Post-translational modifications in activation and inhibition of oct-1–DNA binding complex in H2B and other diverse gene regulation: Prediction of interplay sites
Abstract
Octamer DNA binding transcription factors play important roles in housekeeping and specific gene regulations. Octamer DNA binding transcription factor-1 (Oct-1), expressed ubiquitously, is a multifunctional molecule. The binding sites of Oct-1 are the promoters of H2B gene and the genes of snRNA, U2, U6, and 7SK, yet Oct-1 has been described as constitutively expressed transcription factor regulating the expression of housekeeping genes. Diverse tissue-specific genes regulations by Oct-1 include genes for interleukins (IL) 2, 3, 5; the granulocyte-macrophagal colony-stimulating factor, immunoglobulins α, β, Ly9; the endocrine-associated Pit-1 gene; the genes for gonadoliberin, prolactin, the thyroid transcription factor, and thyrotropin. The most interesting aspect of the gene regulations of Oct-1 includes both activation and inhibition of transcription. These opposite regulations of Oct-1 have been described through presence/absence of a post-translational modification (PTM) in its different domains. We propose a mechanism of interplay of different PTMs or presence/absence of PTMs in the different domains of Oct-1. We also suggest that the absence of phosphorylation and acetylation in G1 and S phases of the cell cycle is associated with interplay of methylation and O-GlcNAc modification. This interplay of O-GlcNAc modification with the phosphorylation and methylation with acetylation in POU sub-domain of Oct-1 may facilitate the formation of Oct-1–DNA complex, consequently activating H2B gene transcription. Whereas, in G2 and M phases these sites are occupied by phosphate resulting in inhibition of Oct-1–DNA complex formation leading to the suppression of H2B gene transcription. J. Cell. Biochem. 114: 266–274, 2013. © 2012 Wiley Periodicals, Inc.
Abbreviations:
Oct-1, octamer binding transcription factor-1; O-GlcNAc, N acetylglucosamine glycosylation; OGT, O-GlcNAc transferase; PTMs, post-translational modifications; IL-2, interleukin 2; POUs, POU specific domain; POUh, POU homeodomain; Ser, serine; Thr, threonine.
Octamer DNA binding transcription factors belong to a family of structurally related POU (pit, oct, unc) factors containing POU domain. The POU factors are widely found in eukaryotes. The POU domain is a bipartite DNA binding structure consisting of a POUspecific (POUs) domain, which is linked by a flexible linker to a POU homeodomain (POUh) [Herr and Cleary, 1995]. This POU domain is highly conserved among all the members of this family, which is a target for several types of regulatory protein–protein interactions. This is the target region required for sequence-specific DNA binding [Stern et al., 1989; Ingraham et al., 1990]. Oct family is comprised of different Oct factors including Oct-1, -2, -3, -4, etc. Octamer DNA binding transcription factor-1 (Oct-1) is a DNA binding transcription factor expressed ubiquitously in eukaryotes including mammals [Sturm et al., 1988; Dong and Zhao, 2007]. A consensus octamer sequence 5′-ATGCAAATNA-3′ is recognized by Oct-1 regulating the activation of promoters of diverse genes. This consensus sequence is found in promoters and enhancers of diverse vertebrate genes [Verrijzer and Van Der Vliet, 1993]. Though, both Oct-1 and Oct-2 recognize the same consensus sequence of DNA, the Oct-1 exhibits the specificity for regulating diverse sets of vertebrate genes [Schaffner, 1989], whereas Oct-2 is specific to bind on promoters and enhancers of immunoglobulin genes in B-cell lineage [Staudt et al., 1988]. The target genes of Oct-1 include promoters of snRNA, mRNA [Tanaka et al., 1992], histone H2B [Fletcher et al., 1987; Labella et al., 1988], interleukins (IL) [Ullman et al., 1991; Wu et al., 1997] and immunoglobulins α, β, and Ly9 [Luo et al., 1992; Strubin et al., 1995].
