Negative regulation of wnt11 expression by Jnk signaling during zebrafish gastrulation†
Jungwon Seo and Yoichi Asaoka contributed equally to this work.
Abstract
Stress-induced Sapk/Jnk signaling is involved in cell survival and apoptosis. Recent studies have increased our understanding of the physiological roles of Jnk signaling in embryonic development. However, still unclear is the precise function of Jnk signaling during gastrulation, a critical step in the establishment of the vertebrate body plan. Here we use morpholino-mediated knockdown of the zebrafish orthologs of the Jnk activators Mkk4 and Mkk7 to examine the effect of Jnk signaling abrogation on early vertebrate embryogenesis. Depletion of zebrafish Mkk4b led to abnormal convergent extension (CE) during gastrulation, whereas Mkk7 morphants exhibited defective somitogenesis. Surprisingly, Mkk4b morphants displayed marked upregulation of wnt11, which is the triggering ligand of CE and stimulates Jnk activation via the non-canonical Wnt pathway. Conversely, ectopic activation of Jnk signaling by overexpression of an active form of Mkk4b led to wnt11 downregulation. Mosaic lineage tracing studies revealed that Mkk4b-Jnk signaling suppressed wnt11 expression in a non-cell-autonomous manner. These findings provide the first evidence that wnt11 itself is a downstream target of the Jnk cascade in the non-canonical Wnt pathway. Our work demonstrates that Jnk activation is indispensable for multiple steps during vertebrate body plan formation. Furthermore, non-canonical Wnt signaling may coordinate vertebrate CE movements by triggering Jnk activation that represses the expression of the CE-triggering ligand wnt11. J. Cell. Biochem. 110: 1022–1037, 2010. © 2010 Wiley-Liss, Inc.
Abbreviations used:
CE, convergent extension; Jnk, c-Jun N-terminal kinase; Mkk, mitogen-activated protein kinase kinase; Dpp, decapentaplegic; MO, morpholino; aa, amino acid; hpf, hours post-fertilization; WT, wild type; slb, silberblick; ppt, pipetail; caMkk4b, constitutively active Mkk4b.
Stress-activated protein kinase/c-Jun N-terminal kinase (Sapk/Jnk) is activated in response to a variety of cellular stresses. Once activated, Jnk phosphorylates downstream targets, including the c-Jun component of the activator protein-1 (AP1) transcription factor. Mkk4 and Mkk7 are two upstream Mapk kinases that interact with downstream kinases and scaffold proteins to activate Jnk. The outcome of the signaling cascades initiated by Mkk4 and Mkk7 is the phosphorylation of the Tyr and Thr residues, respectively, in the Jnk's Thr-Pro-Tyr motif [Davis, 2000; Chang and Karin, 2001]. However, there is evidence that the specific transmission of signals from these upstream kinases to Jnk may rely on different sets of, and/or interactions with, downstream kinases and scaffold proteins [Whitmarsh et al., 1998], such that Mkk4 and Mkk7 have distinct biological functions in vivo. For example, Mkk7 (but not Mkk4) is an essential and specific component of the Jnk signaling pathway activated by proinflammatory cytokines [Tournier et al., 2001]. In addition, although they both die of massive liver cell apoptosis, mkk4−/− mice die on embryonic day 10.5 (E10.5), whereas mkk7−/− mice die on E11.5 [Nishina et al., 1997, 1999; Yang et al., 1997; Ganiatsas et al., 1998; Watanabe et al., 2002; Wada et al., 2004].
Analyses of various jnk knockout mice have revealed much about the physiological role of Jnk signaling in embryogenesis. In mammals, the Jnk family consists of three related genes, jnk1, jnk2, and jnk3 [Derijard et al., 1994; Kallunki et al., 1994; Mohit et al., 1995]. A role for Jnk in tissue morphogenesis was first suggested by the observation that Jnk1−/−Jnk2−/− double mutant mice died at E11 with defective closure of the neural tube in the hindbrain [Kuan et al., 1999]. In this case, Jnk was required to control the survival and apoptosis of neuronal cells. More recently, Jnk has emerged as a critical regulator of cell migration and the morphogenetic movement of epithelial sheets. In Drosophila, a well-orchestrated Jnk signaling pathway is required for the sealing of embryonic epidermis in a process known as dorsal closure [Glise et al., 1995; Riesgo-Escovar et al., 1996; Sluss et al., 1996]. Jnk is activated in the leading edge of the dorsal epidermis at the onset of dorsal closure and drives the expression of the TGFβ homolog Decapentaplegic (Dpp), a secreted morphogen that regulates dorsal closure [Reed et al., 2001]. Little is known as yet about the function of the Jnk pathway in vertebrate morphogenesis, particularly when the body plan is first laid down.
The basic body plan of vertebrate embryos is established during gastrulation by a series of coordinated cell movements that lead to the formation of endoderm, mesoderm, and ectoderm, and overtly shape the embryonic axis [Keller, 2002]. A major driving force of vertebrate gastrulation is convergent extension (CE), a mechanism in which dorsal mesodermal cells polarize, elongate along the mediolateral axis, and intercalate toward the midline (convergence), leading to the extension of the anterior–posterior axis of the embryo [Keller, 2002; Tada et al., 2002; Seifert and Mlodzik, 2007]. CE is well conserved among vertebrate species, including in frog (Xenopus laevis) and zebrafish (Danio rerio) [Solnica-Krezel, 2005].
The study of gastrulation in mammals is difficult because these animals develop in utero, preventing direct observation of the embryos. In contrast, fertilized zebrafish eggs develop ex utero into transparent embryos that can be directly observed and are highly amenable to manipulations such as tissue transplantation and molecular perturbation. There is a high degree of conservation between zebrafish and mammalian genes, and a shared developmental path that results in fundamental similarities in many tissues and organs. In addition, there exists a wide selection of mutant zebrafish lines with developmental abnormalities, including gastrulation defects. Thus, zebrafish provide a very attractive alternative to mammals for studying the molecular and cellular bases of vertebrate morphogenesis.
Genetic analyses of gastrulation mutants in zebrafish and functional studies in Xenopus have revealed that CE is regulated by the non-canonical Wnt signaling pathway, which does not involve β-catenin [Seifert and Mlodzik, 2007]. Wnt11 and Wnt5 have been found to be essential ligands for normal cell movements during vertebrate CE. The zebrafish mutants silberblick (slb) and pipetail (ppt) have a mutation in wnt11 or wnt5, respectively, and slb–ppt double mutants show severe CE defects [Heisenberg et al., 2000; Kilian et al., 2003]. Interestingly, wnt5 mRNA can partially rescue the slb phenotype [Kilian et al., 2003], indicating a partial redundancy of Wnt11 and Wnt5 functions. In Xenopus, Wnt11 and Wnt5 bind to the Frizzled receptor, initiating non-canonical Wnt signaling that leads to activation of RhoA and Rac [Habas et al., 2001, 2003]. This RhoA and Rac activation triggers Jnk signaling critical for CE in Xenopus [Habas et al., 2003; Kim and Han, 2005]. Notably, depletion of Jnk in Xenopus causes defective gastrulation in these animals [Yamanaka et al., 2002]. All these findings suggest that Jnk is an essential component of the non-canonical Wnt pathway involved in vertebrate CE. However, the precise molecular mechanism by which Jnk signaling regulates CE has remained obscure. In an effort to elucidate the downstream targets of Jnk signaling in the non-canonical Wnt pathway that is associated with early embryogenesis, we have analyzed the functions of Mkk4 and Mkk7 orthologs in zebrafish (Mkk4a, Mkk4b, and Mkk7). Using morpholino-mediated knockdown, we demonstrate that Mkk4b is essential for CE movements. Furthermore, we provide the first evidence that wnt11 itself is a downstream target of the Jnk cascade in the non-canonical Wnt pathway associated with early embryogenesis.
