000 rpm to remove any residual F-actin. Pyrene-labeled actin was added to unlabeled actin to attain 12% labeled actin. Polymerization reactions were started by adding 10× initiation buffer (20 mM MgCl2, 10 mM EGTA pH 8.0, 5 mM ATP) to 12% pyrene-labeled actin to a final concentration of 1×. Other components or similar volumes of appropriate buffers were added to the reaction prior to initiation if necessary. When necessary, WA and Arp2/3 (gifts from Dr. Defne Yarar, Whitehead Institute) were diluted to the desired concentration into control buffer (20 mM HEPES pH 7.4, 100 mM KCl, 1 mM EDTA pH 8.0, 1 mM EGTA pH 8.0, 2 mM MgCl2, 0.5 mM DTT, 10% glycerol). Aldolase was diluted in G-buffer. Polymerization was immediately monitored by reading fluorescence emission over time using a Tecan infiniTE platereader (excitation wavelength 366 nm, emission wavelength 407 nm).
Tissue Culture
NIH-3T3 cells were maintained in Dulbecco's Modified Eagle's Medium plus 10% newborn calf serum, 50 units/ml penicillin, and 50 mg/ml streptomycin. A293T cells were maintained in Dulbecco's Modified Eagle's Medium plus 10% fetal bovine serum and 50 units/ml penicillin and 50 mg/ml streptomycin. Cells were incubated at 37°C with 5% CO2 (v/v).
siRNA Design and Transfection
Three siRNAs to mouse aldolase A (NM_007438) were designed and custom synthesized (Invitrogen) for the following target sequences: siRNA 287: 5′-CGCCUGCAGUCCAUUGGCA-3′, siRNA 1171: 5′-CGCUUGUCAAGGAAAGUAU-3′, siRNA 1301: 5′-CUACCCACTUUGCUAUUGAA-3′. The negative control siRNA was from ABI/Ambion. Cells were seeded so that they were approximately 20% confluent on the day of transfection. Each siRNA was tested individually for efficacy and persistence, and pooling showed no additional knockdown as demonstrated by aldolase activity assay (Fig. S2). Therefore, aldolase siRNAs were pooled for all subsequent experiments. siRNAs were transfected at a pooled final concentration of 25 nM into NIH-3T3 cells using Lipofectamine 2000 and OptiMEM media (both from Invitrogen) according to the manufacturer's instructions. Media was changed 24 h after transfection.
Native Cell Lysates and Protein Determination
Cells were washed twice with ice-cold PBS, and collected. Cell pellets were resuspended in 100 µl 20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton-X 100, 1 mM DTT, and 1 µg/µl each leupeptin, pepstatin A, and phenylmethanesulfonylfluoride. Lysates were cleared by centrifugation at 20,000g for 1 h. Protein concentration for each lysate was determined by dye-binding assay [Bradford, 1976].
Enzymatic Activity Assays
Activity of glycolytic enzymes in cleared cell lysates was determined as described previously for aldolase [Morris and Tolan, 1993], glyceraldehyde 3-phosphate dehydrogenase (GAPDH) [Knight et al., 1996], triose phosphate isomerase (TIM) [Brooks, 1976], and enolase [Brooks, 1976] (the latter two did not use added rotenone). Enzyme activity was a preferred method for measuring knockdown as it takes into account both changes in protein content as well as changes in activity.
ATP and Glycolytic Flux Determinations
For [ATP], cells were rinsed with PBS, scraped from their dishes and frozen at −80°C. Cells were lysed and deproteinized by incubation at 80°C for 10 min and cleared by centrifugation at 8,000g. [ATP] was determined using a commercially available luciferase-based assay (Invitrogen). Values were normalized to the number of cells measured. Glycolytic flux was determined by the rate of lactate production as described previously [Gutman and Wahlefeld, 1983; Ritterson Lew and Tolan, 2012].
Construction of Stable Cell Lines
The pMSCV-MycAldolase stable mammalian expression plasmid was constructed using the PCR-amplified rabbit aldolase A open reading frame from pPB14 [Beernink and Tolan, 1992] amplified with primers containing restriction enzyme sites compatible for cloning into pcDNA3.1-Myc. MycAldolase was subsequently subcloned into BglII—EcoRI digested pMSCV (Clontech; pMSCV-MycAldolase). For stable expression of D33S and R42A aldolase in NIH-3T3 cells, the D33S and R42A mutations were subcloned into pMSCV-MycAld from pAM9 and pAM19, respectively [Morris, 1995]. Either the empty pMSCV vector or the pMSCV-MycAldolase plasmid was co-transfected with the pCL-10A1 packaging vector (Imgenex) into A293T cells using polyethylenimine. Media was changed 1 day after transfection. Viral supernatant fractions were collected 2 and 3 days after transfection, and NIH-3T3 cells were infected with viral supernatant containing the desired plasmid in the presence of 4 µg/ml polybrene. Cells that incorporated the appropriate plasmid were selected by puromycin-resistance. Expression of the desired protein was confirmed by immunoblotting.
