Uniaxial mechanical tension promoted osteogenic differentiation of rat tendon-derived stem cells (rTDSCs) via the Wnt5a-RhoA pathway†
Yu Shi and Yujie Fu contributed equally to this work.
Abstract
Chronic tendinopathy is a tendon disorder that is common in athletes and individuals whose tendons are subjected to repetitive strain injuries. The presence of ossification worsened the clinical manifestation of the disorder. The change of tendon loading due to mechanical overload, compression, or disuse have been implicated as the possible etiologies, but the pathological mechanisms of tendinopathy remain unclear. In this study, we demonstrated that ossification in tendon tissue might be due to the osteogenesis of tendon-derived stem cells (TDSCs) induced by uniaxial mechanical tension (UMT) which mimics the mechanical loading in tendon. Rat TDSCs (rTDSCs) could be induced to differentiate into osteogenic lineage after treatment with 2% elongation UMT for 3 days as shown by the increased expression Runx2 mRNA and protein, Alpl mRNA, collagen type 1 alpha 1 (Col1a1) mRNA, ALP activity, and ALP cytochemical staining. RhoA, an osteogenesis regulator, was activated in rTDSCs upon UMT stimulation. Blockage of RhoA activity in rTDSCs by C3 toxin or ROCK activity, a downstream target of RhoA, by Y-27632 inhibited UMT-induced osteogenesis in rTDSCs. UMT up-regulated the mRNA expression of Wnt5a but not the other non-canonical Wnts. The inhibition of Wnt5a expression by siRNA abolished UMT-induced Runx2 mRNA expression and RhoA activation in rTDSCs and the inhibition of Runx2 expression could be rescued by addition of LPA, a RhoA activator. In conclusion, our results showed that UMT induced osteogenic differentiation of rTDSCs via the Wnt5a-RhoA pathway, which might contribute to ectopic ossification in tendon tissue due to mechanical loading. J. Cell. Biochem. 113: 3133–3142, 2012. © 2012 Wiley Periodicals, Inc.
Chronic tendon pathology (tendinopathy), although common, is difficult to treat. Calcifying tendinopathy is a tendon disorder with ossified deposits in the mid-substance presented with symptoms including chronic activity-related pain. It is a special case of tendinopathy and the presence of ossified deposits in calcifying tendinopathy worsens its clinical manifestations [Riley et al., 1996; Hashimoto et al., 2003]. Repeated minor strain injuries are thought to be the major precipitating factor in tendinopathy, although further work is required to determine whether it is mechanical over- or under-stimulation that leads to the change in tenocyte activity [Riley, 2008]. Recently, it has been demonstrated that stem cells naturally reside in tendon tissues [Bi et al., 2007; Rui et al., 2009, 2010; Yin et al., 2010]. Four percent mechanical tension was reported to stimulate BMP-2 expression in tendon-derived stem cells (TDSCs) and BMP-2 could induce osteogenic differentiation of TDSCs in vitro [Rui et al., 2011]. Furthermore, 8% mechanical stretch could also up-regulated the mRNA expressions of osteogenic (Runx2), chondrogenic (Sox9), and adipogenic (PPAR gamma) markers in both rabbit patellar and Achilles tendon stem cells [Zhang and Wang, 2010]. However, the direct evidence of mechanical tension-induced osteogenesis of TDSCs was limited and the mechanotranduction mechanism was still unclear. Runx2 is a master regulator of osteogenic differentiation of mesenchymal stem cells (MSCs) [Ducy, 2000], which can be regulated by small GTP binding proteins such as RhoA, a key regulator of osteogenesis in MSCs [McBeath et al., 2004; Arnsdorf et al., 2009a; Arnsdorf et al., 2009b]. Rho GTPases and its downstream kinase, RhoA-activated kinase (ROCK), play significant roles in regulating cytoskeletal dynamics and have been shown to be crucial for cell proliferation and differentiation [Welsh et al., 2001; McBeath et al., 2004; Arnsdorf et al., 2009b] as well as cell adhesion [Kobune et al., 2007; van Nieuw Amerongen et al., 2007]. Constitutive activation and inhibition of RhoA/ROCK have been reported to promote and inhibit the osteogenic differentiation of MSCs, respectively [McBeath et al., 2004]. Furthermore, it has been reported that mechanical force could promote osteogenesis of bone marrow-derived MSCs by activating RhoA [Arnsdorf et al., 2009b]. Another study also showed that continuous cyclic mechanical tension inhibited osteogenesis of MSCs by down-regulation of RhoA activity [Shi et al., 2011]. There were works focusing on the regulation of RhoA by non-canonical Wnts. The transduction of non-canonical Wnt signals via the small GTPases Rho and Cdc42, followed by the activation of c-Jun N-terminal kinase (JNK) have been demonstrated [Boutros et al., 1998; Yamanaka et al., 2002; Ryu and Chun, 2006]. Based on the