MATERIALS AND METHODS
Zebrafish Strains
The AB and TL wild-type (WT) strains were maintained essentially as described in “The Zebrafish Book” [Westerfield, 1994]. Embryos were produced by natural matings and staged by standard morphological criteria or by hours post-fertilization (hpf), as described [Kimmel et al., 1995; Asaoka et al., 2002].
Cloning of Zebrafish mkk4 and mkk7 Genes
Zebrafish sequences highly homologous to mouse mkk4 and mkk7 cDNAs were identified by database searching. Full-length zebrafish cDNAs were obtained by 5′- and 3′-RACE PCR according to the manufacturer's protocols (Invitrogen). RACE-PCR fragments were purified and subcloned into pGEM-T easy (Promega).
Semi-Quantitative RT-PCR Analysis
Total RNA was isolated from embryos at various developmental stages using TRIzol reagent according to the manufacturer's protocol (Invitrogen). First-strand cDNA was synthesized from 1 µg total RNA using SuperscriptIII reverse transcriptase (Invitrogen) and a random or oligo-dT primer. Semi-quantitative PCR was done essentially as described [Okuda et al., 2006]. Primers used for RT-PCR analysis of mRNA expression in zebrafish extracts were as follows: for mkk4a, 5′-CGTTC AACAG TAGAC GAGCG-3′ and 5′-AATCA CTTCG TCTAA AGAGG-3′; for mkk4b, 5′-CGCTC CACGG TGGAT GAGAA-3′ and 5′-AATGA CGTCA TCTGA CGCAC-3′; for mkk7, 5′-TGGCC ATGTC ATCGC AGTCA-3′ and 5′-GGAAA CTGTC CTGTG GCTAG-3′; for β-actin, 5′-CAGCT TCACC ACCAC AGC-3′ and 5′-GTGGA TACCG CAAGA TTCC-3′; for jnk1a-1, 5′-AGCGT ATGAC CACGT CCTCG-3′ and 5′-GGGCC AGACC GAAAT CCAG-3′; for jnk1a-2, 5′-GGATG CTTAC ACATC GACTT CAC-3′ and 5′-CATCC ATCAG CTCCA TTACT AGG-3′; for jnk2, 5′-ATCTG GACCA TGAGA GGATG TC-3′ and 5′-CTGGG GTTTG TTCAT CACAT AG-3′; for e-cadherin, 5′-ACAAA CTTAG GGCTC ATGCG-3′ and 5′-ACAGA TGCAG TGTAC GAGGA-3′; for stat3, 5′-TGAAT GGAAA CAGCC AGGCA-3′ and 5′-TTTGA TGACA AGGGG TCGGT-3′; for liv1, 5′-CGGTT GCCAA TATGA TTGGC-3′ and 5′-GGTGG ATTCC TGGTT CATCT-3′; for wnt5, 5′-CCGGA GATGT ACATC ATTGG-3′ and 5′-TTCTC ACGTT CACGA GCGTC-3′; for wnt11, 5′-GTAAA CTTCT GGACG GGCTC-3′ and 5′-CGAAG GTTAT CTCCA CATCC-3′.
Zebrafish Embryo Preparation
Embryos were deyolked with deyolking buffer (1/2 Ginzburg Fish Ringer) without calcium [Link et al., 2006]. Subsequently embryos were lysed in buffer containing 100 mM NaCl, 40 mM Tris–HCl (pH 8.0), 1% Nonidet P-40, 0.05% 2-mercaptoethanol, 1 mM EDTA, 1 mM EGTA, 4 µg/ml aprotinin, 100 µM Na3VO4, and 50 mM NaF, and sonicated for 1 min. Lysates were centrifuged at 20,000g to pellet cellular debris. Protein concentration was measured by BCA™ Protein Assay Kit (Pierce).
ES Cell Lines
Mkk4−/− and mkk7−/− murine ES cell lines were generated as previously described [Nishina et al., 1997; Fleming et al., 2000; Kishimoto et al., 2003].
Antibodies
Anti-Jnk1 polyclonal Ab (FL), which recognizes both the 46 and 55 kDa splice variants of Jnk1, was from Santa Cruz Biotechnology, Inc. Anti-phospho-Jnk (9251) and anti-FLAG (M2) Abs were from Cell Signaling Technology and Sigma–Aldrich Co., respectively.
Construction of Plasmids and Transfection
cDNAs encoding FLAG-tagged zebrafish Mkk4a-l, Mkk4b, and Mkk7 were cloned into the mammalian expression vector pCE-IRES2-EGFP [Ura et al., 2007]. ES cells were plated at 1 × 106 cells/35-mm dish and transfected 1 day later with expression construct (4 µg) plus Lipofectamine 2000 reagent (Invitrogen). Transfected cells were stimulated with UV light (1 kJ/m2) and subjected to standard Western blot analysis as described below.
Western Blot Analysis
Proteins were fractionated by SDS–PAGE and transferred to a polyvinylidene difluoride membrane that was then incubated for 1 h in blocking solution (2% or 5% skim milk in Tris-buffered saline [TBS]). The blocked membrane was incubated for 1 h in blocking solution containing anti-phospho-Jnk, anti-Jnk1, or anti-FLAG Ab. The membrane was then washed in TBS–Tween 20 (0.05%), incubated with anti-mouse/rabbit horseradish peroxidase-conjugated Ab (Jackson ImmunoResearch Laboratory) for 30 min, and washed three times in TBS–Tween 20. Proteins were visualized using Immobilon HRP (Millipore) or the SuperSignal West Femto Kit (Pierce) and a ChemiDoc XRS system (Bio-Rad), as described [Kishimoto et al., 2003].
Anti-Sense Morpholino (MO) and mRNA Injections
All MOs were designed to bind to exon–intron junctions and were synthesized by Gene Tools (Philomath, OR). Sequences of splice-blocking MOs were as follows: mkk4a MO, 5′-GATGA AACAG ACGAA CCTCT CTGAA-3′; mkk4b MO, 5′-TGTGT GTGTC TGACC TCTCT GAAGA-3′; mkk7 MO, 5′-AGAGG AACTC ACCAG AGAAA TGCCA-3′; jnk1a-1 MO, 5′-AGACA AATAA CTTAC ACATC CTGGA-3′; jnk1a-2 MO, 5′-ATTTC AGTGT CTTGA CTTAC ATTTT-3′; jnk2 MO, 5′-AAAAA CAGCA TTACC ATTCT CCTTG-3′. Sequences of translation-blocking MOs were: mkk4a-l atgMO, 5′-GCGTC GCCAT TTGGG TTTGA CTCTT-3′; mkk4b atgMO, 5′-AGAGT CACTG CTGGG AGCCG CCATT-3′; mkk7 atgMO, 5′-AGAGT CTCTG CTCCA GCGAC GACAT-3′; mkk4a-s atgMO, 5′-GCCAT CTTGT TGACC GAGCC ATACG-3′. The standard control MO was: 5′-CCTCT TACCT CAGTT ACAAT TTATA-3′.
For knockdown, MO solution was injected into the yolks of one- to four-cell stage zebrafish embryos immediately beneath the cell body. For wnt5 or wnt11 overexpression, a construct designed to produce mRNA encoding full-length zebrafish wnt5 or wnt11 was created by cloning the relevant fragment (amplified by high-fidelity PCR) into the pCS2+ plasmid. A constitutively active form of Mkk4b (caMkk4b) was constructed by replacing Ser275 and Thr279 with aspartic acid and glutamic acid, respectively, and subcloning into the pCS2+ plasmid. Sense strand capped mRNA was synthesized using SP6 RNA polymerase and the mMESSAGE mMACHINE system (Ambion). RNA injections were performed as described [Shinya et al., 2000].