Immunoblotting
Protein from cleared cell lysates (25–50 µg) was separated by SDS–PAGE (12% (w/v)) and transferred to nitrocellulose using a semi-dry transfer apparatus (Bio-Rad). Blots were blocked overnight in 20 mM Tris, pH 7.5, 50 mM NaCl (TBS) containing 0.1% (v/v) Tween-20 and 5% (w/v) Carnation instant milk. For MycAldolase detection, blots were incubated with mouse anti-Myc primary antibody (Santa Cruz sc-40, 1:10,000 in TBS, 0.1% (v/v) Tween-20, and 2% (w/v) Carnation instant milk), followed by HRP-conjugated goat anti-mouse secondary antibody (BioRad 170-6516, 1:2,000 in TBS, 0.1% (v/v) Tween-20, and 2% (w/v) Carnation instant milk) for 1 h each. Bands were visualized using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific).
Cell Migration Assay
Tissue culture dishes (35 mm) were coated with poly-L-lysine (100 µg/ml). Cells were plated at a density of 400,000 cells/dish the day before the assay to ensure a confluent monolayer. “Wounds” were scratched into the cell monolayer using a P200 yellow pipette tip. Cells were washed once with media to rid the plate of any detached cells, and photos of the scratch wound widths were taken at 0, 4, 8, and 12 h after scratching. Measurement of the wound width in micrometer was measured at each timepoint using ImageJ software (http://rsbweb.nih.gov/ij/index.html), and the rate of migration into the wound space was determined.
Cell Spreading Assays
Cells were trypsinized from their plates, then re-plated onto poly-L-lysine-coated coverslips in six-well plates (100,000 cells/well). At various times after plating, spreading was stopped by fixing the cells in 4% formaldehyde and stained with Alexa-488 phalloidin. The degree of spreading for each cell was measured by either length:width ratio or area. The longest possible line drawn through the cell defined the length of a cell. The width was defined as a line drawn through the cell perpendicular to the midpoint of the cell length measurement. Lengths and widths were determined using ImageJ software, and the ratio of length:width was used to determine the degree of cell spreading. Areas of cells were also determined using ImageJ software.
Statistics
Error is expressed as standard deviation (SD) or standard error of the mean (SEM) as indicated in figure legends. Statistical significance was determined using Student's t-test.
RESULTS
Effect of Aldolase on WA/Arp2/3 Stimulated Actin Polymerization
Because of its known interaction with WASP [Buscaglia et al., 2006], the effect of aldolase on WASP activity was tested in the context of an in vitro actin-polymerization assay. In the presence of 1 µM aldolase, the WA-stimulated Arp2/3-dependent actin-polymerization reaction was significantly inhibited (Fig. 1A) and in a concentration dependent manner. At 2 µM aldolase, the slope of the aldolase-inhibited reaction was the same as the un-stimulated polymerization (Fig. 1B). This inhibition was quantified by plotting the maximal rate of polymerization as a function of aldolase concentration and showed an IC50 of 250 nM aldolase (Fig. 1C) when fit to an exponential decay equation.
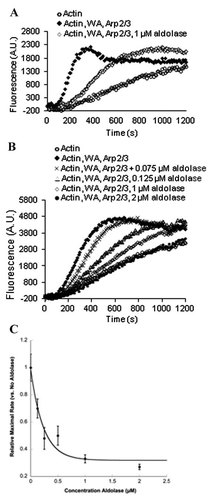
Effect of aldolase on GST-WA and Arp2/3 stimulated actin polymerization. A: Representative actin polymerization curves for actin only (2 µM, 12% pyrene-labeled), actin stimulated with WA (10 nM) and Arp2/3 (6.5 nM), and stimulated actin inhibited with aldolase (1 µM) in an in vitro actin polymerization assay. B: Representative actin polymerization curves with varying concentrations of aldolase (0.075–2 µM, as indicated) showing the effect on WA and Arp2/3 stimulated actin polymerization. Actin, WA, and Arp2/3 concentrations, as in (A). C: Inhibition curve of maximal actin polymerization rates with different concentrations of aldolase normalized to WA and Arp2/3 stimulated polymerization reactions without aldolase. Error is represented as SEM. The data was fit to an exponential decay equation using Kaleidagraph software (solid line).
