Post-transcriptional and post-translational regulation of PTEN by transforming growth factor-β1
Abstract
PTEN is a critical tumor suppressor gene mutated frequently in various human cancers. Previous studies have showed that PTEN mRNA expression is down-regulated by TGF-β1 in various cell lines. In present study, we have found that TGF-β1 down-regulates PTEN mRNA and protein expression in a dose- and time-dependent manner in hepatocarcinoma cell line SMMC-7721. Based on the PTEN promoter dual-luciferase report assay, we have found that PTEN transcription is not affected by TGF-β1. By using transcriptional inhibitor actinomycin D (Act D), the turnover rate of PTEN transcripts appeared to be accelerated during TGF-β1 stimulation, which indicated that down-regulation of PTEN by TGF-β1 was post-transcriptional. What interested us was that transfection of PTEN coding sequence increased TGF-β1-induced degradation of PTEN mRNA, suggesting that PTEN coding region was account for TGF-β1-mediated down-regulation of PTEN. In addition, TGF-β1 down-regulated PTEN expression was blocked by the TβIR inhibitor SB431542 and the p38 inhibitor SB203580, suggesting Smad and p38 MAPK signal pathways played crucial roles in PTEN down-regulation via TGF-β1 stimulation. In this study, we also found TGF-β1 accelerated down-regulation of PTEN through the ubiquitin-proteasome pathway. Collectively, our data clearly demonstrated that TGF-β1-mediated down-regulation of PTEN was post-transcriptional and post-translational, depending on its coding sequence, Smad and p38-MAPK signal pathways were involved in this down-regulation. J. Cell. Biochem. 106: 1102–1112, 2009. © 2009 Wiley-Liss, Inc.
Abbreviations used:
TGF-β1, transforming growth factor β1; PTEN, phosphatase and tensin homologue deleted on chromosome10; UTR, untranslated region; DN, dominant negative; CRD, coding region determinants; Act D, actinomycin D; CHX, cycloheximide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
INTRODUCTION
Phosphatase and tensin homologue deleted on chromosome10 (PTEN, also called MMAC1 or TEP1) is a tumor suppressor gene located on human chromosome 10q23.3 [Li and Sun, 1997; Li et al., 1997; Steck et al., 1997] and is frequently deleted or mutated in various human cancers to promote tumorigenesis. PTEN functions as a dual-specificity phosphatase and a lipid phosphatase in vitro [Myers et al., 1997, 1998], and dephosphorylates phosphatidylinositol 3,4,5-trisphosphate—a product of PI3-Kinase, which plays a crucial role in regulating cell growth, apoptosis, invasion, and metastasis [Maehama et al., 2001; Yamada and Araki, 2001; Leslie and Downes, 2002; Goberdhan and Wilson, 2003].
Despite a crucial role of PTEN in tumorigenesis, the signaling mechanisms by which PTEN expression is regulated in human tumors have been poorly understood. Previous reports indicate that PTEN is regulated by multiple post-translational mechanisms indeed [Maehama, 2007]. PICT-1 [Okahara et al., 2006], NEDD4-1 [Wang et al., 2007b], and DJ-1 [Kim et al., 2005] had been found to interact with PTEN and affect PTEN function. In addition to the post-translational regulation described previously, some transcription factors had been implicated in transcriptional regulation of PTEN and affect PTEN expression level. Transcription factors of P53 [Stambolic et al., 2001], Egr-1 [Virolle et al., 2001], and peroxisome proliferator-activated receptor γ (PPAR-γ) [Patel et al., 2001; Zhang et al., 2006] had been shown to increase PTEN transcription. In contrast, nuclear factor κB (NF-κB) had been known to suppress PTEN transcription [Kim et al., 2004; Vasudevan et al., 2004; Wang et al., 2007a]. Recent studies had identified that NOTCH1 might be able to activate or repress PTEN transcription depending on the cellular context. Micro RNA-214 had been reported to down-regulate PTEN expression through targeting the 3′-UTR of the PTEN [Yang et al., 2008]. Previous studies also demonstrated that TGF-β1 suppressed PTEN mRNA expression, suggesting TGF-β1 was a potent regulator of PTEN in human cancers [Li and Sun, 1997; Ebert et al., 2002; Chow et al., 2007; Vasudevan et al., 2007].
TGF-β1 is a multifunctional growth factor that regulates cell growth, differentiation, apoptosis, migration, adhesion, and extracellular matrix deposition in a context-dependent manner [Massague, 1990; Akhurst, 2004; Zavadil and Bottinger, 2005]. TGF-β1 transduces signaling through transmembrane serine/threonine kinase receptors and intracellular signaling molecules of the Smad family [Massague, 1998; ten Dijke and Hill, 2004]. Upon ligand binding, type II receptors are to phosphorylate the type I receptors, then the activated type I receptors propagate the signal by phosphorylating the downstream effectors, Smad2 and Smad3 [Massague et al., 2000]. The activated R-Smad proteins form a heteromeric complexes with the Co-Smad, Smad4 and translocate into the nucleus and in conjunction with other nuclear cofactors, regulate the transcription of target genes [Moustakas et al., 2002]. Besides the Smad pathway, a series of reports indicate that the Src [Tanaka et al., 2004], the phosphatidylinositol 3-kinase (PI3K) [Bakin et al., 2000], Ras- and Rho-GTPases [Derynck and Zhang, 2003] and several MAP kinases (JNK, p38, and Erk) [Javelaud and Mauviel, 2005] can be rapidly activated by TGF-β1 in a manner which is highly dependent on the cell type and conditions [Massague et al., 2000].