PTMS SWITCHBOARD IN Oct-1
In the context of Oct-1 binding to promoters of diverse genes, the binding of Oct-1 at one specific promoter will require a specific set of conditions that will be different when binding to the promoter of another gene. In most of the cases post-translational modifications (PTMs), including phosphorylation, O-GlcNAc modification, acetylation and methylation, play vital role in regulating the functions of transcription factors and gene expression or repression. Different PTMs can compete on same or neighboring amino acids implicated in regulating different cellular functions. Additionally, presence of one PTM may favor or inhibit the other PTM. A combination of different PTMs on specific amino acids may result in diverse regulations of proteins, hence proteins perform multi-functions. Different amino acids predicted for bearing potential for different PTMs showed that these amino acids are located in different regions of Oct-1, that is, N-terminal, C-terminal, POUs domain, POUh domain, and the linker region of POUs and POUh domain. Switching of one PTM with the other in different transcription factors is the phenomenon that regulates the gene transcription. Moreover, phase-specific PTMs, in cell cycle, seem to be important in regulating the orderly progression of the cell cycle. Phosphorylation of Oct-1 has been documented in regulating the H2B gene expression [Schild-Poulter et al., 2003, 2007]. Involvement of multiple kinases and phosphatases in cell cycle-specific regulation of Oct-1 is evidenced [Roberts et al., 1991]. Sufficient evidences show the involvement of differential phosphorylation of POU domain of different proteins in regulating gene transcription [Tanaka et al., 1992; Caelles et al., 1995; Schild-Poulter et al., 2007]. Additionally, phosphorylation and dephosphorylation is a common mechanism in regulating the functions of different transcription factors [Boulikas, 1995]. This phosphorylation and de-phosphorylation results in regulating the signaling pathways. De-phosphorylation of the Ser/Thr may result in a possibility of an addition of OGlcNAc on the OH function of the same Ser/Thr. Thus, blocking the phosphorylation on those specific sites, consequently resulting in blocking of the function regulated by phosphorylation such as described earlier [Ahmad et al., 2006, 2007].
Similar interplay or interdependence of acetylation and methylation on adjacent Lys residues has also been described in histone H3 [Zhang et al., 2004; Millar and Grunstein, 2006; Taverna et al., 2007] and these acetylated and methylated Lys residues have been documented to function in gene transcription by regulating the interdependent molecular pathways [Lee and Workman, 2007]. Additionally, another interplay of phosphorylation and methylation on vicinal (Lys 9 and Ser 10; Lys 27 and Ser 28) amino acids has also been evident in histone H3 [Wang et al., 2004] and named as “Methylation/Phophorylation Switching.” This complex interplay of different PTMs regulates diverse protein functions and multi-functions performed by the same protein. Therefore, complex networks of combinatorial PTMs are operative in modulating different functions of proteins. These networks of PTMs can be considered as a “PTMs switchboard” [Everett et al., 2009] regulating protein multi-functionality through various functional switches of different proteins in coordinated manner. Therefore, there is a competition of different enzymes of PTMs to modify same or vicinal amino acids by different modifying groups, for instance, kinases and OGT modify the same or neighboring Ser/Thr either by a phosphate group or by O-GlcNAc. These Yin Yang sites predicted to be potential in different domains of Oct-1 may serve as functional switch through differential combination of phosphorylation and O-GlcNAc modification at different Ser/Thr in different domains. However, a competition between OGT and different kinases to modify the Oct-1 with a specific combination of phosphorylated and O-GlcNAc modified Ser/Thr will depend on the cell cycle stage. Same mechanism of PTMs switching for methylation with acetylation and phosphorylation on adjacent amino acids makes the situation more complex to envisage the functional switches in Oct-1 and other proteins. Unrevealing of the novel mechanisms for these PTMs switchboard will lead to explore new possibilities to maneuver different biological systems and processes from gene expression to signal transduction and cell cycle regulation.