downstream signaling mediators, the Wnt ligands are divided into two categories. One is the canonical Wnts which are beta-catenin-dependent, the other is the non-canonical Wnts, such as Wnt5a, Wnt5b, Wnt7a, and Wnt11, which function independently of the beta-catenin signaling [Church and Francis-West, 2002; Katoh, 2002]. As a non-canonical Wnt protein, Wnt5a has been shown to bind to its specific receptor Ror 1 and 2 (receptor tyrosine kinases-like orphan receptor) for signal transduction [Kuhl et al., 2000; Yamanaka et al., 2002; Ryu and Chun, 2006]. It is widely reported that beta-catenin is involved in mechanical loading-induced osteogenesis of MSCs. Sen et al. [2008] demonstrated that mechanical strain promoted osteogenesis while inhibited adipogenesis of C3H10T1/2 via beta-catenin. However, it is not clear that whether non-canonical Wnts also play an important role in mechanical tension-induced osteogenesis of MSCs. Moreover, a recent paper by Arnsdorf et al. indicated that non-canonical Wnt signaling, particularly with respect to Wnt5a, Ror2, and RhoA, mediated fluid flow-induced osteogenesis of C3H10T1/2 [Arnsdorf et al., 2009a]. Therefore, we hypothesized that uniaxial mechanical tension (UMT) could induce osteogenic differentiation of rat TDSCs (rTDSCs) via the Wnt5a-RhoA pathway. This study therefore aimed to investigate the effect of UMT on the osteogenic differentiation of rTDSCs. The role of Wnt5a-RhoA pathway in mediating the UMT-induced osteogenic differentiation of rTDSCs was also examined.
MATERIALS AND METHODS
Isolation and Culture of rTDSCs
rTDSCs were isolated according to the protocol of a previous study [Rui et al., 2010]. Three 8-week-old Sprague-Dawley rats, male, weighing 250–300 g, were used. The whole piece of intact patellar tendon was excised from both limbs of each rat. Care was taken that only the mid-substance tissue but not the tissue in the bone-tendon junction was collected. Peritendinous connective tissue was carefully removed and the samples were stored in sterile phosphate-buffered saline (PBS). The tissues were minced, digested for 2.5 h at 37°C with type I collagenase (3 mg/ml; Sigma-Aldrich, St Louis, MO) and passed through a 70 µm cell strainer (Becton Dickinson, Franklin Lakes, NJ) to yield single-cell suspension. The released cells were washed in PBS by centrifugation at 300g for 5 min and resuspended in growth medium (GM) which contained Dulbecco's Modified Eagle Medium (DMEM) (Gibco), 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin (all from Hyclone). The isolated cells were plated at an optimal cell density (about 50 cells/cm2) and cultured at 37°C, 5% CO2 to form colonies. At day 2 after initial plating, the cells were washed twice with PBS to remove the non-adherent cells. At day 7, they were trypsinized and mixed together as passage 0 (P0). Cells from passage 1 to passage 3 were used for all experiments. Culture medium was changed every 3 days during experiments.
Identification of rTDSCs
Colony assay
To determine the optimal initial plating density for the isolation and culture of stem cells from tendon tissue, nucleated cells derived from patellar tendon of three rats were plated in triplicate at 50, 500, and 5,000 cells/cm2 in 100 cm2 dishes each and cultured for 7 days. The cells were fixed by 4% paraformaldehyde and then stained with 2.5% crystal violet solution (Amresco, Solon OH). The optimal initial cell density was chosen based on the following criteria: (1) The colony size was not affected by colony-to-colony contact inhibition, and (2) the greatest number of colonies per tendon-derived nucleated cells was obtained.
Flow cytometry assay
Tendon-derived cells (5 × 105) at P3 were incubated with 1 µg of phycoerythrin (PE) -conjugated or fluorescein isothiocyanate (FITC) -conjugated mouse anti-rat monoclonal antibodies (R&D systems, Inc., Minneapolis, MN) for 45 min at room temperature. After washing with PBS at 12,000g for 5 min, the stained cells were re-suspended in 300 µl of ice cold PBS (with 10% FBS and 1% sodium azide) and subjected to fluorescent activated cell sorting (FACS) analysis (Becton Dickinson Biosciences, San Diego, CA). 104 events were counted for each sample. The percentage of cells with positive signal was calculated using the FACSCAN program (Becton Dickinson, San Jose, CA). Antibodies used included anti-CD45 (557015; BD Biosciences), anti-CD34 (sc-7324; Santa Cruz Biotechnology, Santa Cruz, CA, US), anti-CD31 (555027; BD Biosciences), anti-CD44 (ab33900; Abcam, Cambridge, UK), and anti-CD90.1 (551401; BD Biosciences). They were replaced with PE- or FITC-conjugated isotype-matched IgG1 in the negative controls (IC002P or IC002F, R&D systems, Inc.). Triplicates of cells from three rats were examined in this assay.