Mechanism of Aldolase Inhibition of In Vitro Actin Polymerization
Testing three hypotheses elucidated the mechanism of aldolase inhibition of actin polymerization: (1) aldolase inhibits polymerization via its interaction with actin; (2) aldolase binds Arp2/3 and inhibits its activity; and (3) aldolase binds the WASP acidic domain, preventing its interaction with Arp2/3. Adding increasing amounts of aldolase (0.25–2 µM) into an un-stimulated polymerization reaction containing 2 µM actin did not affect this reaction (Fig. 2A). Plotting the maximal polymerization rate as a function of aldolase concentration showed little to no effect on actin polymerization at any concentration (Fig. 2B). Next, titration of Arp2/3 (3.25–26 nM) in the F-actin polymerization assay, while holding concentrations of actin (1 µM), WA (10 nM), and aldolase (0.5 µM) constant did not alleviate inhibition of polymerization by aldolase (Fig. 2C–F). Plotting the maximal polymerization rate as a function of Arp2/3 concentration showed that inhibition of the polymerization reaction by aldolase remained constant at approximately 40% (Fig. 2G). Finally, titrating WA (5–50 nM) in the F-actin polymerization assay, while holding actin (2 µM), Arp2/3 (6.5 nM), and aldolase (0.5 µM) constant overcame the inhibition by aldolase (Fig. 2H–K). Plotting the maximal polymerization rate for the reactions as a function of WA concentration with and without aldolase followed a saturation-binding curve better than a linear fit (linear fit not shown; Fig. 2L; R2-values were 0.95 and 0.96 for the hyperbolic curves versus 0.88 for the linear curve for reactions containing aldolase). These results indicated that aldolase modulated in vitro actin polymerization by binding WA and likely sequestered it from Arp2/3, thereby decreasing the rate of polymerization. Consistent with this model, the inhibition of in vitro actin polymerization was alleviated by excess substrate (Fig. S3; the active site is adjacent to the WA binding site [St-Jean et al., 2007]). Taken together with the inhibition of polymerization, it is clear that aldolase affects stimulated actin polymerization through its interaction with WASP. Therefore, WASP-mediated processes were investigated in cells.
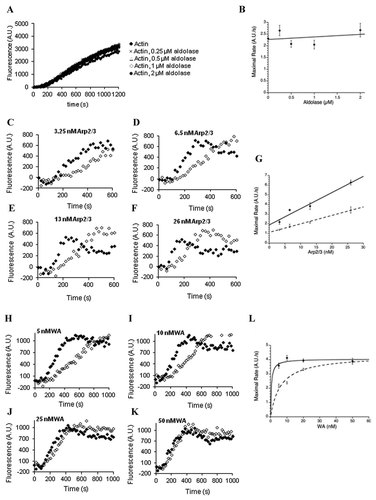
The mechanism of inhibition by aldolase on actin polymerization. A: Representative actin polymerization curves showing effect of varying aldolase concentration (0.25–2 µM, as indicated) on reactions with actin only (2 µM, 12% pyrene-labeled), without WA or Arp2/3. B: The maximal rate of actin polymerization versus aldolase concentration. The experiment was performed 3–12 times at each concentration of aldolase. Error is represented as SEM. The data is fit to a linear equation (solid line). C–F: Representative polymerization curves showing increasing concentration of the Arp2/3 (indicated above each graph) in the absence (black) or presence (white) of 0.5 µM aldolase, holding actin (1 µM) and WA (10 nM) constant. G: The maximal rate of actin polymerization versus Arp2/3 concentration in the absence (solid) or presence (dashed) of 0.5 µM aldolase. Each experiment was done in duplicate, and error is shown as the range of values for each point. The data were fit to lines with R2 = 0.953 (solid) and 0.955 (dashed). H–K: Representative polymerization curves are depicted with increasing concentrations of WA (indicated above each graph) in the absence (black) or presence (white) of 0.5 µM aldolase, holding actin (2 µM) and Arp2/3 (6.5 nM) constant. L: The maximal rate of polymerization versus WA concentration in the absence (solid) or presence (dashed) of 0.5 µM aldolase. Each experiment was done ≥6 times, and error is represented as SD. The data were fit to a hyperbolic equation with R2 = 0.3818 (solid) and 0.9270 (dashed). All fits were performed using Kaleidagraph software.