Whole-Mount In Situ Hybridization
Digoxigenin-labeled RNA probes were synthesized using the DIG RNA Labeling Kit (Roche Diagnostics). Primers used for mRNA expression analysis in whole zebrafish embryos were as follows: for dlx3, 5′-TCCGA CTTCT AAGGA CTCTC-3′ and 5′-TTCAC CTGTG TCTGT GTGAG-3′; for ntl, 5′-AGACG AATGT TTCCC GTGCT-3′ and 5′-CTTCT CTCTT TGGCA TCGAG-3′; for hgg1, 5′-ATGAG GAGTT CAGAC AGGCA-3′ and 5′-ATTAC CGCTG GGAAT GTCCA-3′; for pax2.1, 5′-CCGGC AGTAT TAAAC CTGGA-3′ and 5′-AGGTG CTTCC GTAAA CTCTC-3′; for gsc, 5′-GTCAC TATGA AGGAC ACTCG TGC-3′ and 5′-TTTGT TCCTG TTTTC AGGCG AC-3′; for wnt5, 5′-GTAGC AGACG TGAGC ACTGG-3′ and 5′-CGCAT TCCGA AAGTT CTTAA GAG-3′. Mixtures of three riboprobe pairs were used to detect wnt11, as follows: (1) 5′-GTAAA CTTCT GGACG GGCTC-3′ and 5′-CTAAA GTCCT GTGGG CCTGA-3′; (2) 5′-CCGGA ATTCA TGACA GAATA CAGGA ACT-3′ and 5′-CGAAG GTTAT CTCCA CATCC-3′; (3) 5′-GGTGC TTATG GACTC TCTAG-3′ and 5′-GAGTC GACTC ACTTC GAGAC GTATC TCT-3′. Riboprobes for myoD and krox20 were used as described [Shinya et al., 2000]. Whole mount in situ hybridization procedures were performed essentially as described [Thisse et al., 1993].
Quantitative Real-Time RT-PCR
Quantitative real-time RT-PCR analyses were performed using the Chromo4 real-time detection system (Bio-Rad). The PCR primers used were as follows: for wnt11, 5′-CACAA CAATG CTGTT GGCAG ACAGG TG-3′ and 5′-GGAGA TGGTG CTGAT GTCTT GAAGA CC-3′; for β-actin, 5′-GCAGA TGTGG ATCAG CAAGC AGG-3′ and 5′-CTGAG TCAAT GCGCC ATACA GAG-3′. For a 20 µl PCR reaction, cDNA template was mixed with 10 µl iQ SYBR Green Supermix (Bio-Rad) plus the appropriate primers to a final concentration of 200 nM each. The reaction was first incubated at 95°C for 3.5 min, followed by 41 cycles of 95°C for 12 s, 60°C for 13 s, and 72°C for 18 s.
RESULTS
Cloning and Characterization of Zebrafish mkk4a, mkk4B, and mkk7 Genes
To unravel the role of Jnk signaling in early zebrafish development, we first determined whether zebrafish mkk4 and mkk7 could function as direct activators of Jnk. We performed BLAST searches with mouse mkk4 and mkk7 to enable predictions of zebrafish mkk4 and mkk7 cDNA sequences. The obtained EST sequences were subjected to 5′- and 3′-RACE methodology to acquire the full-length cDNA sequences. The zebrafish has two mkk4 genes, named mkk4a and mkk4b (GenBank accession no. AB438979), but only one mkk7 ortholog (GenBank accession no. AB438980). The predicted amino acid (aa) sequences of the proteins encoded by these zebrafish genes are 81–84% identical to those of the mouse, and the Mkk phosphorylation sites are conserved (Fig. S1A). Two splice variants of zebrafish mkk4a were identified: mkk4a-s and mkk4a-l (GenBank accession nos. AB030901 and AB438978). Mkk4a-s (281 aa) is an N-terminal truncated form of Mkk4a-l (404 aa). A phylogenetic analysis of vertebrate mkk4 genes revealed that zebrafish mkk4a was clustered with mammalian mkk4 genes, whereas zebrafish mkk4b was clustered with those of other teleosts (Fig. S1B). These phylogenetic relationships suggest that the duplication of the mkk4 gene occurred in the common ancestors of teleosts and tetrapods.
We next used semi-quantitative RT-PCR to examine the expression dynamics of mkk4a, mkk4b, and mkk7 during early zebrafish development. All three genes were continuously expressed from the one-cell stage to the blastula (4.7 hpf) (Fig. 1A). Levels of both mkk4a and mkk7 were markedly decreased by the shield stage and became relatively low at the tailbud stage. In contrast, mkk4b expression was essentially constant from the one-cell stage through to the shield stage and relatively high during later gastrula stages.
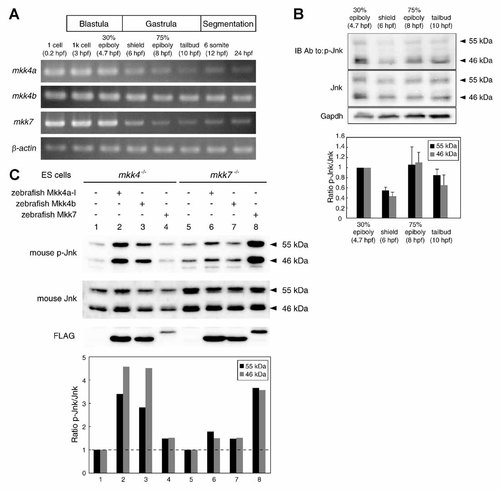
Expression and biochemical properties of zebrafish mkk4a, mkk4b, and mkk7 gene products during early development. A: Temporal expression patterns of zebrafish mkks. Semi-quantitative RT-PCR reveals relatively high expression of mkk4b up to 24 hpf in WT embryos, whereas mkk4a and mkk7 levels abruptly decrease around the shield stage. Lanes 1–8 correspond to template cDNA derived from 0.2 to 24 hpf embryos (n = 30, 30, 50, 20, 20, 20, 30, 50 embryos, respectively). β-Actin, loading control. B: Analysis of Jnk activity during zebrafish embryogenesis. Approximately 50 µg of protein was loaded in SDS–PAGE in each lane. Activated Jnk and total Jnk were detected with anti-phospho-Jnk (p-Jnk) and anti-total-Jnk antibodies, respectively. Image shown is representative of three separate experiments with similar results (top). Graphic results are expressed as a ratio of p-Jnk to total Jnk and the value of 30% epiboly stage is assigned an arbitrary value of 1 (bottom). Data shown are the mean ± SEM of three independent experiments. C: Effects of zebrafish Mkk4 or Mkk7 expression on stress-induced Jnk activation in mkk4−/− or mkk7−/− mouse ES cells. Mkk4−/− or mkk7−/− mouse ES cells were transfected with zebrafish mkk4a-l, mkk4b, or mkk7 expression vectors, cultured for 24 h, and stimulated with UV (1 kJ/m2). Expression levels of zebrafish Mkks (FLAG) and endogenous levels of p-Jnk and total Jnk (46 and 55 kDa isoforms) in cell lysates were detected by Western blotting (top). The histogram shows quantitative representations of the relative activated Jnk, which was expressed as a ratio of p-Jnk to total Jnk (bottom). The values of lanes 1 and 5 are assigned an arbitrary value of 1.
To monitor Jnk activity during zebrafish embryogenesis, we prepared protein extracts from zebrafish embryos collected at each of the 30% epiboly, shield, 75% epiboly, and tailbud stages, and examined the relative amounts of the active, phosphorylated form of Jnk by Western blot analysis. Jnk activity was detected from 30% epiboly and relatively high during later gastrula stages (Fig. 1B), suggesting the possibility that Jnk plays a role in the gastrulation stage.