Multi-Lineage Differentiation Potential
Osteogenic differentiation assays
Tendon-derived cells were plated at 1 × 104 cells/cm2 in a six-well plate and cultured in complete culture medium until the cells reached confluence. They were then incubated in GM or osteogenic medium (Osteo-M), which was GM supplemented with 1 nM dexamethasone, 50 µM ascorbic acid, and 20 mM beta-glycerolphosphate (all from Sigma-Aldrich) for 21 days for the assessment of mRNA expression of Runt-related transcription factor 2 (Runx2) by real-time PCR as well as Alizarin Red S staining. For Alizarin Red S staining, the cultured cells were rinsed with PBS three times and fixed with 4% paraformaldehyde for 10 min at room temperature. The fixed cells were soaked in 0.5% Alizarin Red S (pH 4.1; Sigma-Aldrich) for 30 min at room temperature, washed with PBS, and were then observed under a digital camera (Canon IXUS 115 HS, Japan).
Adipogenic differentiation assays
Tendon-derived cells were plated at the same density as that indicated for the osteogenic assays. Two days later, the medium was replaced with GM or adipogenic medium (Adipo-M), which was GM supplemented with 500 nM dexamethasone, 0.5 mM isobutylmethylxanthine, 50 µM indomethacin, and 10 µg/ml insulin (all from Sigma-Aldrich). After another 2 days, the medium was replaced with GM containing only insulin. The medium was changed at 2-day intervals. The cells were cultured for 21 days for the assessment of mRNA expression of PPARgamma2 by real-time PCR (see below) as well as the presence of oil droplets by Oil red-O staining. The presence of oil droplets was confirmed by staining the cells with 0.3% fresh Oil red-O solution (Sigma-Aldrich) for 30 min after fixation with 70% ethanol for 10 min, and were then observed under a microscope with a CCD camera (Nikon eclipse TE2000, Japan).
Chondrogenic differentiation assays
For chondrogenic differentiation, a modified micromass culture system was applied as described previously [Mello and Tuan, 1999]. Briefly, the cells were harvested and resuspended in chondrogenic medium at 1 × 107 cells/ml. Twenty microliters cell suspension was carefully placed in the interior of each well of a 24-well plate. The cells were allowed to adhere at 37°C, 5% CO2 for 2 h, followed by the addition of 500 µl chondrogenic medium (Chondro-M) which contained GM, supplemented with 10 ng/ml rhTGF-beta1 (recombination human transforming growth factor-beta 1) and 50 ng/ml rhIGF1 (recombination human insulin-like growth factor 1) (R&D Systems, Inc.). After 24 h, the cell droplets coalesced and became spherical. The culture medium was changed every 3 days. After 12 days, the micromass was either fixed for toluidine blue staining or lysed for examination of mRNA expression of Sox9 as described below.
Mechanical Tensile Load Application
rTDSCs were plated at the density of 1 × 104 cells/cm2 in 2 ml of GM in six-well flexible silicone rubber BioFlex™ plates coated with collagen type I (Flexcell International Corporation, Hillsborough, NC). The cells were cultured for 48 h to allow them to attach and reach 80–90% confluence, at which time the GM was replaced, and then uniaxial cyclic mechanical tension (UMT) at 0.5 Hz sinusoidal curve, 2% elongation was applied to the cells for 4 h per day using an FX-4000T™ Flexercell® Tension Plus™ unit (Flexcell International Corporation). The cultures were incubated in a humidified atmosphere at 37°C and 5% CO2 while stretching (UMT group) or in the static state (NC group).
Biochemical Agents and siRNA Transfection
The following biochemical agents were used in this study: 10 µM lysophosphatidic acid sodium salt (LPA) (Sigma-Aldrich), 0.5 µg/ml C3 toxin (Cat. # CT40, Cytoskeleton, Denver, CO, US), 10 µM Y-27632 (Sigma-Aldrich). rTDSCs were exposed to each biochemical agent for 1 h before UMT stimulation. 0.4 µM siRNA against Wnt5a or scrambled siRNA was transiently transfected into rTDSCs by lipofectamine 2000 (Invitrogen, Carlsbad, CA). After 4 h of incubation, it was replaced by GM. The sequences of siRNA targeting Wnt5a (Shanghai GenePharma Co., Ltd., China) were as follows: Sense 5′-GGUGGUCCCUAAGUAUGAATT-3′ and anti-sense 5′-UUCAUACUUAGGGACCACCTT-3′. The sequences of scrambled control siRNA (Shanghai GenePharma Co., Ltd.) were as follows: Sense 5′-UUCUCCGAACGUGUCACG-3′ and anti-sense 5′-ACGUGACACGUUCGGAGAATT-3′.