Aldolase Knockdown Affects Migration and Spreading of NIH-3T3 Cells
Two cellular processes in which WASP family members participate are cell migration [Jimenez et al., 2000] and cell spreading [Misra et al., 2007]. The effect of aldolase knockdown on cell migration was investigated in NIH-3T3 fibroblasts. Four days after transfection, cells treated with siRNAs targeting aldolase A (siAldolase) showed aldolase activity that was maximally diminished by 80–90% compared to mock-transfected cells or cells treated with a negative control siRNA (siNegative; Figs. S2A and 3A). Consistent with a moonlighting function of aldolase, there was no change in total cellular [ATP] (Fig. 3B) or glycolytic flux (Fig. 3C) after aldolase knockdown. These siAldolase cells were subjected to a wound-healing assay that measured cell migration. Aldolase-knockdown cells showed a decrease in overall migration compared to mock- or siNegative-transfected cells after 12 h (Fig. 3D), and the rate of wound healing was significantly slower (25%, P < 0.05) compared to mock-transfected cells (Fig. 3E). The effect of aldolase knockdown on cell spreading in NIH-3T3 cells was investigated in similarly treated cells. At 4 days post-transfection, two measures of cell spreading were performed. Compared to mock transfected, siNegative-transfected, or untreated cells, aldolase knockdown cells showed a much slower rate of spreading after reattachment and were spread over a smaller area (Fig. 3F–H). Together with the cell migration data and in vitro data, these data indicate that the changes in aldolase concentration affect WASP-mediated processes in these cells.
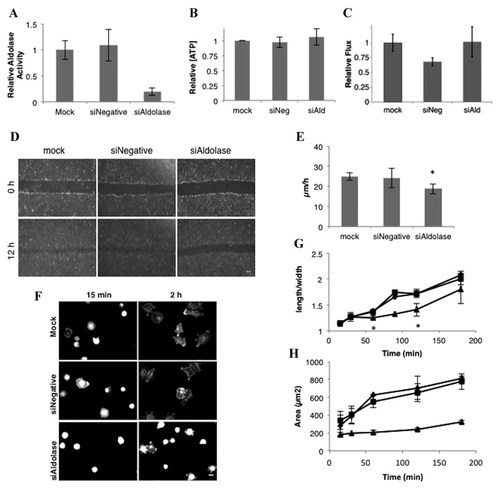
Knockdown of aldolase A in NIH-3T3 cells causes a decrease in cell motility and cell spreading. A: Aldolase activity was measured in native cell lysates from mock-, siNegative-, or siAldolase-transfected NIH-3T3 cells. Relative aldolase activity was normalized to mock-transfected activity (0.014 ± 0.004 U/mg). B: [ATP] from lysed cells was measured using a luciferase-based assay and normalized to mock-transfected control (0.1 fmole/cell). Error is represented as SEM. C: Glycolytic flux as measured by rate of lactate production after a glucose bolus. Rates were normalized to mock-transfected cells (1.84 × 10−6 µmol/h-cell). Error is represented as SEM. D: Representative micrographs of the indicated cell treatments at 0 and 12 h after scratch. Scale bar, 200 µm. E: The space between each edge of the scratch was measured every 4 h for 12 h after scratching, and the rate of recovery was measured. Error bars represent SEM. *, P < 0.05. F–H: Cells for each indicated treatment were trypsinized from their dishes and re-plated on PLL-coated coverslips. At least 30 cells were imaged per coverslip, and experiment was done 3–6 times in triplicate. F: Representative images of cells for each treatment spread for 15 min and 2 h. Scale bar, 10 µm. G: The length:width plotted versus time. H: The area (µm2) of each cell plotted versus time. For both (G) and (H), ♦, mock transfection (G) or untreated NIH-3T3 cells (H); ▪, siNegative; ▴, siAldolase. Error is represented as SEM. *, P < 0.05.
Rescue of Aldolase Knockdown Phenotypes With Exogenous Aldolase A
Aldolase knockdown affected two WASP-mediated cellular processes – migration and spreading. Exogenous untargeted rabbit aldolase A (Figs. S4 and 4B) was tested for its ability to rescue the knockdown phenotype. For this experiment, NIH-3T3 cells were stably transfected with Myc-tagged rabbit aldolase A (MycAld), or an empty vector control (Empty-3T3). The presence of MycAld in NIH-3T3 cells was confirmed by immunoblotting (Fig. 4A). MycAld-3T3 cells were transfected with the siRNAs specific to mouse aldolase A. At 4 days post-transfection, immunoblotting showed exogenous MycAld expression in MycAld-3T3 cells was not affected by siRNA treatment (Fig. 4B).