In the current study, utilizing different prediction methods, the modification potential for phosphorylation, O-GlcNAc modification, acetylation, and methylation on different amino acid in mouse Oct-1, has been predicted. Additionally, we have also tried to develop an association among different PTMs, including inhibitory and favoring affect of one PTM on the other. The conservation status of these potential sites (amino acids) for different PTMs has also been estimated in diverse vertebrate and invertebrate members to substantiate the modification potential of different amino acid residues predicted by different methods. A mechanism of Oct-1 involvement in H2B gene regulation by interplay sites of different PTMs has been proposed. We suggest that a combination of different PTMs and their interplay with each other may activate or inhibit the Oct-1 function for H2B gene transcription at different stages of the cell cycle.
REGULATION BY PHOSPHORYLATION AND DE-PHOSPHORYLATION
Oct-1 binds directly to the specific DNA motif, but in some cases it requires some co-factors that interact with the DNA binding domain. Binding of these co-factors ultimately result in Oct-1 phosphorylation by different kinases such as protein kinase A (PKA), protein kinase C (PKC), and casein kinase 2 [Grenfell et al., 1996; Inamoto et al., 1997]. Phosphorylation and de-phosphorylation of proteins are important mechanisms to alter the activities and interactions of proteins at specific phases of cell cycle. Phosphorylation of Oct-1 by DNA-protein kinase (DNA-PK) takes place in cell-cycle-dependent manner and this phosphorylation is dependent on physical interaction of Oct-1 with Ku antigen [Roberts et al., 1991; Schild-Poulter et al., 2003]. Netphos 2.0 [Blom et al., 1999] has predicted 68 amino acids to have potential for phosphorylation. Amongst the potential predicted 68 sites, the POU sub-domains in mouse Oct-1 include Ser 284, 328, 336, 358, 361, 365, 367, 387, 388, and 409 and Thr 326 and 387 potential phosphorylation sites (Fig. 1, Table I). The conservation status also showed that most of the potential Ser and Thr were conserved in vertebrates (Fig. 3) and invertebrates (Fig. 4). Fifteen specific phosphorylation sites in Oct-1 catalyzed by DNA-PK were already identified [Schild-Poulter et al., 2007]. Out of these, 13 phosphorylation sites lie within the N-terminal domain of Oct-1 protein, a glutamine-rich domain. Moreover, differential phosphorylation in different activation domains of Oct-1 has been described as a mechanism for its activation [Roberts et al., 1991]. A similar control mechanism for Oct-2, a very similar protein to Oct-1 has also been evident [Tanak and Herr, 1990]. It is documented that hyperphosphorylation of Oct-1 during mitosis by p34cdc2 involve in terminating its DNA binding ability [Roberts et al., 1991]. It has been documented that phosphorylation of Oct-1 by PKA also inhibits its DNA binding activity in a sequence-specific manner [Segil et al., 1991]. However, PKA has an interesting feature that it failed to inhibit Oct-1 binding to DNA when there is a simple TAATGARAT motif and an overlapping octamer TAATGRAT motif [Roberts et al., 1991]. Additionally, it has also been reported that regulation of transcription factors mostly takes place by CK-II mediated phosphorylation [Meisner and Czech, 1991]. It has been documented that phosphorylation of Oct-1 results in its failure to bind with DNA during G2 and M-phases [Hwang and Chae, 1989; Schild-Poulter et al., 2003] of the cell cycle.
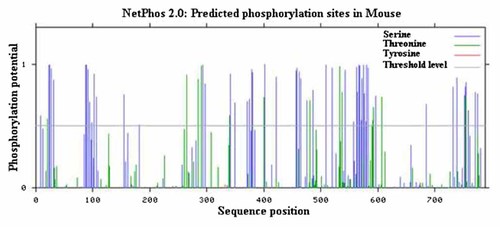
Graph shows prediction results for phosphorylation by NetPhos 2.0. Vertical lines show phosphorylation potential on Ser (blue color), Thr (green color), and Tyr (red color) in mouse Oct-1 protein. Threshold level is represented by horizontal gray line.