Alkaline Phosphatase (ALP) Activity Assay
After being treated with or without 2% UMT for 3, 7, and 14 days, rTDSCs were tested by ALP activity assay. Cells were rinsed two times with ice-cold PBS, scrapped from the dishes and suspended in ddH2O. This was followed by three cycles of freezing and thawing. ALP activity was determined at 405 nm using p-nitrophenyl phosphate (pNPP) (Sigma-Aldrich) as the substrate. A 50 µl of sample was mixed with 50 µl of pNPP (1 mg/ml) in 1 M diethanolamine buffer containing 0.5 mM MgCl2 (pH9.8) and incubated at 37°C for 15 min on a bench shaker. The reaction was stopped by the addition of 200 µl of 2 M NaOH per 200 µl of reaction mixture. Total protein content was determined by the BCA method with a protein assay kit (PIERCE, Rockford, IL, US). ALP activity was calculated as nmol p-nitrophenol per minute per mg protein, and presented as fold changes over the non-loading group at day 3. This experiment was conducted in triplicate.
ALP Staining
After being treated with or without 2% UMT for 14 days, the rTDSCs were tested by ALP staining. The cultured cells were rinsed with PBS three times and fixed with 4% paraformaldehyde for 10 min at 4°C. The fixed cells were soaked in 0.1% naphthol AS-MX phosphate and 0.1% fast red violet LB salt in 56 mM 2-amino-2-methyl-1,3-propanediol (pH9.9, all from Sigma-Aldrich) for 10 min at room temperature, washed with PBS, and were then observed under a digital camera (Canon IXUS 115 HS) or a microscope (Nikon eclipse TE2000).
Active RhoA Detection
rTDSCs were harvested at day 3 with and without 2% UMT, rinsed with ice-cold PBS, and lysed at 4°C with 200 µl of lysis buffer [50 mM Tris pH 7.2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 500 mM NaCl, 10 mM MgCl2, 10 µg/ml aprotinin/leupeptin, and 1 mM PMSF (Sigma-Aldrich)]. Samples were centrifuged for 10 min at 12,000g at 4°C. Twenty microliters of supernatant was used to determine the expression of total RhoA. The remaining supernatant was used to isolate the GTP-bound RhoA (active RhoA) using EZ-DetectTM Rho Activation Kit (catalog number: 89854, Thermo Scientific Pierce), which contained SwellGel® Immobilized Glutathione Disc corresponding to the residues of rhotekin RhoA-binding domain (GST-Rhotekin-RBD), according to the manufacturer's instructions. The expressions of total and active-RhoA were then detected by Western blotting.
Real-Time PCR
Total RNA of cells was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. After reverse transcription of total RNA, real-time PCR was performed by a Roche Applied Science LC480 system using SYBR®Premix Ex TaqTM (Takara, Dalian, China) according to the manufacturer's instructions. The conditions of real-time PCR were as follows: Denaturation at 95°C for 5 min, 50 cycles at 95°C for 5 sec, 60°C for 10 sec. Dissociation stage was added to the end of the amplification procedure. There was no non-specific amplification as determined by the dissociation curve. Gapdh was used as the internal control. Data were analyzed using the comparison Ct (2-ΔΔCt) method [Pfaffl, 2001] and expressed as fold change compared to non-loading group on day 3. Each sample was analyzed in triplicate. The primer sequences used in this study were as follows: Gapdh (Glyceraldehyde-3-phosphate dehydrogenase) (accession no. NM_017008.3) forward 5′-CTCAACTACATGGTCTACATGTTCCA-3′ and reverse 5′-CTTCCCATTCTCAGCCTTGACT-3′; Alpl (Alpl alkaline phosphatase) (accession no. NM_013059.1) forward 5′-CGTCTCCATGGTGGATTATGC -3′ and reverse 5′-TGGCAAAGACCGCCACAT-3′; Runx2 (accession no. NM_053470.1) forward 5′-CGGAGCGGACGAGGCAAGAG-3′ and reverse 5′-AGAGTCATCAAGCTTCTGTCTGTGC-3′; Col1a1 (collagen, type I, alpha 1) (accession no. NM_053304.1) forward 5′-TTCACCTACAGCACGCTTGTG -3 and reverse 5′-GATGACTGTCTTGCCCCAAGTT-3′; Wnt5a (accession no. NM_022631.1) forward 5′-AGTTTCACTGGTGCTGCTA-3′ and reverse 5′-ATATGTGGGTCCTGGGAG-3′; Wnt5b (accession no. NM_001100489.1) forward 5′-GCAGGGTCATGCAGATAGG-3′ and reverse 5′-CGGTAGCCATACTCCACGT -3'; Wnt7a (accession no. NM_001100473.1) forward 5′-GGGAGTCAGCCTTCGTGT-3′ and reverse 5′-GGTTGTCCGCACATCCTC-3′; Wnt11 (accession no. NM_080401.1) forward 5′-GTTCAAACCTCGCCATTA-3′ and reverse 5′-CTTAGAAAGCCTCTTATCCAGT-3′; Ror1 (accession no. NM_001108671.1) forward 5′-ATTGACGGTTCTCATTTAGC-3′ and reverse 5′-TTCCCAACAACATATCATACAC-3′; Ror2 (accession no. NM_001107339.1) forward 5′-ATTGGGAACCGAACTATTTATG-3′ and reverse 5′-TCTGACAGGTGCGTGGAG-3′.