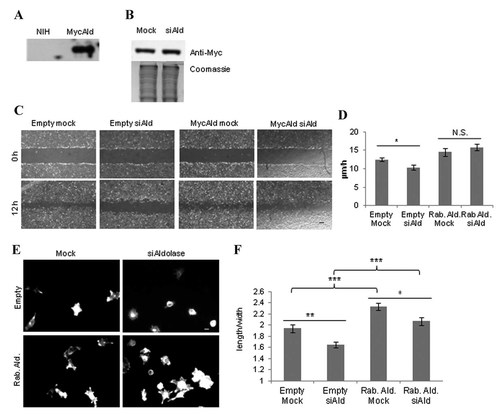
Rescue of aldolase knockdown phenotypes using Myc-tagged rabbit aldolase A in NIH-3T3 cells. A: Immunoblot using anti-Myc antibody in NIH-3T3 and MycAld-3T3 cells. B: Immunoblot for MycAld-3T3 cells that were mock- or siAldolase-transfected and probed for MycAldolase using the anti-Myc antibody. Coomassie-stained samples for each are shown are shown as a loading control. C: Scratch wound healing assays were performed on NIH-3T3 cells stably expressing either empty vector or MycAldolase that were mock- or siAldolase (siAld)-transfected. Times 0 h (immediately after scratch) and 12 h after scratch are shown for comparison. Scale bar, 200 µM. D: Comparison of the rates of wound healing for empty vector and MycAld-3T3 (MycAld = Rab.Ald.) mock- or siAld-transfected cells. The space between each side of the scratch was measured every 4 h for 12 h after scratching, and the rate of recovery was measured. Error is represented as SEM. *, P < 0.05; N.S., not significant. E–F: Mock- and siAldolase (siAld)-transfected empty vector or MycAld-3T3 cells were detached from their plates then allowed to re-attached to PLL-coated coverslips for 2 h. After 2 h, cells were fixed, permeabilized, and stained with Alexa-488 phalloidin. E: Representative micrographs for each treatment. Scale bar, 10 µM. F: Quantification of the degree of spreading for Mock- and siAld-transfected empty- or MycAld-3T3 cells. At least 30 cells were counted per coverslip, and the experiment was repeated three times in triplicate Error is shown as SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The rescue of the migration and spreading defects (see Fig. 3D–H) used mock- or siAldolase-transfected Empty-3T3 and MycAld-3T3 cells. After siAldolase transfection, the Empty-3T3 cells showed a significant cell migration defect (Fig. 4C,D; 20% defect, P ≤ 0.01), similar to wild type NIH-3T3 cells (see Fig. 3D,E), whereas the cells expressing the rescue construct (MycAld-3T3) were unaffected by endogenous aldolase knockdown. Next, cell spreading was measured for mock- and siAldolasetransfected Empty-3T3 and MycAld-3T3 cells 2 h after plating (time point for maximal difference, see Fig. 3G). While siAldolase-treated Empty-3T3 cells were clearly less elongated than the mock-transfected controls, siAldolase-treated MycAld-3T3 cells showed no obvious differences in morphology from mock-treated cells (Fig. 4E). The length:width ratio for each treatment showed the knockdown Empty-3T3 cells had a 35% reduction in spreading compared to mock-transfected cells (P ≤ 0.002). In contrast, knockdown MycAld-3T3 cells showed only a 20% decrease in spreading (P ≤ 0.01), indicating a partial rescue of this phenotype (Fig. 4F). In addition, both mock- and siAldolase-treated MycAld-3T3 cells were significantly more spread than their empty vector counterparts (P ≤ 0.0001), indicating that overexpression of aldolase may affect the rate of cell spreading. Taken together, the rescue of knockdown phenotypes for cell migration and spreading with MycAldolase showed that these phenotypes were associated with expression levels of aldolase, and not due to off-target effects of the siAldolase.
Separating the Moonlighting and Catalytic Functions of Aldolase
The data presented thus far support the hypothesis that modulation of WASP activity is an aldolase moonlighting function. The fact that aldolase knockdown does not affect [ATP] or glycolytic flux in NIH-3T3 cells supports this model (see Fig. 3B,C). This moonlighting hypothesis was tested using aldolase variants defective in either F-actin binding or aldolase catalytic activity in both in vitro and cell-based assays. The R42A variant of aldolase (R42A-aldolase) is catalytically active, but binds F-actin with 20-fold less efficiency than wild type aldolase A [Wang et al., 1996]. Furthermore, the residue Arg-42 interacts with a peptide of WASP in a crystal structure of the complex [St-Jean et al., 2007]. It is likely therefore that WASP binding is affected for R42A-aldolase. If aldolase moonlighting functions are required for modulation of WASP activity, and aldolase activity is irrelevant, then R42A-aldolase should not inhibit WASP-stimulated polymerization in vitro and should not rescue cells from the slower spreading and migration effects following aldolase knockdown. Conversely, the D33S variant of aldolase (D33S-aldolase) is catalytically inactive, but F-actin binding is unaffected [Wang et al., 1996]. If aldolase catalytic activity is not required for the modulation of WASP activity, then D33S should inhibit WASP- and Arp2/3-mediated polymerization in vitro and should rescue the effects seen in cells.