| Amino acid | Conservation | Modification potential | Amino acid | Conservation | Modification potential | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Inver. | Vert. | Net phos 2.0 | YinOyang 1.2 | Yin yang site | Inver. | Vert. | Net Phos 2.0 | YinOyang 1.2 | Yin yang site | ||
| Thr7 | NC | SC | + | + | Pr | Thr264 | C | SC | − | ++ | − |
| Ser10 | NC | NC | + | + | − | Ser268 | SC | SC | − | + | − |
| Ser11 | NC | SC | + | − | − | Ser270 | SC | CS | − | + | − |
| Ser14 | NC | SC | + | − | − | Thr271 | SC | C | + | + | HPr |
| Ser18 | NC | NC | + | − | − | Thr277 | NC | SC | + | − | − |
| Ser74 | NC | SC | + | + | Pr | Ser279 | SC | SC | + | − | − |
| Ser75 | NC | NC | + | − | − | Ser284 | SC | C | + | − | − |
| Ser78 | SC | SC | + | − | − | Thr326 | C | SC | + | − | − |
| Ser81 | SC | SC | + | + | Pr | Ser328 | SC | SC | + | − | − |
| Ser84 | NC | NC | − | +++ | − | Ser336 | C | CS | + | − | − |
| Ser85 | NC | SC | + | +++ | Pr | Ser358 | NC | CS | + | ++ | LPr |
| Ser89 | NC | SC | + | +++ | Pr | Ser359 | CS | C | − | + | − |
| Ser93 | NC | SC | + | + | Pr | Ser361 | NC | SC | + | + | Pr |
| Ser142 | NC | CS | + | + | Pr | Ser365 | SC | SC | + | + | Pr |
| Ser144 | NC | SC | − | + | − | Ser367 | NC | C | + | − | − |
| Ser148 | NC | SC | − | + | − | Thr387 | C | C | + | − | − |
| Thr153 | C | SC | − | + | − | Ser388 | C | C | + | − | − |
| Thr163 | CS | C | − | ++ | − | Ser409 | SC | C | + | + | HPr |
| Ser168 | SC | C | + | − | − | Ser444 | SC | C | + | +++ | HPr |
| Thr200 | NC | SC | − | + | − | Ser445 | NC | SC | + | +++ | Pr |
| Ser233 | NC | C | − | ++ | − | Ser449 | CS | CS | + | − | − |
| Ser243 | NC | C | − | ++ | − | Ser451 | NC | CS | + | − | − |
| Ser248 | NC | CS | − | + | − | Ser462 | NC | SC | + | + | − |
| Thr251 | NC | CS | − | + | − | Thr467 | NC | CS | + | +++ | LPr |
| Thr252 | SC | C | + | ++ | HPr | Ser469 | NC | C | − | ++ | − |
| Thr254 | SC | C | − | + | − | Thr472 | NC | CS | − | ++ | − |
| Ser473 | SC | C | + | + | H | Thr574 | NC | NC | − | ++ | − |
| Thr488 | NC | C | − | + | − | Thr576 | NC | CS | + | ++ | L |
| Ser489 | NC | C | − | + | − | Thr577 | NC | SC | + | ++ | P |
| Ser496 | NC | S | + | − | − | Ser578 | NC | CS | − | + | − |
| Ser507 | NC | N | + | − | − | Thr579 | NC | NC | + | ++ | − |
| Thr519 | NC | N | − | + | − | Ser583 | NC | CS | + | − | − |
| Thr520 | NC | N | + | + | − | Thr593 | NC | CS | − | + | − |
| Thr521 | NC | N | − | + | − | Thr594 | NC | SC | + | − | − |
| Thr524 | NC | C | + | − | − | Ser626 | NC | SC | − | + | − |
| Thr525 | NC | C | − | + | − | Ser671 | NC | C | + | − | − |
| Ser527 | NC | N | − | + | − | Ser672 | NC | SC | − | ++ | − |
| Thr528 | NC | N | − | + | − | Thr699 | NC | NC | − | + | − |
| Ser529 | NC | N | + | − | − | Ser714 | NC | CS | − | + | − |
| Ser538 | NC | C | − | + | − | Ser718 | NC | NC | + | + | − |
| Thr539 | NC | N | − | + | − | Ser726 | NC | CS | + | ++ | L |
| Ser544 | NC | C | + | + | P | Thr727 | NC | NC | − | ++ | − |
| Ser545 | NC | C | − | + | − | Thr733 | NC | NC | − | ++ | − |
| Thr548 | NC | C | − | + | − | Ser735 | NC | SC | − | + | − |
| Ser549 | NC | C | + | + | P | Ser739 | NC | CS | + | − | − |
| Ser551 | NC | C | + | + | P | Ser740 | NC | NC | + | − | − |