In mammals, Mkk4 and Mkk7 have distinct biochemical properties and preferentially phosphorylate the Tyr and Thr residues, respectively, within the Thr-Pro-Tyr motif of Jnk [Lawler et al., 1998; Kishimoto et al., 2003]. To qualitatively determine whether these distinctions applied to the zebrafish orthologs of Mkk4 and Mkk7, and to gauge the functional relatedness of the mammalian and zebrafish enzymes, we analyzed whether zebrafish Mkks could compensate for the lack of mouse Mkk4 and Mkk7 in mkk4−/− and mkk7−/− murine ES cells, respectively. We transfected zebrafish mkk4a-l, mkk4b, and mkk7 expression vectors separately into mkk4−/− or mkk7−/− ES cells, which lack the capacity to activate Jnk in response to UV irradiation [Nishitai et al., 2004]. We then subjected the transfected cells to UV irradiation and assessed Jnk activation. We found that zebrafish Mkk4a-l or Mkk4b, but not Mkk7, rescued UV-induced Jnk activation in mkk4−/− ES cells (Fig. 1C, lanes 2–4). Conversely, zebrafish Mkk7, but not Mkk4a-l or Mkk4b, rescued UV-induced Jnk activation in mkk7−/− ES cells (Fig. 1C, lanes 6–8). These results suggest that zebrafish Mkk4 and Mkk7 are analogous in function to their murine counterparts and play distinct biochemical roles during stress-induced Jnk activation.
Zebrafish Mkk4B Is Indispensable for Normal CE Regulation
To elucidate the physiological roles of Mkk4a, Mkk4b, and Mkk7 during early zebrafish development, we performed anti-sense morpholino (MO)-mediated knockdown of each gene's mRNA and analyzed its residual expression by RT-PCR. Microinjection of mkk4b MO effectively prevented correct splicing of target pre-mRNA from the shield to tailbud stages (Fig. 2A). As a result, mkk4b morphants exhibited severe defects in anterior–posterior extension after gastrulation. At 11 hpf, mkk4b morphants displayed an MO dose-dependent shortening of body length (Fig. 2B, row 1) that did not recover at later developmental stages (Fig. 2B, rows 2 and 4). In addition, mkk4b morphants displayed broader notochords and somites during early segmentation than did control MO-injected embryos (Fig. 2B, row 3), indicating that CE is defective in the absence of Mkk4b. Quantification of the anterior–posterior axis extension and mediolateral convergence at 16 hpf revealed that mkk4b knockdown significantly increased the angle between the anterior and posterior ends of the embryo, as well as its mediolateral distance, in a MO dose-dependent manner (Fig. 2C).
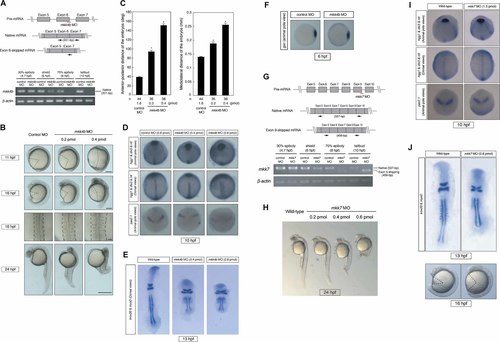
Mkk4b–Jnk signaling is required for proper CE movements. A: Validation of mkk4b MO efficacy. The mkk4b MO targets the exon 6/intron 6 junction of mkk4b pre-mRNA as shown in the diagram (top). WT zebrafish embryos were injected with 0.8 pmol of mkk4b MO or control MO, and total RNA was extracted from the 30% epiboly to tailbud stages and subjected to RT-PCR analysis (bottom). Arrows in the diagram represent the primer pairs used for the RT-PCR analysis. The mkk4b MO efficiently prevents correct splicing of mkk4b pre-mRNA beginning at the shield stage (6 hpf), conceivably leading to a functional impairment of Mkk4b. B: Gross appearance of mkk4b MO-injected embryos. Images of a live control MO-injected embryo and live mkk4b morphants injected with the indicated doses of mkk4b MO were acquired at 11, 16, and 24 hpf (rows 1, 2, and 4, respectively). These embryos are viewed laterally, with anterior to the top. Mkk4b morphants exhibit a shortening of body length and brain degeneration in a MO dose-dependent manner. Row 3 shows that the notochord and somites are wider in the mkk4b morphant than in the control at 16 hpf (dorsal view). Scale bars = 200 µm (row 1); 200 µm (row 2); 50 µm (row 3); 500 µm (row 4). C: Quantification of anterior–posterior axis extension and mediolateral convergence at 16 hpf. Left panel shows the average angle between the anterior and posterior ends of embryos (n = 44, 36, 38) that were injected as indicated. Right panel shows the average mediolateral width of embryos (n = 44, 36, 38) that were injected as indicated. Data shown are the mean ± SEM. *P < 0.01 versus control. D: Impaired CE in mkk4b morphants. Control MO-injected embryos and mkk4b morphants were analyzed by whole-mount in situ hybridization for the expression of tissue-specific genes at the tailbud stage (10 hpf). The position of the prechordal plate (hgg1) is more posterior and broader in mkk4b morphants compared to the control. Expression of dlx3 (neuroectoderm) reveals a broader neural plate, and the notochord (ntl) is shorter and broader. The midbrain/hindbrain boundary (pax2.1) has expanded laterally. E: Normal rhombomeres in mkk4b morphants. WT embryos and mkk4b morphants had the same number and shape of rhombomeres (krox20) and somites (myoD) at the eight-somite stage (13 hpf), although the body axis was shorter and somites were wider in the mkk4b morphants. F: Expression of the dorsoventral patterning marker gene gsc is normal in mkk4b morphants. Control MO (0.8 pmol) or mkk4b MO (0.8 pmol) was injected into 1- to 4-cell embryos, which were analyzed by whole-mount in situ hybridization at the shield stage. Shown are animal pole views with dorsal to the right. For B and D–F, results shown are one experiment representative of at least three trials. G: Validation of mkk7 MO efficacy as for A. Top: The mkk7 MO targets the exon 9/intron 9 junction of mkk7 pre-mRNA as shown in the diagram. Arrows represent primer pairs used in RT-PCR analysis. Bottom: The mkk7 MO (0.8 pmol) causes a marked reduction in mkk7 expression beginning at the shield stage (6 hpf). H: Gross appearance of mkk7 MO-injected embryos. Images of a live untreated WT embryo and mkk7 morphants injected with the indicated doses of mkk7 MO were acquired at 24 hpf. All embryos are viewed laterally, with anterior to the top. Mkk7 morphants exhibit weak, MO dose-dependent defects in the morphogenesis of the somites and a slightly shortened body length. I: Normal CE in mkk7 morphants. WT embryos and mkk7 morphants were analyzed for tissue-specific gene expression as for D. Even at a high dose of mkk7 MO (1.5 pmol), the expression patterns of all tissue-specific genes examined were normal. J: Impaired somitogenesis in mkk7 morphants. Top: The morphologies of rhombomeres (krox20) and somites (myoD) were examined at the eight-somite stage (13 hpf) in WT embryos and mkk7 morphants. The body axis was shorter and the somites were wider in the latter (dorsal view). Bottom: Images of a live WT embryo (left) and live mkk7 morphant (right) were acquired at 16 hpf (lateral view).
With respect to marker expression patterns, mkk4b morphants showed a more posteriorly and broadly positioned prechordal plate (hgg1), a wider neural plate (dlx3 and pax2.1), and a shorter and broader notochord (ntl) at the tailbud stage than did controls (Fig. 2D). Although their body axis was shorter and their somites wider, mkk4b morphants had the same numbers and shapes of rhombomeres (marked by krox20 expression) and somites (myoD) as control embryos at the eight-somite stage (Fig. 2E), implying that Mkk4b has no obvious role in specifying cell fate.
To further confirm that it was loss of Mkk4b that was responsible for the observed CE defects, we co-injected mkk4b MO with in vitro-transcribed mkk4b mRNA and assayed for the rescue of the CE defects. Inspection of live co-injected embryos showed that the synthetic mkk4b mRNA was able to prevent mkk4b MO-induced defects (Fig. S2). Thus, the observed phenotypes are the result of specific knockdown of Mkk4b by the mkk4b MO.