It had been reported that PTEN repressed TGF-β1-mediated cellular migration and invasion whereas loss of PTEN expression increased TGF-β1-induced cellular migration and invasion [Hjelmeland et al., 2005]. Although previous studies had showed that MAP kinases (JNK, Erk) [Vasudevan et al., 2007; Wang et al., 2007a] were involved in down-regulation of PTEN by TGF-β1, the underlying mechanisms of TGF-β1-mediated PTEN down-regulation were still partially understood. In present study, we find that PTEN mRNA and protein expression are decreased with TGF-β1 treatment in a dose- and time-dependent manner in hepatocarcinoma cell line SMMC-7721. Based on the PTEN promoter dual-luciferase report assay and the PTEN mRNA decay analysis, PTEN transcription is not affected by TGF-β1, suggesting that TGF-β1 down-regulates PTEN expression through reducing PTEN mRNA stabilization. After the transfection of PTEN coding sequence, our studies also show that the expression of endogenous and exogenous PTEN mRNA are down-regulated by TGF-β1, indicating that the PTEN coding region is involved in TGF-β1-mediated down-regulation of PTEN. Here, we also find TGF-β1 increases the degradation of PTEN through the ubiquitin-proteasome pathway. It has been showed previously that both PTEN and TGF-β1 play crucial roles in regulating cell growth, apoptosis, migration, and cancer metastasis, unraveling the mechanisms of the down-regulation of PTEN by TGF-β1 seems to be particularly important to understand the effect of TGF-β1 on tumorigenesis.
MATERIALS AND METHODS
Reagents and Antibodies
Recombinant human TGF-β1 was ordered from R&D, the TβIR inhibitor SB431542, the MEK1 inhibitor PD98059, the p38 MAPK inhibitor SB203580, the JNK inhibitor SP600125, the PI3K inhibitor LY294002, the Src inhibitor PP2 were purchased from Calbiochem. Transcription inhibitor Actinomycin D (Act D), translation inhibitor cycloheximide, proteasome inhibitor MG132 and lysosome inhibitor chloroquine were obtained from Sigma and added to cultures for 1 h before the addition of TGF-β1. Anti-PTEN monoclonal antibody, Anti-PTEN polyclonal antibody, Anti-AKT monoclonal antibody, Anti-ERK1 monoclonal antibody, and anti-GAPDH monoclonal antibody were obtained from Santa Cruz. Phospho-AKT473 rabbit mAb, phospho-ERK1/2 rabbit mAb, phospho-p38 rabbit mAb, phosphor-JNK rabbit mAb, p38 rabbit mAb, and JNK rabbit mAb were ordered from CST. Secondary antibody conjugated with HRP or FITC was purchased from Calbiochem and Sigma, respectively.
Cell Culture
SMMC-7721 was cultured in RPMI 1640 (Gibco, Grand Island, NY) supplemented with 10% of newborn calf serum in a 37°C incubator with 5% CO2. Human 293T cells, HepG2 cells, and Huh7 cells were maintained in the Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum in a 37°C incubator with 5% CO2.
Plasmid Constructs
Full-length of PTEN promoter-luciferase report vector pGL3−2768 (−2927/−160) and the core region of PTEN promoter-luciferase construct pGL3−612 (−1389/−778) were described beforehand [Ma et al., 2005]. Human 3′-UTR of PTEN was obtained by 3′-RACE with the 3′-RACE special Primer: 5′-ACAGGCTCCCAGACATGACA-3′ from total mRNA and subcloned downstream of luciferase in the pGL3 basic-luciferase plasmid. pEGFP-C3-PTEN (full-length, amino acids 1-403), pEGFP-C3-PTENΔC (amino acids 1-186, C2 domain and PDZ binding motif were deleted completely) expression vectors were obtained by PCR amplification from human genomic DNA with the special primers (5′-CTTAAGCTTATGACAGCCAT-3′ and 5′-ACGAATTCTCAGACTTTTGTA-3′ for PTEN, 5′-CTTAAGCTTATGACAGCCAT-3′ and 5′-GGGAATTCTCACAGATGATTCTT-3′ for PTENΔC) and then subcloned into pEGFP-C3 vector.
Transient Transfection and Luciferase Reporter Assay and SiRNA
Cells were seeded into 24-well plates 1 day before transfection. The transfection was performed with the Lipofectamine™ 2000 Transfection Reagent (Invitrogen) according to the manufacturer's directions. Typically, 0.9 µg of pGL3 vector and 0.1 µg pRL-SV40 were used per well. After 24 h, cells were treated with 10 ng/ml TGF-β1 or buffer for another 24 h. Luciferase activities were assessed using the Dual-Luciferase Assay System (Promega). Activities of firefly (experimental) and Renilla (control) luciferases were measured in a luminometer Lumat LB 9507. PTEN promoter activity was normalized by calculating the ratio of firefly/Renilla luciferase activity of the same lysate sample. The analysis of each promoter construct was done in three independent experiments, which were repeated at least three times. For SiRNA knockdown experiment, 293T cells were transfected with SiRNA using Lipofectamine™ 2000 Transfection Reagent (Invitrogen) according to the manufacturer's directions. Si-Smad4, Si-p38 and Si-control were ordered from Invitrogen.
