Acidic pH stimulates the production of the angiogenic CXC chemokine, CXCL8 (interleukin-8), in human adult mesenchymal stem cells via the extracellular signal-regulated kinase, p38 mitogen-activated protein kinase, and NF-κB pathways
Abstract
Blood vessel injury results in limited oxygen tension and diffusion leading to hypoxia, increased anaerobic metabolism, and elevated production of acidic metabolites that cannot be easily removed due to the reduced blood flow. Therefore, an acidic extracellular pH occurs in the local microenvironment of disrupted bone. The potential role of acidic pH and glu-leu-arg (ELR+) CXC chemokines in early events in bone repair was studied in human mesenchymal stem cells (hMSCs) treated with medium of decreasing pH (7.4, 7.0, 6.7, and 6.4). The cells showed a reciprocal increase in CXCL8 (interleukin-8, IL-8) mRNA levels as extracellular pH decreased. At pH 6.4, CXCL8 mRNA was induced >60× in comparison to levels at pH 7.4. hMSCs treated with osteogenic medium (OGM) also showed an increase in CXCL8 mRNA with decreasing pH; although, at a lower level than that seen in cells grown in non-OGM. CXCL8 protein was secreted into the medium at all pHs with maximal induction at pH 6.7. Inhibition of the G-protein-coupled receptor alpha, Gαi, suppressed CXCL8 levels in response to acidic pH; whereas phospholipase C inhibition had no effect on CXCL8. The use of specific mitogen-activated protein kinase (MAPK) signal transduction inhibitors indicated that the pH-dependent increase in CXCL8 mRNA is due to activation of ERK and p38 pathways. The JNK pathway was not involved. NF-κB inhibition resulted in a decrease in CXCL8 levels in hMSCs grown in non-OGM. However, OGM-differentiated hMSCs showed an increase in CXCL8 levels when treated with the NF-κB inhibitor PDTC, a pyrrolidine derivative of dithiocarbamate. J. Cell. Biochem. 104: 1378–1392, 2008. © 2008 Wiley-Liss, Inc.
Tissue trauma including bone fractures can lead to an interruption of blood supply in the local environment. This results in hypoxic conditions with a shift in cellular metabolism to an anaerobic pathway resulting in the accumulation of lactic acid. Thus, a local acidosis with a low tissue pH can develop at the fracture site [Newman et al., 1985, 1987]. An early inflammatory response also occurs as the first step in tissue repair after hemostasis has been established. This inflammatory response, required for wound healing in general, is largely orchestrated by chemotactic cytokines (chemokines) that attract polymorphonuclear leucocytes and monocytes/macrophages and are also elaborated by these inflammatory cells [Charo and Ransohoff, 2006].
ELR+ CXC chemokines, members of the α family of chemokines having a glu-leu-arg (ELR) motif just N-terminal of the C-X-C amino acids, have been shown to be angiogenic [Strieter et al., 1995]. CXCL8, also known as interleukin-8 (IL-8) the prototypic representative of the ELR+ CXC chemokines family, has been shown to be present in the inflammatory milieu during tissue repair. Thus, CXCL8, the other ELR+ CXC chemokines such as CXCL1, CXCL2, and CXCL3 (also known as GROα, GROβ, and GROγ, respectively), as well as other angiogenic growth factors such as fibroblast growth factor-2 could potentially be responsible for the vascularity of the granulation tissue seen in skin wound repair or the early callus formation in fracture repair.
The induction of CXCL8 expression from monocytes/macrophages and other cell types has been previously described [Rothe et al., 1998; Jung et al., 2002; Parhar et al., 2003; Pae et al., 2005; Romero et al., 2006]. Constitutive expression of CXCL8 mRNA has also been observed in human mesenchymal stem cells (hMSCs) [Majumdar et al., 1998, 2000; Kim et al., 2005], and we have shown induction of CXCL8 during differentiation of hMSCs towards the osteoblastic lineage [Bischoff et al., 2007]. While it has been noted that IL-1α stimulation of hMSC cultures can increase the steady-state mRNA levels of CXCL8 [Majumdar et al., 1998], it has not previously been systematically investigated if CXCL8 can be induced by hMSCs subjected to changes in extracellular proton concentration that would be expected during the initial phase of bone repair. hMSCs that migrate into the area of disrupted bone would encounter a microenvironment that is both hypoxic and having an acidic pH before differentiating into osteoblasts, producing new bone proteinaceous matrix, vasularization, and finally mineralization of the bone matrix to restore structural integrity of the bone.