To rule out the possibility that the observed phenotypes in mkk4b knockdown embryos were secondary to a patterning alteration, we evaluated the distribution of goosecoid (gsc), a dorsal mesoderm “organizer” marker at the shield stage. No alterations to the gsc expression pattern were observed in mkk4b morphants (Fig. 2F), indicating that dorsoventral specification is not disturbed by mkk4b MO treatment.
The data in Figure 2D suggested that mkk4b morphants developed slightly slower than controls. To determine if Mkk4b influences epiboly, we monitored the development of live embryos over a time course of 6–10 hpf. As was true for control embryos, gastrulation began at 6 hpf in mkk4b morphants and 75% epiboly was reached at 8 hpf (Fig. S3). However, whereas control embryos had completed their epiboly movements by 10 hpf, mkk4b morphants exhibited a mild delay in epiboly that extended it to 11 hpf (Figs. S3 and 2B). Thus, Mkk4b activity is not required for the initiation or progression of epiboly but does influence its late phase.
When parallel experiments were used to examine the importance of Mkk4a in early zebrafish development, a very different result was obtained. Microinjection of 0.8 pmol of mkk4a MO reduced WT mkk4a mRNA beginning at the shield stage (Fig. S4A). Unlike mkk4b morphants, however, mkk4a morphants had no gross morphological abnormalities (Fig. S4B), and the expression patterns of hgg1, dlx3, ntl, and pax2.1 were completely normal (Fig. S4C).
In morphants injected with 0.8 pmol of mkk7 MO, expression of the WT mkk7 transcript declined by the shield stage and was undetectable at the tailbud stage (Fig. 2G). Instead, a shorter amplification product was detected. Like mkk4b morphants, mkk7 morphants exhibited an MO dose-dependent shortening of body length at 24 hpf (Fig. 2H). However, the expression patterns of hgg1, dlx3, ntl, and pax2.1 were normal in mkk7 morphants, even at a high dose of MO (1.5 pmol) (Fig. 2I). These data suggest that mkk7 is dispensable for proper gastrulation. Nevertheless, during segmentation, the mkk7 morphants displayed somites of abnormal patterning and morphology (Fig. 2J). Thus, Mkk7 function is essential for normal somite morphogenesis.
Because the mkk4a, mkk4b, and mkk7 genes are maternally expressed, it was possible that their expression products might persist throughout gastrulation. To rule out this possibility, we used translation-blocking MOs (atgMOs) that would specifically target maternal and zygotic mkk transcripts and prevent their translation. We injected embryos with a specific translation-blocking MO (mkk4a-l atgMO, mkk4b atgMO, or mkk7 atgMO) and examined their development using gross morphological analysis and in situ hybridization. By 23 hpf, mkk4b atgMO- or mkk7 atgMO-injected embryos showed reduced body length compared to control-injected embryos, whereas few or no morphological differences were apparent in mkk4a atgMO-injected embryos (Fig. S5A). Mkk4b atgMO-injected embryos also showed altered expression of several marker genes (hgg1, dlx3, ntl, and pax2.1; Fig. S5B), highlighting the morphological abnormalities associated with defective CE movements at 10 hpf. This phenotype was strikingly similar to that of embryos subjected to mkk4b splice-blocking MO knockdown (Fig. 2D). Taken together, these results demonstrate that Mkk4b but not Mkk4a is essential for co-ordinated CE. In the following analyses, we focused on the specific role of Mkk4b–Jnk signaling in proper CE movements during gastrulation.
Zebrafish Jnk2 Is Necessary for Correct CE Movements
It has previously been reported that non-canonical Wnt signaling regulates CE movements in Xenopus through the activation of Jnk [Yamanaka et al., 2002]. We therefore asked whether overexpression of wnt11 or wnt5 could activate Jnk during zebrafish gastrulation. Extracts from zebrafish embryos overexpressing wnt11 or wnt5 mRNA were prepared at 75% epiboly (8 hpf) and subjected to Western blotting with anti-phospho-Jnk antibody. This analysis confirmed that wnt11 or wnt5 overexpression significantly increased levels of the active, phosphorylated form of Jnk (Fig. 3A). Furthermore, analysis of extracts from embryos injected with 0.4 pmol mkk4a MO, mkk4b MO, or mkk7 MO showed that Jnk phosphorylation was significantly decreased in mkk4b morphants but still detected in mkk4a and mkk7 morphants (Fig. 3B). These data indicate that Mkk4b (exclusively) has a profound effect on CE, and that this effect is mediated through activation of the Jnk pathway. Thus, Mkk4b–Jnk signaling plays a critical role in proper CE movements during gastrulation.
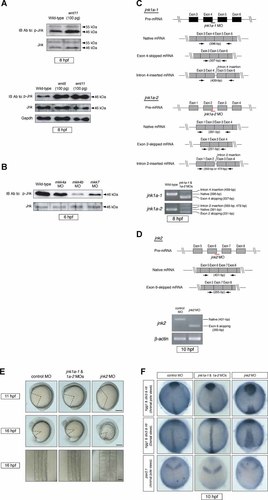
Jnk2 is required for proper CE movements in zebrafish. A: Jnk is activated by wnt11 or wnt5 overexpression. Pooled extracts of 25 embryos overexpressing wnt11 were analyzed by Western blotting (top). Levels of phospho-Jnk (p-Jnk) were increased compared to the control but total Jnk (55 and 46 kDa isoforms) was unchanged. Pooled extracts of 25 embryos overexpressing wnt5 were analyzed by Western blotting (bottom). Gapdh, internal control. B: Markedly reduced Jnk phosphorylation in mkk4b morphants. Twenty-five embryos injected with 0.4 pmol of mkk4a MO, mkk4b MO, or mkk7 MO were analyzed at the shield stage by Western blotting to detect Jnk and p-Jnk. C: Validation of jnk1a MOs. Schematic illustrations of the target sites of the jnk1a-1 MO and jnk1a-2 MO are shown in the top and center panels, respectively. Arrows represent the primer pairs used in the RT-PCR evaluation of MO efficacy. The bottom panel shows an RT-PCR analysis of the effects of these MOs on jnk1a-1 and jnk1a-2 expression. Total RNA was extracted at 8 hpf from untreated WT embryos, or WT embryos co-injected with jnk1a-1 MO and jnk1a-2 MO (0.8 pmol each). Jnk1a-1 MO and jnk1a-2 MO prevent correct splicing of jnk1a-1 and jnk1a-2 pre-mRNAs, respectively, resulting in a reading-frame shift and production of mutant proteins. D: Validation of jnk2 MO. Top panel depicts the location of the target site of jnk2 MO and the primer pairs used for RT-PCR. The bottom panel shows an RT-PCR analysis of the effect of jnk2 MO. Total RNA was extracted from WT embryos injected with control MO (0.8 pmol) or jnk2 MO (0.7 pmol). Jnk2 MO induces defective splicing. E: Gross appearance of jnk morphants. Images of live embryos injected with control MO (0.8 pmol), jnk1a-1 MO plus jnk1a-2 MO (0.8 pmol each), or jnk2 MO (0.7 pmol) were acquired at 11 and 16 hpf. These embryos are viewed laterally, with anterior to the top. Jnk1 MOs cause no gross morphological defects, whereas jnk2 morphants exhibit a shortening of body length. At 16 hpf, the notochord and somites are wider in the jnk2 morphant than in the control (dorsal view). Scale bars = 200 µm (row 1); 200 µm (row 2); 50 µm (row 3). F: Impaired CE in jnk2 morphants. Jnk morphants and controls were analyzed by whole-mount in situ hybridization for the expression of tissue-specific genes at the tailbud stage (10 hpf). Jnk1a morphants showed normal marker expression. In jnk2 morphants, the position of the prechordal plate (hgg1) is more posterior and broader than in the control. Expression of dlx3 (neuroectoderm) reveals a broader neural plate, and the notochord (ntl) is shorter and broader. The midbrain/hindbrain boundary (pax2.1) has expanded laterally.