Semi-Quantitative Transcription Polymerase Chain Reaction
Total RNA was isolated using the trizol system according to the manufacturer's guidelines. Oligo(dT)18 primer and M-MLV reverse transcriptase were used for first strand synthesis. For semi-quantitative RT-PCR, the cDNA products were amplified by PCR using primers as follows, PCR primer pairs for total PTEN from 117 to 741 bp were as described before [Teng et al., 1997]. The primers designed to amplify the endogenous PTEN comprised a region from −270 to 459 bp (5′-GCCGTTCGGAGGATTATTCGTC-3′ and 5′-GCCGTTCGGAGGATTATTCGTC-3′), which contained a Hha I restriction site at the 255 bp in the PTEN pseudo-gene, which was not in the products. The primers for pEGFP-C, which are used to amplify the exogenous PTEN (GFP-PTEN) are 5′-CATGGTCCTGCTGGAGTTCGTG-3′ and 5′-TATGGCTGATTATGATCAGT-3′. Primers for β1-actin [Takano et al., 2000] were used as the internal control. The samples were amplified for 27 cycles at cyclic temperatures of 94°C for 45 s, 55°C for 30 s, and 72°C for 1 min. PCR products were analyzed through 1% agarose gel electrophoresis. The band area was measured by using TotalLab software (Nonlinear Dynamics Ltd).
Western Blot Analysis
Cells were lysed in 1× sodium dodecyl sulfate (SDS) lysis buffer (50 mM Tris–HCl, pH6.8, 2% SDS, 10% glycerol, 100 µg/ml PMSF, 10 µg/ml leupeptin, and 5 mM Na3VO4). Protein concentration was determined with Hartree assay. Cells lysates were separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. The membranes were blocked in 5% non-fat milk. After incubation of primary and secondary antibodies, membranes were developed by enhanced chemiluminescence (Amersham). The bands were scanned and quantified by TotalLab2.01 (Nonlinear Dynamics Ltd).
Immunoprecipitation
Cells were washed with ice-cold PBS, and lysed in lysisbuffer (containing 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 15 mM EGTA, 0.5% (w/v) Nonidet P-40, 1 mM PMSF, 1 mM DTT, 1 mM Na2VO3, 100 mM NaF, 5 µg/ml leupeptin, and 5 µg/ml aprotinin). The cells lysates were centrifuged at 12,000 rpm for 15 min at 4°C. The supernatants were collected, and protein concentration was determined by means of Lowry protein assay. Equal amounts of protein samples (1 mg) were incubated with 1 µg of each antibody for 1.5 h and then 20 µl of protein A/G plus-agarose for incubation at 4°C overnight. The immunoprecipitates were washed four times with lysis buffer. Samples were resuspended in 15 µl of 2× SDS sample buffer and boiled 3 min at 95°C prior to analyses by Western blotting.
Statistical Analysis
Statistical analysis of data was done with t-test or one-way ANOVA, statistical significance was set at P < 0.05.
RESULTS
TGF-β1 Decreases PTEN Expression in Hepatocarcinoma Cell Lines
Li and Sun 1997 had showed by Northern blot assay that PTEN transcription was down-regulated by TGF-β1 in human keratinocytes. Previously we had also demonstrated that TGF-β1 down-regulated PTEN mRNA [Zhang et al., 2004] and protein expression [Cai et al., 2000] in hepatocarcinoma cell line SMMC-7721. To further investigate the interrelation between TGF-β1 and PTEN, we examined the status of PTEN mRNA and protein expression in SMMC-7721 cells with the treatment of different amount of TGF-β1. TGF-β1 down-regulated PTEN mRNA (Fig. 1A) and protein (Fig. 1B) expression in a dose-dependent manner, 10 ng/ml TGF-β1 decreased about 50% PTEN expression and was used throughout the study. Then we performed the time course experiment to measure the TGF-β1 effect on PTEN mRNA and protein levels. Both PTEN mRNA (Fig. 1C) and protein (Fig. 1D) levels were down-regulated by TGF-β1 in a time-dependent manner. Surprisingly, the PTEN protein is reduced prior to the reduction in mRNA at 2 h time point, implying that there are translational or post-translational regulation mechanisms that reduce PTEN protein levels even before the PTEN mRNA is potentially destroyed. To distinguish it, we performed a CHX chase protein decay assay between TGF-β1-treated and -untreated cells (Fig. 4C,D). In TGF-β1-untreated cells, PTEN levels were decreased with time, and its half-life was about 10 h. On the contrary, the accelerating degradation of PTEN was observed in TGF-β1-treated cells where the PTEN half-life was about 4 h. Taken together, these findings suggested that TGF-β1 promoted PTEN degradation at the post-translational level. Here we also found that down-regulation of PTEN by TGF-β1 was also existed in other hepatocarcinoma cell lines (Fig. 1E). In all, these findings indicate that TGF-β1 acts as a transcriptional repressor and also a post-translational modifier in hepatocarcinoma cell lines.