Western Blotting
The cells were lysed on ice for 30 min in lysis buffer containing 50 mM Tris-HCl, pH7.4, 150 mM NaCl, 1%Nonidet P-40, and 0.1%SDS supplemented with protease inhibitors (10 µg/ml leupeptin, 10 µg/ml pepstatin A, and 10 µg/ml aprotinin). For Western analysis, 15 µg of sample was resolved on 12% SDS–PAGE and electro-transferred onto nitrocellulose membranes (Whatman, Piscataway, NJ). The primary antibodies used were anti-Runx2 (rabbit polyclonal anti-Runx2 (M-70), Santa Cruz Biotech, Santa Cruz Biotechnology) at a dilution of 1:1000; anti-Wnt5a (rabbit polyclonal anti-Wnt5a (ab72583), Abcam, Cambridge, MA) at a dilution of 1000; anti-RhoA (89854), Thermo Scientific Pierce) at a dilution of 1:1000. For the normalization of protein loading, anti-beta-actin antibody (mouse monoclonal anti-beta-actin antibody (A1978), Sigma-Aldrich) was used at a dilution of 1:5000. HRP-conjugated secondary antibodies were used at a dilution of 1:2000. The antigen–antibody complexes were visualized using the enhanced chemiluminescence detection system (Millipore, Billerica, MA) as recommended by the manufacturer. The immunoreactive bands were semi-quantitatively analyzed by normalizing the band intensities to their respective controls on scanned films with Quantity One® software (Bio-Rad Laboratories, Inc., CA).
Statistical Analysis
The comparison between the induction and GM groups was made by performing Student's t-test. The comparisons between NC and UMT groups as well as vehicle versus biochemical agents (LPA and C3-toxin) were made by performing ANOVA, followed by Student–Newman–Keuls post-hoc test. All the data analyses were done using SPSS software (SPSS Inc, Chicago, IL, version 16.0). Statistical significance was denoted by P < 0.05.
RESULTS
Identification and Characterization of Rat Tendon-Derived Stem Cells
The rat tendon-derived stem cells (rTDSCs) showed fibroblast-like morphology from P0 to P3 (Fig. 1A) and formed colonies after in vitro culture for 7 days as indicated by 2.5% crystal violet staining. To determine the optimal initial plating density that gave the maximum number of cell colonies, we plated the cells isolated from tendon tissue at different densities. Colonies at 50 cells/cm2 seeding densities formed well according to our assessment criteria (Fig. 1B). These cells expressed MSC markers CD44 and CD90.1 but were negative for hematopoietic marker CD34, leukocyte marker CD45, and endothelial cell marker CD31 (Fig. 1C). Osteogenic differentiation assays showed that most of the cells formed mineralized calcium deposits, as confirmed by Alizarin Red S staining, after 21 days of induction. Meanwhile, there was significant increase in Runx2 expression after osteogenic induction (Fig. 1D). The chondrogenic differentiation of rTDSCs was verified by positive toluidine blue staining after 12 days of micromass culture and induction. The micromass after chondrogenic induction in the chondrogenic medium (Chondro-M) showed higher toluidine blue staining than micromass in GM. There was significantly higher expression of Sox9 after chondrogenic induction (Fig. 1E). The cells also had the capacity to undergo adipogenic differentiation. Lipid droplets were formed after incubating the cells in complete medium with adipogenic supplements for 21 days. This was not observed in the GM group (Fig. 1F). The mRNA expression of PPAR gamma 2 was significantly up-regulated at day 21 after adipogenic induction (Fig. 1F).
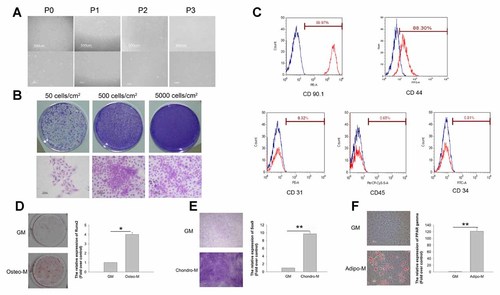
Identification and characterization of rat tendon derived stem cells (rTDSCs). A: The morphology of rTDSCs was observed from passage 0 to passage 3. In the upper panel, scale is 500 µm; in the lower panel, the scale is 100 µm. B: Formation of cell colonies of rTDSCs after culturing for 7 days at different initial seeding densities. Distinctive colonies were stained by 2.5% crystal violet and observed at the initial seeding density of 50, 500, and 5,000 cells/cm2. In the upper panel, the magnification is 1×; while in the lower panel, scale is 100 µm. C: Graphs showing the expression of rat MSC markers (CD44 and CD90.1), endothelial cell marker (CD31), leukocyte marker (CD45), and hematopoietic stem cell marker (CD34) on rTDSCs. Red traces show TDSCs stained with tested antibodies, while