To examine Mkk4b-Jnk signaling in zebrafish embryos, it was first necessary to identify zebrafish Jnk enzymes. To isolate zebrafish jnk genes, we used cDNA sequences for mammalian Jnks to perform BLAST searches of the zebrafish genomic database and identified four zebrafish jnk genes. A phylogenetic analysis based on the aa sequences of vertebrate Jnks revealed that the four zebrafish Jnk homologs closely resembled members of the three mammalian Jnk subfamilies, Jnk1, Jnk2, and Jnk3. We therefore designated the zebrafish proteins as Jnk1a-1, Jnk1a-2, Jnk2, and Jnk3 (Fig. S6). During gastrulation, mRNAs for Jnk1a-1, Jnk1a-2, and Jnk2 (but not Jnk3) were transcribed (data not shown).
To ascertain the functional importance of these zebrafish Jnk(s) for normal CE movements, we performed MO-mediated knockdown of the jnk1a-1, jnk1a-2, and jnk2 genes and analyzed residual expression by RT-PCR. Co-injection of jnk1a-1 MO and jnk1a-2 MO efficiently prevented the correct splicing of jnk1 pre-mRNAs during gastrulation (Fig. 3C). However, unlike the Xenopus Jnk1 morphant [Yamanaka et al., 2002], jnk1a-1 and jnk1a-2 morphants showed no gross morphological abnormalities (Fig. 3E, center), and the expression patterns of hgg1, dlx3, ntl, and pax2.1 were completely normal (Fig. 3F, center). On the other hand, injection of jnk2 MO not only prevented correct splicing of jnk2 pre-mRNA (Fig. 3D) but also induced severe defects in anterior–posterior extension and mediolateral convergence, as assessed by analysis of morphology and molecular markers (Fig. 3E,F, right). These results indicate that Jnk2, but no form of Jnk1, is required for normal CE movements during zebrafish gastrulation.
Transcription of Zebrafish wnt11 Is Repressed by mkk4b–Jnk Signaling
The molecular mechanism by which Jnk regulates CE movements is poorly understood. To identify relevant transcriptional targets downstream of the Mkk4b–Jnk signaling cascade associated with zebrafish CE, we used RT-PCR to survey mRNA levels of candidate genes in control and Mkk4b morphants at 75% epiboly (8 hpf). Because normal gastrulation requires modulation of cadherin-mediated cell–cell adhesion, we first investigated e-cadherin expression but found that its mRNA levels were unchanged in our morphants (Fig. 4A, row 1). Similarly, although stat3 and liv1 are reportedly essential for zebrafish gastrulation [Yamashita et al., 2004], the expression of these mRNAs was also normal in our mkk morphants (Fig. 4A, rows 2 and 3). We next screened components of the non-canonical Wnt pathway and found that, although wnt5 expression was normal (Fig. 4A, row 4), wnt11 expression was highly upregulated in Mkk4b-depleted embryos at 75% epiboly (8 hpf) (Fig. 4A, row 5), as well as at the 30% epiboly, shield, and tailbud stages (Fig. 4B, left). In contrast, upregulation of wnt11 expression was not detected in Mkk4a- or Mkk7-depleted embryos (Fig. 4A). When we analyzed the expression of wnt11 transcripts using quantitative real-time RT-PCR, we confirmed that levels of wnt11 mRNA were increased in mkk4b morphants (Fig. 4C). On the other hand, Mkk4b knockdown did not affect wnt5 expression at any stage (Fig. 4B, right). These data indicate that Wnt11 itself is one of the molecular components downstream of the Jnk cascade in the non-canonical Wnt pathway associated with early embryogenesis.
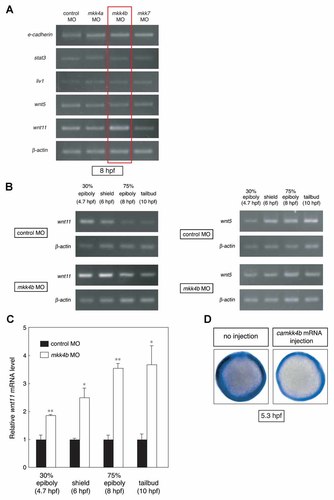
Identification of wnt11 as a downstream component of Mkk4b–Jnk signaling in the non-canonical Wnt pathway. A: Specific upregulation of wnt11 in mkk4b morphants. Total RNA was isolated from control, mkk4a, mkk4b, and mkk7 morphants at the 75% epiboly stage (8 hpf) and subjected to RT-PCR to detect expression of the indicated mRNAs. β-Actin, internal control. B: Upregulation of wnt11 but not wnt5 in mkk4b morphants throughout early embryogenesis. Total RNA was isolated from control and mkk4b morphants at the indicated stages and subjected to RT-PCR to detect expression of wnt11 (left) and wnt5 (right) mRNAs. C: Quantitative RT-PCR analysis of wnt11 mRNA expression in mkk4b morphants. For all quantitative PCR experiments, wnt11 cDNA amplification was standardized to the amplification of β-actin cDNA. Wnt11 expression in mkk4b morphants was normalized to that in controls (assigned an arbitrary value of 1). Data shown are the mean ± SEM of three independent experiments (*P < 0.05; **P < 0.01 vs. control). D: Mkk4b–Jnk signaling represses wnt11 expression. WT embryos were left untreated or injected with camkk4b mRNA (40 pg) and subjected to whole-mount in situ hybridization to detect wnt11 at the 50% epiboly stage (5.3 hpf; animal pole views). Decreased wnt11 expression was observed in camkk4b-overexpressing embryos (right) compared with uninjected embryos (left).
We next examined whether activation of the Mkk4b–Jnk signaling pathway was sufficient to repress wnt11 expression. Overexpression of a constitutively active form of Mkk4b (caMkk4b) markedly reduced the level of wnt11 expression at 50% epiboly (Fig. 4D), reinforcing the notion that wnt11 is a transcriptional target downstream of the Mkk4b–Jnk signaling cascade.
Overexpression of wnt11 Induces Abnormal CE Movements
To ascertain whether the increased wnt11 expression in mkk4b morphants induced CE defects, we injected WT embryos with wnt11 mRNA. Embryos overexpressing wnt11 mRNA showed a shortening of body length at 11, 16, and 24 hpf and exhibited broader notochords and somites at 16 hpf (Fig. 5A), reminiscent of the mkk4b morphant phenotype (Fig. 2B). With respect to marker expression patterns, the expression of the organizer marker gsc was not changed in wnt11-overexpressing embryos compared to control embryos injected with egfp mRNA (Fig. 5B, top). At the tailbud stage, wnt11-overexpressing embryos showed a more posteriorly positioned prechordal plate (hgg1), a wider neural plate (dlx3), a shorter and broader notochord (ntl), and a laterally expanded midbrain/hindbrain boundary (pax2.1) than did controls (Fig. 5B, bottom). These observations imply that the level of wnt11 mRNA present critically influences CE.