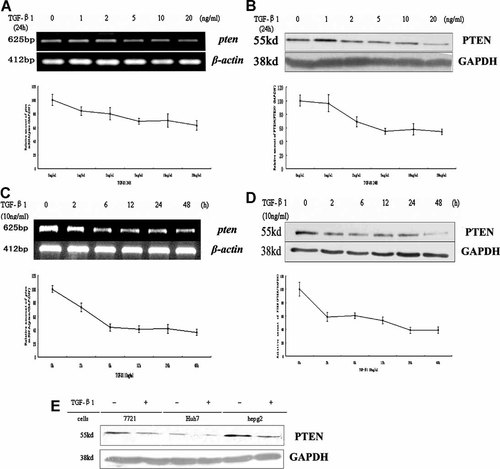
TGF-β1 decreases PTEN expression in hepatocarcinoma cell line SMMC-7721. A: Dose-dependent regulation of PTEN mRNA expression by TGF-β1 in SMMC-7721 cells. Cells were treated with the indicated concentrations of TGF-β1 for 24 h and total RNA was subjected to semi-quantitative RT-PCR analysis with special primers. B: Dose-dependent regulation of PTEN protein expression by TGF-β1 in SMMC-7721 cells. Cells were treated with the indicated concentrations of TGF-β1 for 24 h and total cell lysates were subjected to Western blot analysis with specific antibodies. C: Time-dependent regulation of PTEN mRNA expression by TGF-β1 in SMMC-7721 cells. Cells were incubated with 10 ng/ml TGF-β1 for the indicated times and total RNA was subjected to semi-quantitative RT-PCR analysis with special primers. D: Time-dependent regulation of PTEN protein expression by TGF-β1 in SMMC-7721 cells. Cells were incubated with 10 ng/ml TGF-β1 for the indicated times and total cell lysates were subjected to Western blot analysis with specific antibodies. E: Expression of PTEN protein levels in three liver carcinoma cell lines was evaluated by Western blotting. Cells were incubated with 10 ng/ml TGF-β1 for 24 h and total cell lysates were subjected to Western blot analysis with specific antibodies. The values represent means of triplicate assays. Bars; standard deviations.
Smad and p38-MAPK Pathways Are Involved in TGF-β1-Mediated Down-Regulation of PTEN
It is well known that Smads are key mediators of TGF-β1-Smad signaling pathway [Massague, 1998; Massague et al., 2000; Akhurst, 2004]. Here we observed that 24 h after TGF-β1 stimulation, phosphorylated Smad2, and Smad3 significantly increased with the down-regulation of PTEN (Fig. 2A), suggesting that Smad pathway may be involved in the down-regulation of PTEN. To evaluate whether Smad pathway is involved, Si-Smad4 is introduced to the following experiments. With the knockdown of TGF-β1-Smad signal pathway by Si-Smad4, we observed that Si-Smad4 restored PTEN expression despite of TGF-β1 treatment (Fig. 2B), indicating that Smad signal pathway was involved in TGF-β1-induced PTEN down-regulation. In addition to the canonical Smad signal pathway, some non-Smad signal pathway including the mitogene-activated protein kinases (ERK, JNK, and p38), the PI3k and Ras- and Rho-GTPases have been described in mediating cellular effects of TGF-β1. To explore the possible involvement of signaling pathways in PTEN down-regulation by TGF-β1, the pharmacological inhibitors were employed to study. As shown in Figure 2C, TGF-β1-mediated down-regulation of PTEN was blocked by the TβIR inhibitor SB431542, the MEK1 inhibitor PD98059 and the p38 inhibitor SB203580 but not rescued by the PI3K inhibitor LY294002, the Src inhibitor PP2, and the JNK inhibitor SP600125. Then, we examined the expression of MAPK proteins, phospho-p38 was increased with TGF-β1 stimulation, but phospho-ERK and phospho-SAPK/JNK were not changed with TGF-β1 treatment (Fig. 2D). So Si-p38 was used to sure whether p38 was involved in the suppression of PTEN by TGF-β1, and TGF-β1-mediated down-regulation of PTEN was recovered by Si-p38. This result indicated that p38-MAPK was also involved in the suppression of PTEN by TGF-β1.
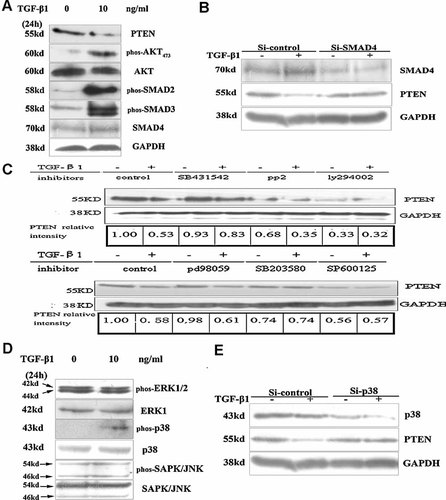
Smad and p38 MAPK pathways are involved in TGF-β1-mediated down-regulation of PTEN. A: Induction of Smad proteins by TGF-β1 in SMMC-7721 cells. Cells were treated with 10 ng/ml TGF-β1 for 24 h, and total cell lysates were subjected to Western blot analysis with specific antibodies. B: Effect of the Si-Smad4 on TGF-β1-mediated down-regulation of PTEN in SMMC-7721 cells by Western blotting analysis. Cells were transfected with the Si-Smad4 or Si-control followed with or without the treatment of TGF-β1 for 24 h. Expression of PTEN protein was examined by Western blot assay. C: Effect of various pharmacological inhibitors of signaling pathways on TGF-β-induced down-regulation of PTEN. SMMC-7721 cells were preincubated with the TβRI inhibitor SB431542 (10 µM), the PI3K inhibitors LY294002(50 µM), the Src inhibitor PP2 (10 µM), the p38 inhibitor SB203580(10 µM), and the JNK inhibitor SP600125(10 µM), and the MEK1 inhibitor PD98059 (25 µM) for 1 h before TGF-β treatment (10 ng/ml, 24 h). PTEN protein level was determined by Western blotting. D: Induction of MAPK proteins by TGF-β1 in SMMC-7721 cells. Cells were treated with 10 ng/ml TGF-β1 for 24 h, and total cell lysates were subjected to Western blot analysis with specific antibodies. E: Effect of the Si-p38 on TGF-β1-mediated down-regulation of PTEN in SMMC-7721 cells by Western blotting analysis. Cells were transfected with the Si-Smad4 or Si-control followed with or without the treatment of TGF-β1 for 24 h. Expression of PTEN protein was examined by Western blot assay.