When R42A-aldolase was added to the in vitro polymerization assay, the resulting polymerization curve was indistinguishable from one without aldolase (Fig. 5A). These data clearly showed that actin and/or WASP-binding activity of aldolase was required for inhibition of actin polymerization in vitro. In contrast, when D33S-aldolase was used in vitro, it inhibited the polymerization (Fig. 5B). In fact, at equimolar concentrations, D33S inhibition of actin polymerization was greater than wild-type aldolase.
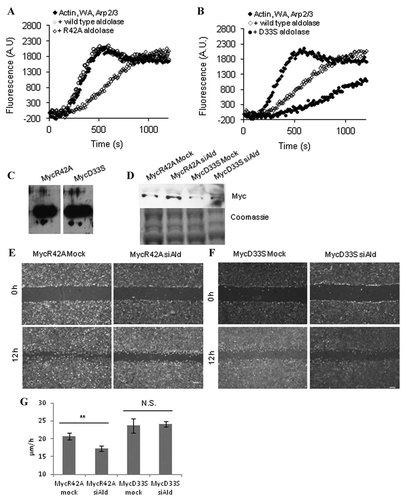
Effect of D33S and R42A aldolase variants on in vitro actin polymerization and cell motility. A–B: Representative actin polymerization curves for actin (2 µM and 12% pyrene labeled), WA (10 nM), and Arp2/3 (6.5 nM; ♦) with wild type aldolase A (◊) or R42A (○; A) and wild type aldolase A (◊) or D33S (●; B) aldolase (1 µM) were compared for their effects on in vitro actin polymerization. These variants had no effect on polymerization of actin alone (no WA, Arp2/3; data not shown). C: Immunoblot of MycR42A- and MycD33S-expressing 3T3 cells. NIH-3T3 cells were stably transduced using MSCV-MycR42A or MSCV-MycD33S. Native cell lysates were made from puromycin-resistant cells from each transduction and immunoblotted for MycAldolase using the anti-Myc antibody. D: Immunoblot with anti-Myc antibody (1:2,000 dilution) of NCLs harvested 4 days after transfection from MycR42A- and MycD33S-3T3 cells that were mock- or siAldolase transfected. Coomassie-stained samples serve as a loading control. E: MycR42A- and (F) MycD33S-3T3 cells were mock- or siAldolase (siAld)-transfected and subjected to a scratch-wound healing assay 4 days post-transfection. Representative micrographs at 0 and 12 h after scratch are shown. Scale bar, 200 µM. G: Quantification of the rate of recovery into the wound area. The space between each side of the scratch was measured every 4 h for 12 h after scratching, and the rate of recovery was measured. Error is represented as SEM. **, P < 0.001; N.S., not significant.
The importance of aldolase moonlighting functions in WASP-dependent processes was investigated using these aldolase variants for rescue of cell migration upon endogenous aldolase knockdown. To this end, R42A- or D33S-aldolase were stably expressed in NIH-3T3 cells as described above for pMSCV-MycAld. Immunoblotting of cell lysates from these cells confirmed the expression of these aldolase variants (Fig. 5C). Mock- and siAldolase-treated MycR42A- and MycD33S-expressing 3T3 cells were immunoblotted with the anti-Myc antibody 4 days post-transfection (Fig. 5D). As with the MycAld-3T3 cells (see Fig. 4B), transfection of siAldolase had no effect on the expression of MycR42A- or MycD33S-aldolase expression in 3T3 cells. The ability of either R42A- or D33S-aldolase to rescue the aldolase knockdown effect on cell migration was measured in a scratch-wound healing assay. Cell migration into the wound area was measured in mock- and siAldolase-transfected R42A- and D33S-aldolase expressing cells. Cells expressing R42A-aldolase treated with siAldolase showed a significant decrease in the rate of migration into the wound area (Fig. 5E,G), indicating that R42A-aldolase was unable to rescue the migration defect of aldolase knockdown in cells. This result was comparable to aldolase knockdown in wild type NIH-3T3 cells (see Fig. 3D) or cells stably transfected with the empty vector (Fig. 4C). On the other hand, cells expressing D33S-aldolase treated with siAldolase showed no significant difference in the rate of migration compared to mock-transfected cells (Fig. 5F,G). These data clearly indicated that aldolase catalysis was not required for its function in cell migration.