In this report, we demonstrate that CXCL8 mRNA and protein can be stimulated by low extracellular pH in hMSCs (bone marrow stromal cells). Furthermore, we demonstrate that the low pH-stimulated increase in CXCL8 occurs through the mitogen-activated protein kinase (MAPK), extracellular signal-regulated kinase (ERK) and p38 pathways, and also via the nuclear factor-κB (NF-κB) signaling pathway.
MATERIALS AND METHODS
Cell Culture
hMSCs (bone marrow stromal cells), hMSC complete growth medium (HMSCGM), and growth supplements were obtained from Cambrex Bio Science Walkersville, Inc. (now Lonza, Walkersville, Inc., Walkersville, MD). These cells have the potential to differentiate to bone, cartilage, and adipose using protocols provided by Cambrex. The supplier has also tested these hMSCs for positive expression of CD105, CD166, CD29, and CD44, found on multipotential bone marrow stromal cells, and to be negative for CD34 and CD45, markers of hematopoietic stem cells. hMSCs from several donors were used (Table I). Cells were subcultured at 60–70% confluence at 37°C under 95% air/5% CO2 atmosphere, and used between passages 2 and 7. For differentiation towards the osteoblastic lineage, hMSCs were treated every 3–4 days with osteogenic medium (OGM) which consisted of HMSCGM supplemented with 50 mM ascorbic acid-2-phosphate, 10 mM β-glycerophosphate, and 10−7 M dexamethasone (Sigma–Aldrich, St. Louis, MO). We have previously shown that treatment of the hMSCs with OGM induces differentiation towards the osteogenic lineage as assayed by up-regulation of alkaline phosphatase (mRNA and activity) and bone sialoprotein (mRNA) and by von Kossa and alizarin red staining for mineralization [Bischoff et al., 2007].
| Lot # | Age (years) | Race | Sex |
|---|---|---|---|
| 1F2155 | 24 | A | M |
| 1F1061 | 19 | C | F |
| 3F0551 | 26 | B | M |
| 4F0218 | 21 | A | M |
| 4F1560 | 23 | B | F |
| 6F4085 | 33 | B | M |
- A, Asian; C, Caucasian; B, Black, M, Male; F, Female.
pH Experiments
hMSC cells were plated at 5,000 cells/cm2 in HMSCGM in 24-well plates and allowed to attach for 4 h prior to treatment with OGM medium or continued culture in HMSCGM. After 3 or 7 days in culture, the OGM or HMSCGM medium was replaced for 24 h with similar medium in which the pH was adjusted between 6.4 and 7.4. This medium was made using reconstituted powdered αMEM (Sigma–Aldrich) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Ogden, UT) and 1% glutamine, penicillin, and streptomycin (Sigma–Aldrich). The pH of the medium was adjusted using 7.5%w/v NaHCO3 and then filter-sterilized. After treatment in pH-adjusted medium for 24 h, total RNA was isolated using the Qiagen RNeasy Miniprep kit (Qiagen, Inc., Valencia, CA). Each experiment was done at least three times in triplicate and the supernatants saved for ELISA and pH determination. CXCL8 protein levels in supernatants were measured with the human CXCL8/IL-8 DuoSet ELISA Development System (R&D Systems, Minneapolis, MN) using protocols supplied with the kit. Each sample was run in duplicate and compared to CXCL8 standard curves. In some experiments, cells were isolated and analyzed for cell viability using trypan blue staining and microscopic quantification for viable (white) and non-viable (blue) cells.
mRNA Stability Study
hMSCs were cultured for 5 days in 24-well plates in HMSCGM or OGM. The cells were then treated for 24 h with medium at pH 7.4 or 6.7. The transcription inhibitor actinomycin D was added (5 µg/ml) and total RNA isolated at various times up to 24 h and analyzed by real time RT-PCR. Relative levels of mRNA at each time point were compared to the 0 h time point to determine mRNA decay rates in relationship to 18S levels.