TGF-β1 Down-Regulates PTEN in Post-Transcriptional and Post-Translational Level
To make it clear whether TGF-β1 down-regulated PTEN at a transcriptional level, we applied a dual-luciferase assay by using full-length and core legion of PTEN promoter reporter vectors which were reported before [Ma et al., 2005]. However, reporter activity of PTEN promoter-luciferase constructs was not affected by TGF-β1 stimulation in SMMC-7721 cells (Fig. 3A). The result indicated that TGF-β1-mediated down-regulation might be under the post-transcriptional control.

TGF-β1 down-regulates PTEN in a post-transcriptional control. A: Dual-luciferase activity assay of PTEN promoter-luciferase activity in TGF-β1 treated or untreated SMMC-7721 cells. Cells were transfected with PTEN full or core promoter-luciferase constructs as described. Twenty-four hours after transfection, cells were treated with or without TGF-β1 for 24 h, followed by dual-luciferase assay. Results are representative of three independent experiments. B: Dual-luciferase activity assay of PTEN 5′- and 3′-UTR-luciferase activity in TGF-β1-treated and -untreated SMMC-7721 cells. Cells were transfected with PTEN 5′- and 3′-UTR luciferase constructs. Twenty-four hours after transfection, cells were treated with or without TGF-β1 for 24 h, followed by dual-luciferase assay. The values represent means of triplicate assays. Bars; standard deviations. P > 0.05 using one-way ANOVA to compare TGF-β1-treated with -untreated groups.
To explore whether TGF-β1 treatment modulates the stability of PTEN mRNA and leads to the decreased PTEN expression, the mRNA decay level of PTEN was compared between TGF-β1-treated and -untreated cells (Fig. 4A). It revealed in a time course of PTEN mRNA decay that the stability of PTEN mRNA was decreased with TGF-β1 treatment. Time required for a 50% loss of PTEN mRNA decreased from 3 h to 1 h by TGF-β1 in comparison of control (Fig. 4B). These results suggested that TGF-β1 stimulation increased the turnover rate of PTEN mRNA and further supported the conclusion that TGF-β1-mediated down-regulation was post-transcriptional.
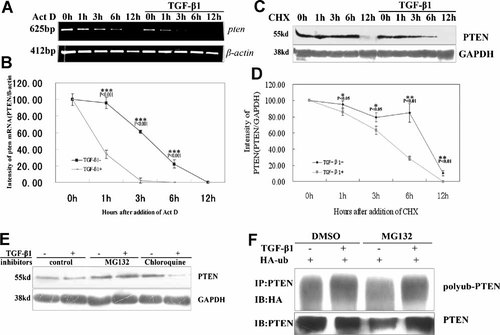
TGF-β1 increases the turnover rate of PTEN mRNA and protein in SMMC-7721 cells. A: Decay time course of PTEN mRNA levels in TGF-β1-treated or -untreated SMMC-7721 cells. Total mRNA was collected at various time periods after Act D treatment and PTEN mRNA levels were analyzed by Semi-quantitative RT-PCR. B: Quantitative representation of (A). C: Decay time course of PTEN levels in TGF-β1-tretaed or -untreated SMMC-7721 cells. Total protein was collected at various time periods after CHX treatment and PTEN levels were evaluated by Western blotting analysis. D: Quantitative representation of (C). E: Effect of MG132 and chloroquine on the suppression of PTEN by TGF-β1. SMMC-7721 cells were treated with 10 ng/ml TGF-β1 for 2 h first, then MG132 or chloroquine were added to the medium for 6 h, and total cell lysates were subjected to Western blot analysis with specific antibodies. F: Polyubiquitination of PTEN. HA-tagged ubiquitin (HA-Ub) was transfected to 293T cells. Twenty-four hours after transfection, cells are treated with or without 10 ng/ml TGF-β1 for 2 h, following by the addition of 25 µM MG132 for 6 h. After immunoprecipitation using anti-PTEN antibody, anti-HA, and PTEN were detected in the precipitates by immunoblotting. The values represent means of triplicate assays. Bars; standard deviations. *P < 0.05; **P < 0.01; ***P < 0.001 by t-test.
The untranslated regions (5′- or 3′-UTR) play an important role in the post-transcriptional regulation of gene expression, especially in the modulation of mRNA stability [Bashirullah et al., 2001; Tourriere et al., 2002; Yang et al., 2008]. To investigate whether UTR of PTEN mRNA is responsible for TGF-β1-mediated down-regulation, luciferase report vectors pGL3-2768 (containing 5′-flanking region and 5′-UTR) and pGL3-3′-UTR (containing the 3′-UTR) were used in dual-luciferase activity assay (Fig. 3B). However, no luciferase activity change was observed between TGF-β1-treated and -untreated cells. So we reached the conclusion that TGF-β1-induced PTEN down-regulation was through post-transcriptional control, but the UTRs of PTEN were not essential for mediating its down-regulation under TGF-β1 stimulation.