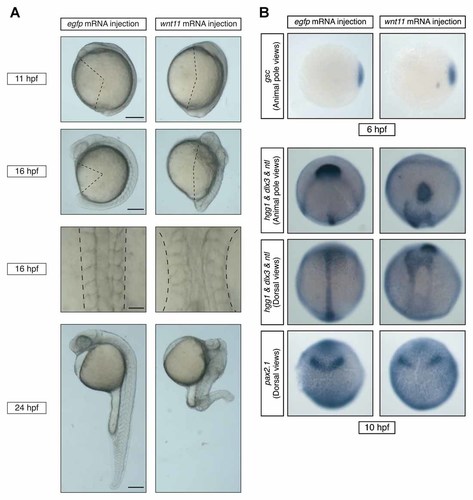
Overexpression of wnt11 induces CE abnormalities. A: Altered gross appearance. WT embryos were injected with either egfp mRNA (100 pg; control) or wnt11 mRNA (75 pg), and the gross phenotype of the embryos was examined at 11, 16, and 24 hpf (rows 1, 2, and 4, respectively). These embryos are viewed laterally, with the anterior to the top. A shortening of body length can be seen as early as 11 hpf in the wnt11-overexpressing embryos. Row 3 shows that the notochord and somites are wider in the wnt11-overexpressing embryos than in the controls at 16 hpf (dorsal view). Scale bars = 200 µm (row 1); 200 µm (row 2); 50 µm (row 3); 200 µm (row 4). B: Impaired CE in wnt11-overexpressing embryos. WT embryos were injected with egfp or wnt11 mRNA (100 pg), and analyzed by whole-mount in situ hybridization for the expression of tissue-specific genes at the shield stage (6 hpf) and the tailbud stage (10 hpf). The expression pattern of the organizer marker gsc is similar in the wnt11-overexpressing embryos and controls. However, the position of the prechordal plate (hgg1) is more posterior in wnt11-overexpressing embryos compared to the control. Expression of dlx3 (neuroectoderm) reveals a broader neural plate, and the notochord (ntl) is shorter and broader. The midbrain/hindbrain boundary (pax2.1) has expanded laterally.
Mkk4B Morphants Show Increased wnt11 Expression in the Organizer and Margin Regions
We next used whole-mount in situ hybridization to localize the increased wnt11 expression in Mkk4b-depleted embryos. During gastrulation in control MO-injected embryos, wnt11 expression became prominent in the lateral and ventral germ ring (margin region) but was downregulated within the region of shield formation (i.e., the organizer) (Fig. 6A; 6 hpf). Wnt11 expression near the margin persisted throughout gastrulation, although the staining intensity gradually decreased in the ventral region. Wnt11 expression remained high in the dorsal region, which converges with the tailbud (Fig. 6A; 8 and 10 hpf). In contrast, during gastrulation in mkk4b morphants, wnt11 expression was significantly increased in both the organizer and margin regions (Fig. 6A; 6 hpf). By late gastrulation, wnt11 expression was strongly upregulated in all domains (Fig. 6A; 8 and 10 hpf). On the other hand, the expression pattern of wnt5 in mkk4b morphants matched that of controls (Fig. 6B).
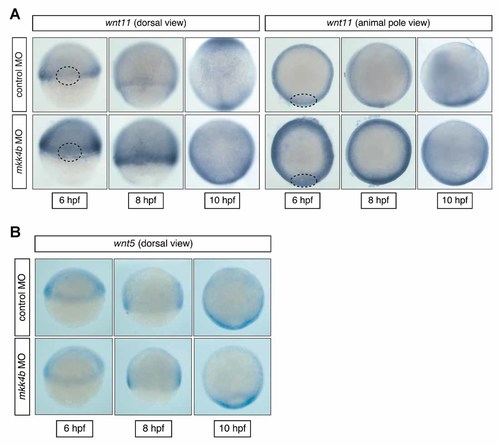
Elevated expression of wnt11 in the organizer and margin regions of Mkk4b-depleted embryos. A: Localization of increased wnt11 expression in mkk4b morphants. Whole-mount in situ hybridization was used to monitor wnt11 expression in developing control and mkk4b morphants from the shield stage (6 hpf) to tailbud stage (10 hpf). Two views are shown as indicated, with dashed lines outlining the boundaries of the organizer region. B: Localization of wnt5 expression in mkk4b morphants. Whole-mount in situ hybridization was used to monitor wnt5 expression in developing control and mkk4b morphants from the shield stage (6 hpf) to tailbud stage (10 hpf). Wnt5 showed the same expression pattern in control and mkk4b morphants.
Impaired Mkk4B Function Promotes wnt11 Transactivation in a Non-Cell-Autonomous Manner
To determine whether wnt11 was a direct transcriptional target of Mkk4b–Jnk signaling, we generated small clones of mkk4b-deficient zebrafish cells within whole embryos by co-injecting mkk4b MO together with egfp mRNA into one blastomere of a 32- or 64-cell embryo (Fig. 7A). We subsequently examined the location of the EGFP-labeled descendant cells at the shield stage (Fig. 7B, top row) and analyzed wnt11 expression pattern by whole-mount in situ hybridization (Fig. 7B, bottom row). Wnt11 expression was significantly elevated even in the embryonic region that did not overlap with the lineage label marking the progeny of mkk4b MO-injected cells (Fig. 7B). Thus, activation of the Mkk4b–Jnk signaling pathway in zebrafish embryos represses wnt11 expression in a non-cell-autonomous fashion.
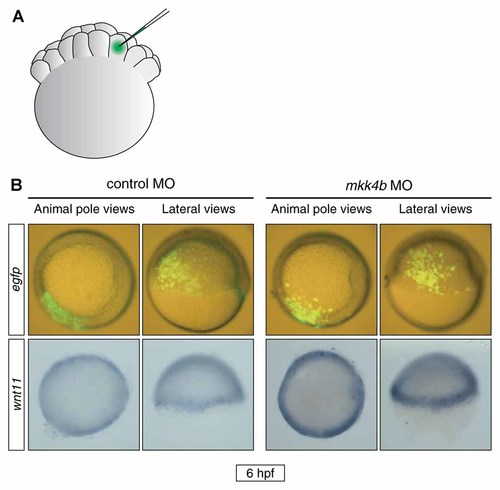
Abrogation of Mkk4b function induces elevated expression of wnt11 in a non-cell-autonomous manner. A: Schematic representation of MO microinjection into one cell of a 32- or 64-cell zebrafish embryo. B: Control MO (0.4 pmol) or mkk4b MO (0.4 pmol) was injected into one cell of a 32- or 64-cell embryo to generate randomly distributed mkk4b-knockdown cell clones. Egfp mRNA was co-injected as a cell lineage tracer. Top, EGFP-labeled cells were observed at the shield stage by fluorescence microscopy using a halogen lamp and blue light. Bottom, wnt11 expression was monitored in developing control and mkk4b morphants at the shield stage by whole-mount in situ hybridization. Two views are shown, with dorsal to the right.
Taken together, our findings suggest that the suppression of wnt11 transcription by Jnk activation is important for the precise regulation of vertebrate CE, and establish a model in which non-canonical Wnt signaling leading to Jnk activation represses expression of the CE-triggering ligand Wnt11 (Fig. S7).
DISCUSSION
In this study, we examined the role of Jnk signaling during the early embryogenesis of zebrafish by carrying out MO-mediated knockdown of the orthologs of the mkk4 and mkk7 genes. We found that mkk4a MO-injected zebrafish embryos had no phenotype (Fig. S4B,C), whereas mkk4b MO-injected embryos exhibited axial tissues that were abnormally short and wide due to defective CE (Fig. 2B–E). These observations suggest that Mkk4b is functionally redundant with Mkk4a and can compensate for its loss during gastrulation, but that Mkk4a cannot compensate for a lack of Mkk4b. Mkk7 morphants had no phenotype during gastrulation but showed abnormal somite morphologies during segmentation (Fig. 2H–J). Mkk7 is thus critical for a slightly later stage of development.
Recently, Rui et al. 2007 demonstrated that zebrafish mkk4a-s plays an important role in dorsoventral patterning in zebrafish blastulas. These authors reported that mkk4a-s knockdown with a translation-blocking MO prior to gastrulation reduced the expression of dorsal markers but expanded the expression of ventral markers. In our study, we performed mkk4a-l knockdown with translation-blocking MO but could not detect any alteration to the expression of the dorsal marker gsc (Fig. S8). This discrepancy implies that there is functional diversity between the two splicing isoforms of mkk4a. The results of our study, together with those of Rui et al., reinforce the idea that Jnk signaling plays multiple roles in major developmental processes, including the specification of the dorsoventral axis, cell movements required during gastrulation, and somite morphogenesis.