| Ser553 | NC | C | + | + | L | Thr741 | NC | NC | + | ++ | − |
| Ser555 | NC | C | + | + | P | Ser742 | NC | CS | + | ++ | L |
| Ser557 | NC | N | + | + | − | Ser745 | NC | C | + | − | − |
| Ser559 | NC | N | + | + | − | Ser755 | NC | NC | − | + | − |
| Ser561 | NC | N | + | − | − | Ser757 | NC | CS | + | + | L |
| Thr562 | NC | N | − | + | − | Ser761 | NC | SC | + | + | P |
| Ser563 | NC | C | + | + | L | Thr762 | NS | CS | − | + | − |
| Ser566 | NC | C | + | − | − | Thr763 | NC | NC | − | ++ | − |
| Ser567 | NC | S | + | − | − | Thr764 | NC | C | − | ++ | − |
| Ser569 | NC | S | + | − | − | Ser767 | NC | C | − | ++ | − |
- C, conserved; NC, non-conserved; CS, conserved substitution; SC, semi-conserved; Pr, probable as Yin Yang site; HPr, highly probable as Yin Yang site; LPr, lower probale; +, positive prediction; ++, highly positive prediction; +++, very highly positive prediction; –, negative.
INTERPLAY OF PHOSPHORYLATION AND O-GlcNAcYLATION
De-phosphorylation on OH function of Ser/Thr provides the chances for other modifications like O-GlcNAc modification. It has been observed that alternation of O-GlcNAc modification with phosphorylation can occur on the same or neighboring Ser/Thr residues (known as Yin Yang sites) [Comer and Hart, 2000]. This interplay between two modifications on the same or neighboring residues is expected to modulate the specific function(s) of Oct-1 protein either by enhancing or inhibiting its function.
To investigate this competition of glycosylation and phosphorylation, a total of 81 Ser/Thr positive potential O-GlcNAc modification sites including 49 Ser and 32 Thr were predicted in different domains of Oct-1 using YinOYang 1.2 (Table I; Fig. 2). Some of these Ser/Thr residues were found conserved in vertebrates (Fig. 3) while others in invertebrates (Fig. 4). The C-terminal part showed a heavy potential for O-GlcNAc modification on various Ser and Thr residues. Amongst the potential phosphorylation and glycosylation sites in Oct-1 37 Ser/Thr sites were also predicted by YinOYang 1.2 to have potential for both phosphorylation and glycosylation (Yin Yang sites) (Fig. 2). From these 37 probable sites for Yin Yang interplay, five sites including Ser 409, 444, 473, and Thr 252, 271 were screened as highly probable sites to act as possible Yin Yang sites on the bases their complete conservation in vertebrate members and semi conservation in invertebrate members (Table I). Additionally, Ser 361, 365 are also the moderately probable Yin Yang sites in the linker region of POUs and POUh sub-domains. Whereas Thr 252 and 271 in N-terminal region and Ser 409 is in POUh domain and other Ser 444 and 473 in the proximal C-terminal domain one of the most probable Yin Yang site. As it has been proposed earlier that differential set of PTMs in DNA binding domain of Oct-2 play vital role in its binding to different DNA octamer motifs, whereas interplay of phosphorylation and O-GlcNAc modification in proximal C-terminal residues can regulate the activation of Oct-2 in mouse and human [Ahmad et al., 2004, 2006]. All the sites described above are fully conserved in mammalian and vertebrate members (Fig. 3), whereas, some of them are also conserved even in invertebrate members (Fig. 4).