Our data have suggested that TGF-β1 increased PTEN degradation at the post-translational level. To make sure of it, proteasome inhibitor MG132 and lysosome inhibitor chloroquine were used to detect the effect of TGF-β1 on PTEN degradation. MG132 but not chloroquine was found to restore the down-regulation of PTEN by TGF-β1 (Fig. 4E). It meant that TGF-β1 accelerated PTEN through a proteasome dependent pathway. Then we examined whether PTEN is ubiquitinated in cells. 293T cells were transfected with plasmids encoding for HA-tagged ubiquitin. PTEN protein was precipitated from cell lysates with anti-PTEN polyclonal antibody and protein A/G plus-agarose. Subsequently, immunoblotting against HA-tag was performed to detect ubiquitinated PTEN (Fig. 4F). We found that treatment of TGF-β1 caused an increase of PTEN polyubiquitination (Fig. 4F). These results further demonstrated that TGF-β1 suppressed the expression of PTEN at a post-translational level.
PTEN Coding Sequence and De Novo Protein Synthesis Are Required for TGF-β1-Mediated Down-Regulation of PTEN mRNA
We have testified that TGF-β1-mediated PTEN mRNA instability is not associated with the UTR of PTEN transcripts. To make sure whether the coding sequence of PTEN is related to TGF-β1-mediated PTEN mRNA instability, PTEN full-length coding sequence was cloned into multiple Cloning Site and expressed as fusions to the C terminus of EGFP (Fig. 5A). EGFP-C Sequencing Primers were used to measure the mRNA level of fused complex, which was marked as the exogenous PTEN, and special primers were designed from −270 to 459 to detect the expression of endogenous PTEN by semi-quantitative RT-PCR (Fig. 5A). Here, we found that TGF-β1 stimulation down-regulated both exogenous and endogenous PTEN mRNAs (Fig. 5B), and the transfection of PTEN full-length coding sequence increased TGF-β1-induced degradation of PTEN mRNA. But in transfectants expressing pEGFP-C3 alone, the GFP transcript was not down-regulated by TGF-β1 (Fig. 5B), suggesting that TGF-β1-mediated down-regulation of exogenous PTEN transcript was not related to the transcriptional regulation of pEGFP-C3 vector. These results revealed that the coding sequence of PTEN was necessary and sufficient for TGF-β1-mediated down-regulation. To figure out which coding region of PTEN mRNA is involved in the down-regulation of PTEN by TGF-β1, pEFGP-PTEN and one delete mutant pEFGP-PTENΔC (aa 1-186) were transfected to 293T cells, it was found that TGF-β1 no longer inhibited the PTEN mRNA expression in pEFGP-PTENΔC (aa 1-186) transfected cells (Fig. 5C). Deletion of the PTEN C-terminus increased the half-life of PTEN mRNA (Fig. 5D), suggesting that the C terminus of PTEN coding sequence affected its stability. Previous studies indicated that the de novo protein synthesis was involved in TGF-β1-mediated mRNA regulation [Beauchamp et al., 1992; Zhang et al., 1999; Chou and Yang, 2006]. To further investigate whether de novo protein synthesis was involved in PTEN mRNA down-regulation by TGF-β1, cycloheximide, a protein synthesis inhibitor, was used to measure the stability of PTEN mRNA. As was shown in Figure 6, the cycloheximide treatment blocked the suppression of PTEN by TGF-β1. Therefore, we conclude that de novo protein synthesis is required for PTEN mRNA down-regulation by TGF-β1. These data support the conclusion that C terminus of PTEN coding sequence and the de novo protein synthesis are essential for its down-regulation by TGF-β1, and also strongly support our earlier data that TGF-β1-mediated down-regulation is through post-transcriptional control.
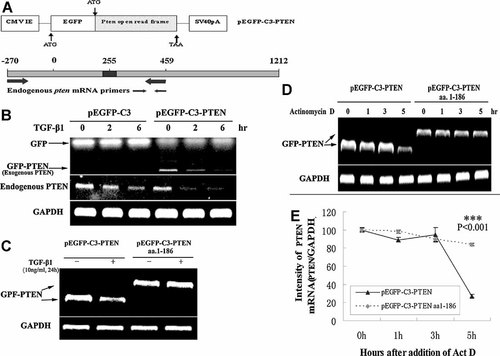
PTEN coding sequence is necessary for TGF-β1-mediated down-regulation of PTEN. A: Schematic expression of pEGFP-C3-PTEN plasmid and special primers for endogenous PTEN. B: Expression of endogenous and exogenous PTEN mRNA levels in pEGFP-C3 or pEGFP-C3-PTEN transfected 293T cells followed with or without the treatment of TGF-β1 for the indicated times by Semi-quantitative RT-PCR. C: Exogenous PTEN mRNA levels are determined by Semi-quantitative RT-PCR in transfected 293T cells. Cells were transfected with pEGFP-C3-PTEN (full-length, amino acids 1-403) or pEGFP-C3-PTENΔC (amino acids 1-168) constructs, 24 h after transfection, cells were treated with or without the TGF-β1 for 24 h. D: Decay time course of exogenous PTEN mRNA levels in transfected 293T cells by Semi-quantitative RT-PCR. Cells were transfected with pEGFP-C3-PTEN or pEGFP-C3-PTENΔC plasmids, 24 h after transfection, cells were treated with 10 ng/ml actinomycin D for the indicated times. E: Quantitative representation of (D). The values represent means of triplicate assays. Bars; standard deviations. ***P < 0.001 by t-test.