Signal Transduction Inhibitors
For some pH experiments signal transduction inhibitors (Calbiochem, San Diego, CA) and their respective control compounds when available (or vehicle) were added at the following concentrations: Gαi pathway, pertussis toxin (PTX) at 100 ng/ml; phospholipase C pathway, U73343 and U73122 (100 ng/ml); extracellular signal-regulated kinase (ERK) pathway, U0124 and U0126 (15 µM) and PD98059 (19 µM); p38 pathway, SB202474, SB202190, SB203580 (15 µM); jun N-terminal kinase (JNK) pathway, JNK(−) and JNK II (10 µM); NF-κB pathway, 1-pyrrolidinecarbodithioic acid (PDTC, 100 µM) which blocks NF-κB activation, (E)3-[(4-methylphenyl)sulfonyl]-2-propenenitrile (BAY11-7082) an inhibitor of IκBα phosphorylation (100 µM), carbobenzoxy-L-leucyl-L-leucyl-L-leucinal Z-LLL-CHO (MG132) a proteasome inhibitor (20 µM), caffeic acid phenethyl ester (CAPE, 50 µM) which inhibits NF-κB translocation but not IκBα phosphorylation via an antioxidant mechanism, 8-(N,N-diethylamino)-octyl-3,4,5-trimethoxybenzoate (TMB-8), an intracellular Ca2+ antagonist (50 µM) and 6-amino-4-(4-phenoxyphenylethylamino) quinazoline (QNZ, 10 µM), a novel NF-κB inhibitor of unknown mechanism.
Quantitative RT-PCR
Relative mRNA levels were quantitated by real-time RT-PCR using the Opticon Continuous Fluorescence System (MJ Research, Inc., Incline Village, NV) and the SYBR Green RT-PCR kit from Qiagen. Primers used for RT-PCR: human CXCL8 sense 5′-GCCTTCCTGATTTCTGCAGC-3′ and antisense 5′-TCCAGACAGAGCTCTCTTCC-3′, human 18S rRNA sense 5′-CCGCAGGTTCACCTACTG-3′ and antisense 5′-CGGGTCATAAGCTTGCCTG-3′. Reactions were performed in triplicate: reverse transcription at 50°C for 30 min, 95°C for 15 min; followed by 50 cycles of denaturing at 94°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Relative changes in gene expression were calculated in relation to 18S ribosomal RNA levels and the reference time point using the 2−ΔΔC(T) method [Livak and Schmittgen, 2001].
Western Blot Analysis
hMSCs were plated at 5,000–10,000 cell/cm2 in 35 mm diameter dishes and cultured in HMSCGM or OGM medium for 3 days before being treated with medium of pH 6.7 or 7.4. Cells were washed with cold PBS and lysates harvested by solubilization in 200 µl PhosphoSafe Extraction Reagent (EMD Chemicals, Gibbstown, NJ) for 5 min at room temperature at the specific time points indicated. Cell lysates were analyzed by SDS–PAGE and transferred to polyvinylidene difluoride membranes. Blots were probed with specific antibodies (Cell Signaling Technology, Danvers, MA) and immunoreactive proteins detected with the ECL kit (GE Healthcare Bio-sciences, Piscataway, NJ). Protein levels were quantitated using the BIOQUANT system (BioQuant Image Analysis Corp., Nashville, TN).
Statistics
Data are reported as the mean value ± SD. Values were analyzed by ANOVA with post-hoc analysis by the Bonferroni method for multiple comparisons between pairs or by Student's t-test using the GraphPad Prism statistical software and are considered significant if P < 0.05 in comparison to negative controls.