blue traces indicate TDSCs stained with relative IgG1 as the negative controls. D: Osteogenic differentiation potential of cells derived from the rTDSCs colonies in vitro. Alizarin Red S staining of cells and Runx2 expression levels were detected after culturing rTDSCs for 21 days in osteogenic medium (Osteo-M) or growth medium (GM) (D). Magnification: 1×. The bars represent mean +/− SD. (n = 3,*P < 0.05) (E) Chondrogenic differentiation potential of micromass of rTDSCs in vitro. Toluidine blue staining of cells and Sox9 expression levels were detected after culturing rTDSCs for 12 days in chondrogenic medium (Chondro-M) or growth medium (GM) (E). Scale: 100 µm. The bars represent mean +/−SD. (n = 3, **P < 0.01) (F) Adipogenic differentiation potential of cells from rTDSCs colonies in vitro. Oil Red O staining of cells and PPAR gamma expression level after culturing the cells for 21 days in adipogenic medium (Adipo-M) or growth medium (GM). Scale: 50 µm. The bars represent mean +/− SD. (n = 3, **P < 0.01). [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/jcb]
UMT Induced Osteogenic Differentiation of rTDSCs
Compared with the non-loading (NC) group, there was significant increase in ALP activity in rTDSCs after 2% UMT stimulation for 3, 7, and 14 days (Fig. 2A). The expressions of osteogenic genes such as Runx2, Alpl, and Col1a1 in rTDSCs with and without UMT stimulation were also detected by real-time PCR. Compared with the NC group, there was significant up-regulation of Runx2 in rTDSCs upon 2% UMT treatment at day 3, 7, and day 14 (Fig. 2B). The expressions of Col1a1 (Fig. 2C) and Alpl were also up-regulated in rTDSCs upon UMT treatment at day 3, 7, and 14 (Fig. 2D). The protein level of Runx2 in the UMT group also increased significantly to almost 2 and 2.5-folds over that in the control group at day 7 and day 14, respectively (Fig. 2E). At day 14, rTDSCs in the UMT group also showed higher level of ALP staining compared to that in the NC group (Fig. 2F).
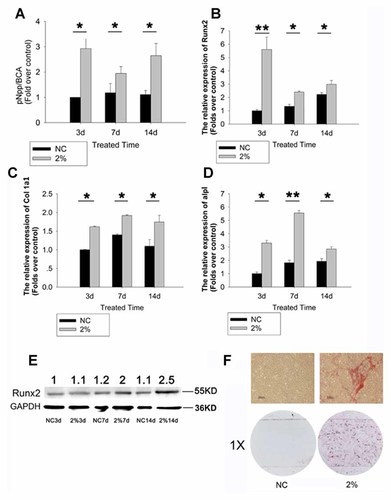
UMT induced the expression of osteogenic genes in rTDSCs. Graphs showing the (A) ALP activity; (B) Runx2 mRNA expression; (C) Col1a1 mRNA expression; and (D) Alpl mRNA expression in rTDSCs in the UMT treated group (2%) and non-loading control group (NC) at day 3, 7, and 14. Gapdh expression was used as an internal control for mRNA expression. Data were normalized to the sample at day 3 in the NC group and presented as mean ± SD (n = 5; *P < 0.05; **P < 0.01). E: Runx2 protein level was also assessed. The densitometric analysis of Runx2 protein expression was normalized to GAPDH. The fold change of Runx2 expression relative to that in the NC group at day 3 was shown on the top. F: Photomicrographs showing ALP staining in rTDSCs in the UMT treated group (2%) and non-loading control group (NC) at day 14. The scale of the upper panel is 500:m; the magnification of the lower panel is 1×. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/jcb]
UMT Up-Regulated Runx2 Expression in rTDSCs Via RhoA
To examine if RhoA was involved in the regulation of osteogenic differentiation in rTDSCs upon 2% UMT stimulation, we first compared the activity of RhoA in rTDSCs with and without 2% UMT at day 3 and day 7. Our results showed that there was significant increase in the activity of RhoA in the UMT group compared to that in the NC group on both day 3 (4.6-fold; Fig. 3A) and day 7 (1.8-fold; Fig. 3B). Inhibition of RhoA and ROCK expression in rTDSCs, by C3 toxin and Y-27632, respectively, prior and during UMT stimulation inhibited the expression of Runx2, Col1a1, and Alpl at day 3 and day 7 (Fig. 3.C–E). As there was significant change of Runx2 protein level at day 7 in the UMT group compared with that in the NC group (Fig. 2), we investigated the alternation of Runx2 protein expression after 7 days' UMT with or without C3 toxin and Y-27632. These data indicated that, similar to the results shown in Figure 2, 2% UMT (2% + Veh) increased Runx2 protein level by 1.6-folds compared with that in the NC group. Inhibition of RhoA/ROCK activities by C3 toxin/Y-27632 abrogated the up-regulation of Runx2 protein expression and reduced Runx2 expression to 0.9-fold (2% + C) or 0.5-fold (2% + Y), respectively, compared to those in the NC group (Fig. 3.F).