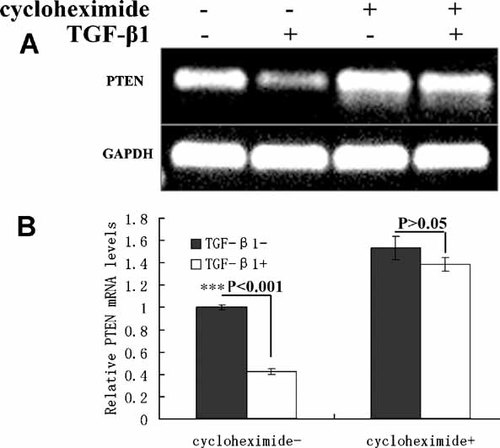
De novo protein synthesis is required for TGF-β1-mediated down-regulation of PTEN. A: Effect of protein synthesis inhibitor Cycloheximide on TGF-β1-mediated down-regulation of PTEN mRNA in SMMC-7721 cells. Cells were pretreated with 10 ng/ml cycloheximide for 90 min, followed by 10 ng/ml TGF-β1 for another 6 h. Total RNA was isolated, and Semi-quantitative RT-PCR was performed with specific primers. B: Quantitative representation of (A). The values represent means of triplicate assays. Bars; standard deviations, ***P < 0.001; P > 0.05 by one-way ANOVA.
DISCUSSION
TGF-β1 is a multifunctional growth factor and works as a tumor suppressor at early stages of tumorigenesis, but at later stages accelerating cancer progression [Akhurst and Derynck, 2001; Roberts and Wakefield, 2003]. PTEN is a tumor suppressor, which plays a novel role in inhibiting cell proliferation, adhesion, cell migration, and cell invasion [Tamura et al., 1998, 1999]. It has been reported that PTEN represses TGF-β1-mediated cellular migration and invasion whereas loss of PTEN expression increases TGF-β1-induced cellular migration and invasion [Hjelmeland et al., 2005]. Meanwhile, PTEN mRNA suppression by TGF-β1 has been observed in keratinocytes and pancreatic cells [Li and Sun, 1997; Ebert et al., 2002; Chow et al., 2007], and modulating role of RAS/ERK in TGF-β1-regulated PTEN expression was described in human pancreatic adenocarcinoma cells [Chow et al., 2007]. It suggests that the down-regulation of PTEN by TGF-β1 may play a crucial role in accelerating cancer progression. However, the molecular mechanism of PTEN down-regulation by TGF-β1 is still undefined. The present study shows that TGF-β1 down-regulates both PTEN mRNA and protein expressions in a dose- and time-dependent manner in human SMMC-7721 hepatoma cell lines. Interestingly, PTEN protein is reduced prior to the reduction of PTEN mRNA at 2 h time point, supposing that translational or post-translational mechanism may exist in the down-regulation of PTEN by TGF-β1. CHX chase protein decay assay show that TGF-β1 increases the turnover rate of PTEN protein, which suggesting that TGF-β1-mediated PTEN degradation may be partly at the post-translational level. The proteasome inhibitor MG132 recovers PTEN expression indicating that TGF-β1 may down-regulate PTEN through an ubiquitin-proteasome dependent pathway. TGF-β1 increases the polyubiquitination of PTEN in 293T cells further supporting that TGF-β1-mediated down-regulation of PTEN is post-translational.
Based on the dual-luciferase activity assay of PTEN promoter reporter, TGF-β1 has little effect on the PTEN gene transcripts. It is supposed that TGF-β1-induced decrease in PTEN mRNA levels might primarily appear in the post-transcriptional level. The decrease of PTEN mRNA induced by TGF-β1 stimulation correlates with an increase in the turnover rate of PTEN mRNA as it is examined by Act D chase experiments. These results further support that TGF-β1-mediated down-regulation of PTEN is also post-transcriptional. Post-transcriptional regulation under TGF-β1 stimulation had been found in some genes, such as TGF-β1,2, IL-6, albumin, AFP, elastin, and cite2 genes [Bascom et al., 1989; Beauchamp et al., 1992; Zhang et al., 1999; Park et al., 2003; Chou and Yang, 2006]. It means that post-transcriptional regulation of mRNA stability by TGF-β1 may also be as important as transcriptional control of TGF-β1 for a number of target genes. Our results suggest that post-transcriptional regulation of PTEN mRNA stability by TGF-β1 may also be important. Many post-transcriptional studies have been focus on the UTRs especially the 3′-UTR of mRNAs containing AU-rich elements (AREs), which bind with the trans-acting protein factors [Hollams et al., 2002; Tourriere et al., 2002; Barreau et al., 2005; Eberhardt et al., 2007]. It has been reported that TGF-β1 increases the ribonucleotide reductase R2 mRNA stability through its 3′-UTR, and 3′-UTR is also account for the down-regulation of the PTEN by micro RNA-214 [Yang et al., 2008]. But in our present study, TGF-β1 treatment has no effect on UTR of PTEN (Fig. 3B).