DISCUSSION
Although aldolase is well characterized in its metabolic functions and has been implicated in binding a number of cellular partners, little is known about how it may function in any actual moonlighting role in the cell. In particular, one well-established interaction is with activated WASP [Buscaglia et al., 2006], but the function of this interaction has not been elucidated. Here, it was shown that aldolase likely sequesters WASP during in vitro actin polymerization and mediates actin dynamics in the cells, likely through its binding of the WA domain of WASP [St-Jean et al., 2007]. Knockdown of aldolase affected the WASP-mediated processes of cell migration and spreading. Moreover, these functions of aldolase were independent of aldolase catalytic activity, as the inactive aldolase variant D33S inhibited actin polymerization in vitro and rescued the effect of aldolase knockdown on cell migration, clearly indicating that a moonlighting function of aldolase was involved in this process.
The concentrations of aldolase used in these studies are consistent with other reports. The in vitro inhibition of actin polymerization by aldolase had an IC50 value of 250 nM, which was similar to the IC50 value for the aldolase-SNX9 interaction (100 nM) [Lundmark and Carlsson, 2004], and to the inhibition of actin polymerization by other proteins such as WASP interacting protein (WIP; 200 nM) [Martinez-Quiles et al., 2001] and coronin (71 nM) [Cai et al., 2008]. In addition, whether the concentrations of proteins that affect actin polymerization in the in vitro polymerization assays are physiological is often an issue (in this case, aldolase, 0.075-2 µM). In fact, the concentrations of aldolase in the polymerization assays in this study were actually less than reports of intracellular aldolase concentrations (6–37 µM [Ottaway and Mowbray, 1977; Orosz et al., 1988b; Lundmark and Carlsson, 2003]). Moreover, local concentrations of aldolase may be even higher in structures such as the lamellipodia [Wang et al., 1997]. Therefore, the in vitro observations are well within physiological concentrations. The effect of aldolase levels on WASP-dependent processes in vivo has been reported in HaCaT keratinocytes, where knockdown of aldolase inhibited lamellipodia formation and cell motility, whereas epidermal growth factor treatment increased both aldolase mRNA expression and cell motility [Tochio et al., 2010]. In addition, changes seen in cell morphology and actin-cytoskeleton organization followed aldolase knockdown in HeLa cells [Merkulova et al., 2011] and mouse fibroblasts [Ritterson Lew, 2012]. These reports are consistent with the positive and negative effects on both migration and spreading in NIH-3T3 cells (see Figs. 3 and 4) due to changes in intracellular aldolase concentrations.
Presuming that these in vitro and in vivo effects are due to WASP binding, the stoichiometry of the interaction between WASP and aldolase is of interest. This unknown stoichiometry is important because of the very strong (Kd ∼ 10−28)[Tolan et al., 2003] native tetrameric structure of the enzyme [Penhoet et al., 1967] and the fact that aldolase can crosslink F-actin filaments [Wang et al., 1996; Sukow and DeRosier, 2003]. In the crystal structure of aldolase bound to the WASP peptide, each subunit of aldolase has the 15-residue C-terminal peptide bound [St-Jean et al., 2007]. However, the actual WASP protein is >500 amino acids long (55 kDa), and physical constraints could prevent 1:1 binding between WASP and the aldolase monomer, particularly if already bound to F-actin. More studies are needed to investigate the stoichiometry of this interaction. The strength of interaction between WASP and aldolase likely affects both in vitro and in vivo processes. This is exemplified by the D33S variant, which inhibited the in vitro polymerization reaction better than wild type aldolase A (see Fig. 5B). Wild-type aldolase A and D33S are comparable in their actin-binding capacities [Wang et al., 1996], so the D33S substitution likely does not enhance actin binding in the reaction; however, it is possible that the D33S substitution enhances WASP binding. The Asp-33 residue does not have any direct interactions with the WASP peptide as seen in the crystal structure [St-Jean et al., 2007]. However, it is only 4.3 Å from Glu-499 of WASP, and the lack of the negatively charged carboxyl group could account for tighter binding to WASP.
At first glance, the in vitro and in vivo data are in opposition. Based on in vitro data, aldolase overexpression should inhibit WASP-dependent processes in cells, and aldolase knockdown should cause an increase in spreading and motility—the opposite was observed. However, the phenomenon of a protein inhibiting actin polymerization in vitro, while stimulating actin-depended processes in cells, is not novel. WIP binds to WASP at the WH1 domain of WASP and stabilizes the WASP protein. WIP inhibits polymerization in vitro, yet overexpression of WIP in cells results in an increase in total F-actin and filopodia [Martinez-Quiles et al., 2001]. For both WIP and aldolase, there are important factors for controlling actin dynamics in cells that can explain their effects, where these factors are not present in vitro.