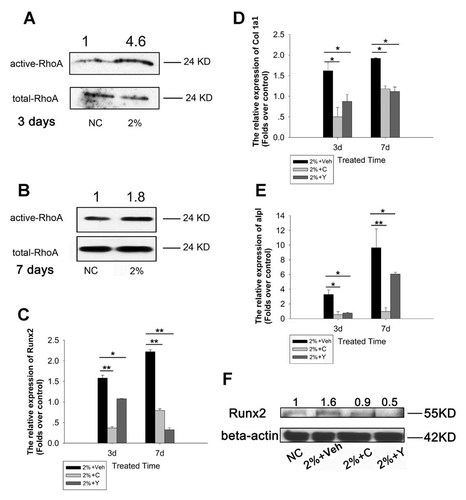
UMT induced Runx2 expression in rTDSCs via the RhoA-ROCK pathway. rTDSCs were treated with or without UMT for 3 days (A) or 7 days (B). The active and total RhoA were detected by Western blotting. The densitometric analysis of active-RhoA protein expression was normalized to total-RhoA which was used as the loading control. The data were shown as the folds over NC group. rTDSCs were pre-treated with or without 10 µM Y-27632 or 0.5 µg/ml C3 toxin for 1 h and then were kept being treated with LPA or C3 toxin during UMT stimulation. Runx2 mRNA expression (C), Col1a1 mRNA expression (D) and Alpl mRNA expression (E) were detected after UMT treatment for 3 days or 7 days. Data were showed as fold changes relative to non-loading (NC) group at day 3 and presented as mean ± SD (n = 3; *P < 0.05; **P < 0.01). Runx2 protein level was also detected at day 7 (F). The densitometric analysis of Runx2 protein expression was normalized to beta-actin which was used as the loading control. The data were shown as the fold changes over the NC group. NC stands for the non-loading group. 2%+Veh stands for the 2% UMT only group. 2%+C stands for the UMT with C3-toxin treatment. 2%+Y stands for the UMT with Y-27632 treatment.
UMT Up-Regulated the Expression of Wnt5a and Ror in rTDSCs
Next, we examined the expression of non-canonical Wnt5a, Wnt5b, Wnt7a, and Wnt11 in UMT induced-osteogenesis of rTDSCs. The mRNA expression of Wnt5a was up-regulated after UMT treatment at day 3 and 7 (Fig. 4A). However, there was no significant change in the mRNA expression of Wnt5b (Fig. 4B), Wnt7a (Fig. 4C), and Wnt11 (Fig. 4D) in the UMT-treated group compared with that in the NC group at day 3 and 7. The expression of Wnt5a's receptor, tyrosine kinases-like orphan receptor Ror1 (Fig. 4E), and Ror2 (Fig. 4F) was up-regulated in the UMT-treated group compared with that in the NC group at day 3. Moreover, the expression of Ror2, but not Ror1, remained high at day 7 after UMT stimulation (Fig. 4E,F).
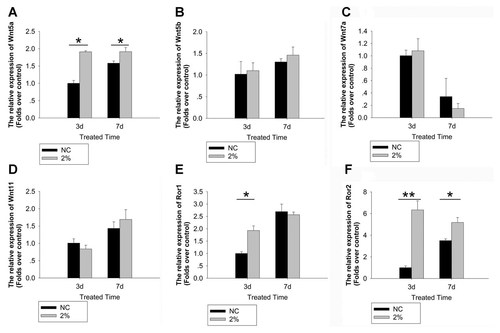
UMT induced Wnt5a and its receptor expression. rTDSCs were treated with or without UMT for 3 days or 7 days. Wnt5a (A), Wnt5b (B), Wnt7a (C), Wnt11 (D), Ror1 (E), and Ror2 (F) mRNA expression were detected by real-time PCR. Data were showed as folds changes relative to the respective NC group at day 3 and presented as mean ± SD (n = 3; *P < 0.05; **P < 0.01). NC stands for the non-loading group, while 2% stands for the 2% UMT group.
Wnt5a Regulated UMT Induced-Runx2 Expression in rTDSCs Via RhoA
To investigate whether Wnt5a mediated UMT-induced ostegenesis in rTDSCs via RhoA pathway, we used specific siRNA to knock down the Wnt5a expression. The protein expression of Wnt5a was significantly decreased to 0.3-fold over the siRNA-control group (Fig. 5A). After treatment of rTDSCs with UMT for 3 days, RhoA activity in Wnt5a knocking-down group was lower compared with that in the siRNA-control group (about 0.6-fold; Fig. 5B). The transfection of siRNA-Wnt5a also abolished Runx2 up-regulation in rTDSCs induced by UMT and this abolishment could be rescued by LPA, a RhoA activator (Fig. 5C).