The regulatory sequence of mRNA stability are often located in the 5′- or 3′-UTRs or within the protein-coding region [Tourriere et al., 2002]. To further elucidate the mechanisms of TGF-β1-mediated down-regulation of PTEN, we analyzed the mRNA expression of the recombinant PTEN gene in SMMC-7721 cells. Protein coding sequences are essential for the mRNA stability of IL-11, c-myc, c-Fos, elastin, albumin, AFP, yeast MAT1α, and cite2 [Shyu et al., 1989; Parker and Jacobson, 1990; Beauchamp et al., 1992; Bernstein et al., 1992; Yang et al., 1996; Zhang et al., 1999; Chou and Yang, 2006]. The stability of these mRNA has appeared to require the de novo protein synthesis. Our results suggest that PTEN protein coding region is essential for its down-regulation by TGF-β1, and TGF-β1-induced PTEN suppression appears to require de novo protein synthesis too. Protein synthesis inhibitor cycloheximide blocks translation elongation through direct interaction with the 60S subunit of ribosomes and results in polysome aggregation. Because CHX could potentially block translation of PTEN transcript or inhibit synthesis of short-lived trans-factors involved in the down-regulation of PTEN mRNA stability by TGF-β1, CHX treatment blocked the TGF-β1-mediated suppression of PTEN mRNA. These two possible mechanisms need to be distinguished in the future research. mRNAs bearing such coding region determinants (CRD) include c-fos, c-myc, β1-tubulin, Drosophila fushi tarazu, MAT1α, and Cite2 [Tourriere et al., 2002]. Recent experiments using the c-fos mRNA as a model system have identified a major destabilizing region, termed mCRD which must be 450 nt away from the poly A tail in the mRNA sequence to exert this function [Chen et al., 1992; Schiavi et al., 1994; Grosset et al., 2000]. C-terminus of PTEN coding sequence increases the turnover rate of PTEN mRNA (Fig. 5D), suggesting that a CRD may be located in the C-terminus of PTEN coding sequence. This is the first demonstration that C-terminus of PTEN coding sequence is required for the PTEN mRNA stability. Whether there exist trans-factors binding to the CRD of PTEN to impend deadenylation and to stabilize PTEN mRNA or there remain other mechanisms need to be further investigated.
With the treatment of TβIR inhibitor SB432541, we have found that SB432541 partially block the down-regulation of PTEN by TGF-β1 in SMMC-7721 cells, further study has indicated that Smad4 is involved in the PTEN suppression by TGF-β1. TGF-β1 signal pathways include Smad-dependent and Smad-independent signal pathways. Mechanistically, many of the pro-oncogenic responses to TGF-β1 are either Smad-independent, or require cooperation between the Smad and non-Smad signal such as MAPK pathways, RhoA, and PI3K/Akt pathways [Wakefield and Roberts, 2002]. In our research, we found that the TβRI inhibitor SB432541, the MEK1 inhibitor PD98059 can rescue the down-regulation of PTEN by TGF-β in SMMC-7721 cells partly, but we had not observed recover of PTEN expression with the treatment of other inhibitors including the JNK inhibitor SP600125. Because of no change of phospho-ERK and phospho-SAPK/JNK, ERK and SAPK/JNK signal pathway might not be involved in the down-regulation of PTEN by TGF-β in SMMC-7721 cells. But Chow et al. found that TGF-β1-induced PTEN suppression was reversed by PD98059 and DNK-RAS in pancreatic adenocarcinoma [Chow et al., 2007]. It was also found that PTEN expression was regulated by c-Jun NH2-terminal kinase and nuclear factor-kappaB in intestinal epithelial cells [Wang et al., 2007a]. So we think that TGF-β1 may down-regulate PTEN in a cellular dependent manner. The MEK1 inhibitor PD98059 can rescue the down-regulation of PTEN by TGF-β in SMMC-7721 cells partly, indicating that other signal pathways may be regulated by PD98059, which need to be further investigated. The PI3K inhibitor LY294002 and the Src inhibitor PP2 can inactivate PI3K/PKB signal pathway which can also inhibited by PTEN, but PTEN was also found to be decreased by the treatment of LY294002 or PP2. These results imply there might be a feedback regulation of PTEN in SMMC-7721 cells and this feedback regulation of PTEN depends on inhibition of PI3K/Akt pathway.
In summary, we find that PTEN is down-regulated by TGF-β1 in SMMC-7721 cells through the Smad and p38 MAPK signal pathways. The down-regulation of PTEN by TGF-β1 is post-transcriptional and partly through the accelerated turnover rate of PTEN mRNA. Accelerated turnover rate of PTEN and increased polyubiquitination of PTEN by TGF-β1 indicate a post-translational regulation mechanism. The present study demonstrates that the C terminus of the PTEN coding sequence is necessary for the down-regulation of PTEN mRNA by TGF-β1, and TGF-β1-mediated PTEN mRNA stability requires de novo protein synthesis. Future research will be focused on the multiple signaling pathways, which may have synergistic collaboration effects on the down-regulation of PTEN by TGF-β1. Nevertheless, future studies will also be carried on to identify the elements in the C terminus of the PTEN coding sequence which lead to the acceleration of PTEN mRNA decay and to define trans-factors that mediate its down-regulation by TGF-β1.
Acknowledgements
This investigation was supported by Shanghai Leading Academic Discipline Project (Project No. B110).