Any model for the role of aldolase in WASP-mediated actin dynamics must accommodate this contradiction between in vitro and in vivo data. One such mechanism would involve aldolase in localization of WASP during changes in cytoskeleton organization. In unstimulated cells, WASP exists in the nucleus and perinuclear region [Vetterkind et al., 2002]. Upon stimulation by growth factors, the activated form of WASP is localized to lamellipodia and filopodia where its activity is required [Fukuoka et al., 1997; Jimenez et al., 2000; Vetterkind et al., 2002]. The overexpression of WIP can stimulate N-WASP localization to sites of actin polymerization [Vetterkind et al., 2002], and focal adhesion kinase can phosphorylate N-WASP and prevent its nuclear localization [Wu et al., 2004]. However, the exact mechanism of how WASP gets from the nucleus/perinuclear region to the edge of the cell is unknown. It is possible that aldolase plays a role in this translocation by functioning as a molecular adaptor between activated WASP and F-actin. In addition, the in vitro actin polymerization assay does not take into account the energy requirements for actin dynamics in the cell. Although aldolase knockdown does not affect [ATP] in NIH-3T3 cells (see Fig. 3B), it is possible that the catalytic activity of aldolase may still be necessary to provide energy locally for polymerization reactions, for example, in the context of a localized “glycosome” complex. Consistent with both of these hypotheses, aldolase localizes to lamellipodia in moving NIH-3T3 cells [Wang et al., 1997].
A model depicting one such mechanism is shown in Figure 6. Extracellular signaling activates WASP exposing its WA domain to which aldolase binds. This inhibits the interaction between WASP and Arp2/3 until it is localized to the site of lamellipodium formation. Subsequently, an unknown signal releases aldolase from WASP, allowing Arp2/3 binding and stimulation of branching polymerization. A similar model has been proposed to explain aldolase involvement with SNX9 during clathrin-mediated endocytosis [Lundmark and Carlsson, 2004]. In this model for aldolase localization of WASP activity, overexpression of aldolase would promote migration and lamellipodia formation, similar to what was shown here. Knockdown of aldolase would inhibit proper localization of WASP and WASP-dependent processes such as migration. On the other hand, overexpression would accelerate these processes as shown here and in other studies [Tochio et al., 2010]. Without cellular signals for localization, this aldolase interaction merely sequesters WASP, as seen in vitro.
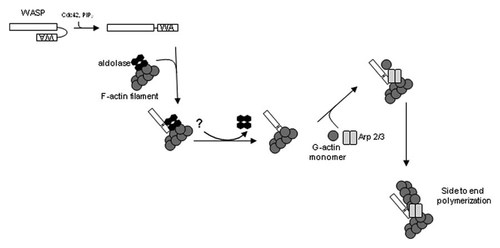
A model for the role of the aldolase-WASP interaction in cells. WASP (white) is activated by cdc42 and PIP2. Upon activation, WASP is bound via the WA domain by aldolase (black), which may already be bound to F-actin (dark gray), and subsequently localizing WASP to the cytoskeleton. In this complex, initiation of branching-polymerization activity by WASP activation of the Arp2/3 complex is blocked by the unavailability of the acidic domain of WASP. After WASP is properly localized, aldolase is released by an as-of-yet undetermined signal, and WASP is free to bind Arp2/3 (light gray), and actin polymerization is initiated.
In addition to the effects reported here, aldolase is likely an important player in many cellular functions that depend on actin dynamics. Many of the purported aldolase moonlighting functions involve the actin cytoskeleton and these studies provide a starting point for understanding these functions. Given its strong tetrameric structure, aldolase could easily bind to cellular proteins and F-actin as a molecular adaptor protein, while maintaining catalytic activity of at least one subunit. In certain processes, this binding may allow correct localization in the cell, as implicated here for activated WASP.
Acknowledgements
We are indebted to Dr. Defne Yarar (Whitehead Institute), who provided us with reagents, access to equipment, and much insight, along with critical reading of this manuscript, all of which were instrumental in the publication of this work. We also thank Dr. Frank Gertler, Michele Balsamo (Koch Institute/MIT), and Dr. Yaniv Erlich (Whitehead Institute) for providing continued access to equipment and reagents, and Sarah Oppelt for critical reading of the manuscript.