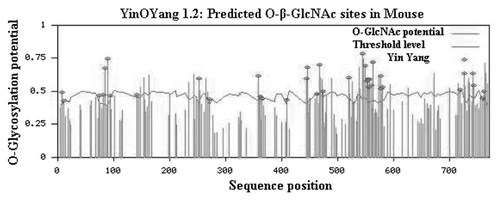
Graphic presentation of potential for O-GlcNAc modification on Ser and Thr (vertical green lines) predicted by YinOYang 1.2 in Mouse Oct-1 protein. The Yin Yang sites (Ser/Thr bearing potential both for phosphorylation and O-GlcNAc modification) are represented by an asterisk on the top of vertical lines. The threshold level is represented by horizontal variable blue line.
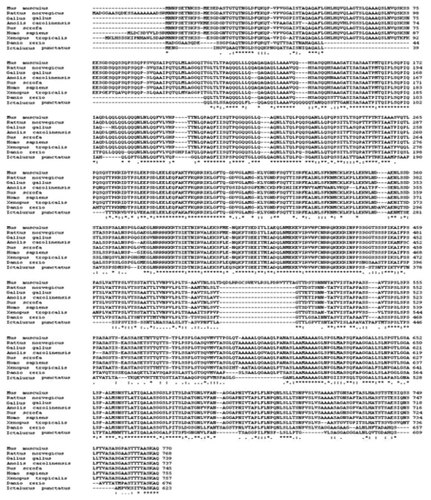
ClustalW [Thompson et al., 1994] results show the alignment of Oct-1 protein of Mus musculus (P25425) retrieved from the Swiss-Prot database [Boeckmann et al., 2003] with eight orthologues from different vertebrates including Homo sapiens (GenBank AAM77920.1), Sus scrofa (Swiss-Prot Q29076.1), Rattus norvegicus (Swiss-Prot P31503) Gallus gallus (Ref Seq NM_205472.1), Xenopus tropicalis (EMBL CR942771.2), Danio rerio (Ref Seq NP_001082798), and Ictalurus punctatus (EMBL CAA03984.1) selected by BLAST search [Altschul et al., 1997]. Conservation status of amino acids in different vertebrates is shown by asterisk for conservation in all members, double dot represent conserved substitution, whereas single dot represents semi conserved amino acid residue.

A multiple alignment of mouse Oct-1 with two invertebrate orthologs selected by BLAST search utilizing ClustalW. Conservation level of amino acids in the two invertebrates [Drosophilla melanogaster (P31368) and Caenorhabditis elegans (Ensembl K02B12.1)] is shown by asterisk for conservation in all members, double dot represent conserved substitution, whereas single dot represents semi-conserved amino acid residue.