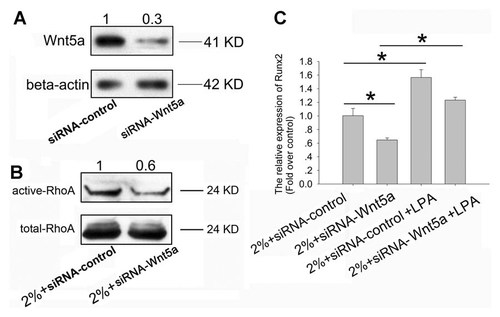
Wnt5a regulated UMT induced-Runx2 expression in rTDSCs via RhoA. Wnt5a protein level was detected after transfection of siRNA against Wnt5a for 3 days. The densitometric analysis of Wnt5a protein expression was normalized to beta-actin which was used as the loading control. The data were shown as the fold over scrambled siRNA transfection group (siRNA-control) (A). After transfected with the siRNA for 24 h, rTDSCs were treated with 2% UMT. Three days later, the cells were harvested for detecting the expression of active-RhoA and total-RhoA. The densitometric analysis of active-RhoA protein expression was normalized to total-RhoA which was used as the loading control. The data were shown as the fold over 2%+siRNA-control transfection group (B). After transfection with siRNA for 24 h, rTDSCs were incubated with or without LPA for 1 h. UMT was then applied for 3 days. The expression of Runx2 was measured by real-time PCR (C). Data were shown as fold changes over the 2%+siRNA-control group and presented as mean ± SD (n = 3; *P < 0.05).
DISCUSSION
The mechanisms of tendinopathy and calcifying tendinopathy remain largely unclear and there is no successful treatment. Recently, MSCs were reported to reside in tendon tissue [Bi et al., 2007; Rui et al., 2010] which were called TDSCs. The TDSCs could be induced to undergo osteogenesis when mechanical loading was applied [Arnsdorf et al., 2009a; Arnsdorf et al., 2009b; Yourek et al., 2010]. It was also reported that mechanical tension could up-regulate expression of BMP2 in TDSCs [Rui et al., 2011]. This study was therefore conducted to understand the molecular mechanisms underlying mechanical tension-induced osteogenesis in rTDSCs.
In this study, by applying UMT on TDSCs, we showed that UMT promoted osteogenic differentiation of rTDSCs as indicated by the increase in Runx2 expression, ALP activity, ALP staining, mRNA expression of Alpl and Col1a1. This observation fitted well with the hypothesis of erroneous differentiation of TDSCs as a result of changes of tendon loading, to non-tenocyes, might result in histopathological changes such as ectopic ossification, as seen in tendinopathy [Lui and Chan, 2011; Rui et al., 2011].
Interestingly, in Wang's article [Zhang and Wang, 2010], it was indicated that in the low mechanical stretching at 4% (“clamp-to-clamp” engineering strain) promoted differentiation of tendon stem cells (TSCs) into tenocytes, whereas large stretching at 8% induced differentiation of some TSCs into adipogenic, chondrogenic, and osteogenic lineages. The apparent discrepancy between our data and theirs might be due to different loading systems and loading regimens. Besides, cyclic stretching of 4 or 8% at 0.5 Hz was applied to cells once for only 12 h in Zhang and Wang's study while we applied 2% UMT 4 h per day for at least 3 days.
It has been reported that Wnt5a mediated fluid shear loading-induced osteogenesis of C3H10T1/2 [Arnsdorf et al., 2009a]. After application for 35 min, oscillatory fluid flow induced beta-catenin translocation and TCF/LEF-dependent gene transcription in C3H10T1/2. When Wnt5a was knocked down by siRNA, the osteogenesis of C3H10T1/2 cells was abolished. Therefore, we investigated the mRNA expression of Wnt5a and its receptors Rors in rTDSCs after UMT stimulation. Interestingly, Wnt5a, but not other non-canonical Wnts, such as Wnt5b, Wnt7a, and Wnt11, was specifically up-regulated after UMT treatment. The knocking down of Wnt5a expression abolished Runx2 expression induced by UMT, indicating that Wnt5a was an up-stream regulator mediating UMT induced-Runx2 expression. Our results also showed that knocking down of Wnt5a inhibited RhoA activity after UMT treatment while RhoA activation by LPA could rescue the abrogation of UMT-triggered Runx2 expression induced by Wnt5a siRNA transfection in rTDSCs. These data suggested that the effect of Wnt5a on the osteogenic differentiation of rTDSCs upon UMT stimulation was mediated via RhoA.
Recently, it has been shown by Rui et al. [2011] that after cyclic uniaxial stretching for 4 h, BMP2 expression was highly expressed. There might be interaction between BMP2 and RhoA pathway in the induction of osteogenesis by TDSCs upon mechanical loading. In the human pulmonary artery smooth muscle cells, BMP2 induced pulmonary angiogenesis via Wnt3a-beta-catenin as well as Wnt3a-RhoA-Rac1 pathways [de Jesus Perez et al., 2009]. However, in the osteogenesis of MSCs, the relationship between BMP2 and Wnt-RhoA pathway is still not clear and should be investigated further.
In conclusion, our results showed that UMT promoted the osteogenic differentiation of rTDSCs through the Wnt5a-RhoA pathway. This work thus provided insight about the possible molecular mechanism of ectopic ossification in tendon tissue due to mechanical loading.