Our previous studies of disruption of the mkk4 and mkk7 genes in murine ES cells revealed that Mkk4 and Mkk7 preferentially phosphorylate the Tyr and Thr residues, respectively, within the Thr-Pro-Tyr motif of Jnk [Kishimoto et al., 2003]. In this study, we expressed zebrafish Mkk4 and Mkk7 in mouse ES cells and obtained results indicating that these Mapks also differ in their biochemical properties (Fig. 1C). These data strongly suggest that the use of Jnk activation as a molecular switch has been evolutionarily conserved from teleosts to mammals. In zebrafish, loss of Mkk4b leads to severe defects in CE movements during gastrulation, whereas our mkk7 morphants had no phenotype during gastrulation (Fig. 2D,I). These observations indicate that mkk4 has a more prominent role than mkk7 in early embryogenesis. Consistent with this hypothesis, mkk4−/− mouse embryos die earlier than mkk7−/− embryos [Nishina et al., 1999; Wada et al., 2004]. This conserved functional diversity of Mapks may be related to the need to activate Jnk at different times and locations during early development. Indeed, we found that Mkk4b depletion reduced Jnk phosphorylation more dramatically than did Mkk7 depletion, at least at the shield stage (Fig. 3B). Alternatively, the functional properties of Mkk4b–Jnk and Mkk7–Jnk signaling modules may be governed in part by scaffold proteins such as the Jnk-interacting proteins (JIPs) that confer specificity to kinase actions [Whitmarsh, 2006].
Jnk1−/−jnk2−/− mice die at about E11 with defective neural tube morphogenesis [Kuan et al., 1999]. Based on our results in zebrafish, we speculate that the failure to close the neural tube in jnk1−/−jnk2−/− mice may be due to defective CE in the neural plate. In support of this idea, disruption of non-canonical Wnt pathway genes (such as dishevelled) in Xenopus inhibits CE in the neural plate and results in abnormal, widely spaced neural folds that do not close [Copp et al., 2003]. Invertebrate development may also rely on Jnk-mediated events that share molecular similarities with vertebrate CE. In Drosophila, dJnk (Basket) is activated by dJnkk (Hemipterous), a homolog of vertebrate Mkk7 [Glise et al., 1995; Riesgo-Escovar et al., 1996; Sluss et al., 1996]. Basket and hemipterous are important for morphogenesis, and loss of function of either gene inhibits dorsal closure [Glise et al., 1995; Riesgo-Escovar et al., 1996; Sluss et al., 1996]. These findings emphasize the notion that Jnk activation is utilized in parallel morphogenetic events among widely divergent species.
Signals initiated by non-canonical Wnt ligands such as Wnt11 and Wnt5 are important for: vertebrate morphogenetic processes that require directed cell movements; cell–cell adhesion; and the establishment of cell polarity. Zebrafish with mutations in wnt5 (ppt) or wnt11 (slb) exhibit defects that reduce CE without affecting cell fate [Heisenberg et al., 2000; Kilian et al., 2003]. The slb mutant shows more anterior CE defects, including a shortened and broadened body axis at the end of gastrulation and a slight fusion of the eyes (cyclopia) at later developmental stages. In contrast, the Wnt5 protein functions in posterior regions. The ppt mutant exhibits a shortened body axis and compressed tail, whereas the position of the eyes is only mildly affected. These observations indicate that Wnt11 and Wnt5 possess distinct and non-redundant functions in regulating zebrafish gastrulation, which may be accounted for by differences in spatiotemporal expression. Although both wnt5 and wnt11 are expressed in the germ ring at the shield stage, wnt11 is specifically expressed in anterior tissues at the end of gastrulation, whereas wnt5 is specifically expressed in posterior mesendodermal tissues (Fig. 6) [Heisenberg et al., 2000; Kilian et al., 2003]. Our study demonstrates that Mkk4b–Jnk signaling precisely regulates wnt11 transcription. This Mkk4b–Jnk-mediated control may contribute to the mechanisms segregating wnt11 and wnt5 expression domains during late gastrulation.
In Drosophila, a role for Dpp in dorsal closure during early morphogenesis has been clearly established [Hou et al., 1997; Riesgo-Escovar and Hafen, 1997]. During dorsal closure, Jnk activates the expression of dpp in leading-edge epithelial cells, and Dpp subsequently acts as a secreted signal that controls the elongation of lateral epidermis in a paracrine fashion [Reed et al., 2001]. Our analyses demonstrate that Mkk4b–Jnk signaling regulates wnt11 expression during zebrafish gastrulation in a non-cell-autonomous manner (Fig. 7). Our results raise the possibility that, like Drosophila dorsal closure, normal vertebrate CE movements depend on Jnk signaling that regulates the expression of a secreted signaling molecule capable of promoting concerted movements of neighboring cells (Fig. S7).
Our proposed model of CE movement regulation by wnt11 expression can be summarized as follows: In normal zebrafish embryos, activated Mkk4b transmits a signal leading to Jnk activation. This activated Jnk may induce secreted factor X, which exerts moderate suppression of wnt11 transcription in neighboring organizer and margin cells, and results in proper co-ordination of CE. In mkk4b morphants, however, JNK activation is blocked such that the expression of factor X is downregulated. As a result, wnt11 is expressed at an abnormally high level that may impair CE (Fig. 7). We undertook preliminary studies to examine whether the mkk4b morphant phenotype could be suppressed by a low dose of wnt11 MO, but found that rescue did not occur (data not shown). This inability to rescue the CE defect may be due to a failure to strictly control the expression pattern and amount of wnt11 expression, since Wnt11 levels and localization must be tightly regulated for normal embryonic development [Heisenberg et al., 2000].
A previous study demonstrated that Bmp activity regulates CE movements during zebrafish gastrulation [Myers et al., 2002]. This report prompted us to investigate whether zebrafish Bmp2 and Bmp4 (homologs of Drosophila Dpp) function downstream of the Mkk4b–Jnk signaling cascade. However, expression levels of bmp2 and bmp4 were not downregulated in Mkk4b-depleted embryos (data not shown). Future identification of secreted factor(s) responsible for the transmission of Jnk signals to neighboring cells may provide insight into the complex mechanisms controlling vertebrate CE.
Our proposed model postulates that wnt11 expression is under precise spatiotemporal control during zebrafish development. Previous studies have shown that Wnt11 expression in zebrafish starts in the dorsal part of the blastoderm margin at the oblong stage, and that the entire margin becomes Wnt11+ by the late blastula stage [Makita et al., 1998]. Our present work shows that, at the onset of gastrulation, wnt11 is highly expressed around the circumference of the germ ring, with slightly reduced expression in the region of the organizer (Fig. 6A). It has been established that wnt11 is expressed predominantly in epiblast cells of the germ ring (ectodermal germ layer), whereas ingressing hypoblast cells (mesendodermal germ layer) show no detectable wnt11 expression [Makita et al., 1998; Ulrich et al., 2003]. This profile of early wnt11 expression in zebrafish is virtually identical to that seen in Xenopus [Ku and Melton, 1993]. Taken together, these results suggest an evolutionary conservation of wnt11 function during gastrulation, and a prominent role for the control of wnt11 expression by Jnk signaling.
Acknowledgements
We thank numerous members of the Nishina and Katada Laboratories for their helpful discussions and critical comments on the manuscript. This work was partly supported by a Grant-in-Aid for Scientific Research on a Priority Area from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also supported by grants from the Japan Society for the Promotion of Science, the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Ministry of Health, Labor and Welfare of Japan.