Additionally, structural model of mouse POU domain was generated by homology modeling utilizing SwissModel [Kiefer et al., 2009] taking human Oct-1 structure 1cqt as template. The model showed that the amino acids Ser 358, 361, 365, and 409 in POU domain of mouse Oct-1 are in the structural positions, which can affect their binding with DNA octamer motif (Fig. 5) differently with different PTMs. Amongst these four sites the three sites including Ser 358, 361, and 365 are located in the linker region of two POUh and POUs sub-domains, since the linker region of these sub-domains is involved in producing flexibility resulting in differential binding patterns of the two sub-domains leading to a capability of Oct-1 to bind on promoters and enhancers of different genes. A phosphate group being small and bearing negative charge is expected to produce different structural and/or conformational changes compared to GlcNAc, a bulky and multiple OH group containing molecule. A differential combination of phosphorylation and OGlcNAc modification of these three sites (Ser 358, 361, and 365) in developing flexibility by different is quite probable (Fig. 5). Similarly, phosphorylated/O-GlcNAc modified Ser 409 located in distal part of POUh sub-domain can also produce the structural/conformational changes that can favor/disfavor Oct-1 binding with DNA. Consequently, these possible Yin Yang sites provide the basis for experimental investigations for their role in Oct-1 binding and activation. On the basis of the findings that phosphorylation and de-phosphorylation is a common mechanism in regulating the functions of different transcription factors [Boulikas, 1995], and phosphorylated Oct-1 results in repression of binding of Oct-1 with DNA [Roberts et al., 1991], it can be logical to propose that de-phosphorylation of Oct-1 facilitates its binding to DNA and ultimate gene expression (Fig. 6). We propose the occupation of the de-phosphorylated sites with O-GlcNAc resulting in facilitation of binding of Oct-1 with DNA. Moreover, phosphorylation of Oct-1 results in its failure to bind with DNA during G2 and M-phases [Hwang and Chae, 1989; Schild-Poulter et al., 2003] of the cell cycle (Fig. 6). Therefore, we propose a mechanism for H2B gene transcription that during G1 and S-phase the OGlcNAc modification on dephosphorylated amino acids may result in activation of the transcription machinery (Fig. 6). We also suggest that during G1 and S phases the Oct-1 may be modified by GlcNAc on one or more dephosphorylated Ser/Thr, in DNA binding domain, acting as Yin Yang sites. Thus, three potential Yin Yang sites, including Ser 361, 365 and 409, in DNA binding domain, two Yin Yang sites, Thr 252 and 271, in N-terminal region and three Yin Yang sites, Ser 444, 445, and 473, in proximal C-terminal domain can serve as the interplay sites for phosphorylation and O-GlcNAc modification for H2B gene regulation. When the transcription process is terminated, an enzyme OGlcNAcase (OGN) removes the O-GlcNAc and these Ser/Thr residues in DNA binding domain, are available for kinase action to phosphorylate them (Fig. 6). Phosphorylation of Oct-1 in its DNA binding domain results in its removal from DNA [Hwang and Chae, 1989; Schild-Poulter et al., 2003] and the cell enters in G2 phase spanning the M-phase (Fig. 5), consequently, inhibition of transcription process of H2B gene during G2 and M phases of the cell cycle may result (Fig. 5).
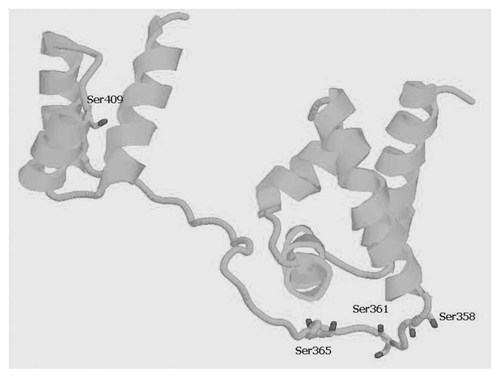
Homology model of POU domain of mouse Oct-1 showing the four sites (Ser358, 361, 365, and 409) in stick style with CPK color scheme. Three of them (Ser 358, 361, and 365), in the linker region of POUs and, POUh are in a position that can affect the flexibility of linker region by differential combinations of different PTMs resulting in diverse gene regulations by Oct-1. The fourth amino acid, Ser 409, is located in distal part of POUh sub-domain and different PTMs can also result in structural/conformational changes leading to diverse gene regulations.
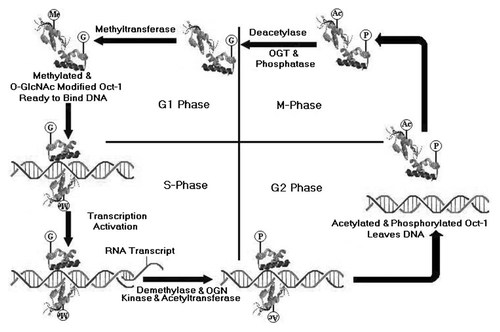
A scheme of the proposed mechanism for activation and inhibition of DNA–Oct-1 complex formation regulated by alternative and cell cycle phase specific phosphoyrlation, acetylation with O-GlcNAc modification and methylation, respectively. During G2 and M phase phosphorylation and acetylation of Oct-1 in its DNA binding domain result in inhibition of DNA–Oct-1 complex formation, whereas in G1 and S phases of the cell cycle, the O-GlcNAc modification and methylation result in activation of DNA–Oct-1 complex formation.
REGULATION BY METHYLATION AND ACETYLATION
Similarly, the competition of methylation with acetylation on same or adjacent Lys can be as promising as that for phosphorylation and O-GlcNAc modification. Prediction of acetylation by PAIL [Li et al., 2006] have shown 15 residues with a higher potential for acetylation including the Lys 9, 273, 297, 302, 316, 338, 342, 344, 384, 385, 399, 436, 438, 454, and 768. While prediction of methylation on Lys residues by MeMo [Chen et al., 2006] resulted in only two potential sites including Lys 73 and 385. Only Lys 385 located in POU domain, also conserved completely in all vertebrates (Fig. 3) and invertebrates (Fig. 4), has been predicted to be potential switch site for methylation and acetylation. Acetylation of basal transcription factors on Lys has been documented to regulate the activity of transcription factors [Imhof et al., 1997]. Additionally, YY1, a sequence-specific DNA binding transcription factor, is also differentially regulated by reversible phosphorylation and acetylation in its different domains involving both activation and repression of different genes during development and differentiation processes [Yao et al., 2001]. Therefore, acetylation of murine Oct-1 Lys 385 located in POUh subdomain can result in providing further strength for repressing the binding of Oct-1 with DNA octamer motif with a mechanism similar to phosphorylation as both bearing negative charges on them (Fig. 6). Similar to phosphorylation, reversible acetylation and deacetylation also act as a mechanism of protein regulations. Methlayion of deacetylated Lys 385 can result in inhibition of acetylation mediated binding repression of Oct-1 to DNA octamer motif (Fig. 6).
REGULATION BY METHYLATION AND PHOSPHORYLATION
Moreover, Lys methylation and Ser/Thr phosphorylation switch on vicinal amino acids can also result in some diverse regulations of Oct-1. The possible methylation and phosphorylation switch sites in Oct-1 can be Lys 73 with Ser 74 and Lys 385 with Ser 387 and 388. Methylation of these Lys 74 and 385 can result in reduced or complete absence of phosphorylation of the vicinal Ser residues (including Ser 74 and Ser 387 and 388).
CONCLUSIONS
In essence, we suggest that different PTMs and their interplay can regulate gene expression by controlling the transcriptional activation and repression in different phases of the cell cycle. Switching of one PTM with the other, in general, and interplay of phosphorylation and O-GlcNAc modification on OH function of Ser/Thr, in particular, is quite probable in different domains of murine Oct-1 and can be investigated experimentally as a possible mechanism for modulation of H2B and other genes regulation. Here, we suggest an involvement of acetylation for producing similar affects as those of phosphorylation and those of methylation similar to those of O-GlcNAc modification (Fig. 6). The PTMs switching with each other can act as a functional switch of proteins resulting to produce an ability in proteins to perform multifunctions. This can serve as a general mechanism to regulate gene transcription initiation and inhibition in different phases of the cell cycle for different transcription factors including Oct-1.
Specifically, interplay of phosphorylation and O-GlcNAc modification on Ser 358, 361 and 365 in linker region of POUs and POUh sub-domains can result in changing its flexibility resulting in binding of Oct-1 to different genes promoters and enhancers. Similarly, Ser 409 in POUh domain can also play a same role through interplay of phosphorylation and O-GlcNAc modification by inducing structural/conformational changes resulting in differential binding of POUh sub-domain to different gene promoters and/or enhancers.
Acknowledgements
A.R.S. and Nasir-ud-Din acknowledge financial support from Pakistan Academy of Sciences and EMRO-WHO for this work.