RESULTS
pH Induced Expression of CXCL8
A progressive increase of CXCL8 mRNA expression in hMSCs was observed as the extracellular pH was decreased from pH 7.4 to 6.4. Reduction of the medium pH to 7.0 for 24 h after the cells were cultured in HMSCGM for 3 days resulted in a small increase (1.5×) in CXCL8 mRNA levels, whereas after 7 days of culture reducing the medium pH to 7.0 for 24 h resulted in a sixfold increase in CXCL8 expression (Fig. 1A; each graph is normalized to pH 7.4 which is set as 1× for each condition). Changing to more acidic pHs, either pH 6.7 or 6.4, for 24 h resulted in significant increases in expression to 33× and 61×, respectively, at Day 3; and 62× and 183×, respectively at Day 7. This induction at pH 6.7 and 6.4 was also detected when the cells were cultured in OGM for 3 days prior to the 24 h pH treatment, although at a reduced level compared to that seen in HMSCGM (3.5× for pH 6.7, and 4.3× for pH 6.4). No change in CXCL8 expression levels was detected after culturing for 7 days in OGM prior to the 24 h treatment in varying pHs. When CXCL8 levels in OGM are normalized to CXCL8 levels in HMSCGM at each pH, CXCL8 expression was shown to be significantly inhibited at pH 6.7 and 6.4 in OGM, whereas at pH 7.0 no effect on CXCL8 levels in OGM medium were found. At pH 7.4, levels of CXCL8 mRNA were highest in OGM in comparison to HMSCGM medium at both Day 3 and Day 7 (Fig. 1B). CXCL8 mRNA stability was examined using actinomycin D chase experiments after treatment of the cells with pH 7.4 or 6.7 medium for 24 h. For cells cultured in either HMSCGM or OGM, no significant difference in the mRNA decay rates was detected for cells treated with either pH over a 4 h time course (data not shown).
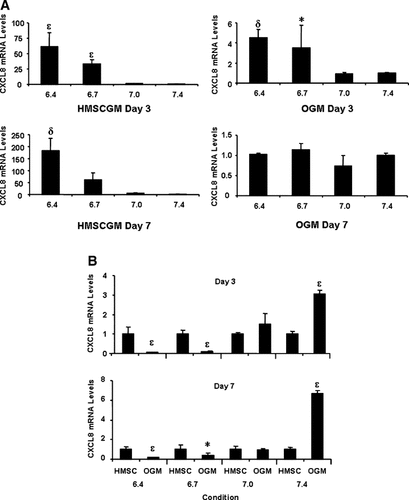
Acidic pH induces expression of CXCL8. hMSC cells were grown in basal growth medium (HMSCGM) or OGM for 3 or 7 days and then treated with medium of decreasing pH for 24 h. Relative levels of CXCL8 mRNA were detected by real-time RT-PCR, normalized to 18S ribosomal RNA levels, and expressed in relation to the reference condition using the 2−((C(T) method. A: Values and significance differences are expressed in relation to pH 7.4 levels (1×) at each day in each medium (HMSCGM or OGM). *P < 0.05, δP < 0.01, εP < 0.001. B: Values and significance differences expressed in relation to HMSCGM levels at each pH (1×). *P < 0.05, εP < 0.001.
There also was no significant change in the pH of the medium over the 24 h treatment time (Table II); although, the pH of the supernatants were slightly more acidic in comparison to the starting medium pH. To verify that the more acidic extracellular pHs did not adversely affect cell viability, cells were isolated after the 24 h treatment period, stained with trypan blue, and quantified for viable or non-viable cells. Cell viability was decreased in the OGM medium in comparison to the HMSCGM medium at both 3 and 7 days and less at 7 days than at 3 days for each condition. Treatment with more acidic pHs did not significantly affect cell viability in comparison to pH 7.4 levels for each of the conditions (Table II).
| Condition | pH treatmenta | pH measuredb | % Cell viabilityc |
|---|---|---|---|
| HMSCGM 3 day | 7.40 | 7.29 ± 0.02 | 84.7 ± 8.1 |
| 7.00 | 6.98 ± 0.03 | 80.0 ± 3.1 | |
| 6.70 | 6.75 ± 0.13 | 80.4 ± 2.6 | |
| 6.40 | 6.26 ± 0.01 | 80.0 ± 7.4 | |
| OGM 3 day | 7.40 | 7.35 ± 0.03 | 74.1 ± 9.2 |
| 7.00 | 6.99 ± 0.02 | 73.4 ± 1.2 | |
| 6.70 | 6.68 ± 0.04 | 74.9 ± 1.9 | |
| 6.40 | 6.37 ± 0.01 | 76.2 ± 0.7 | |
| HMSCGM 7 day | 7.40 | 7.26 ± 0.01 | 81.1 ± 2.7 |
| 7.00 | 6.92 ± 0.01 | 72.5 ± 3.1 | |
| 6.70 | 6.63 ± 0.03 | 72.6 ± 8.9 | |
| 6.40 | 6.28 ± 0.01 | 65.8 ± 11.7 | |
| OGM 7 day | 7.40 | 7.31 ± 0.03 | 62.6 ± 5.6 |
| 7.00 | 6.95 ± 0.03 | 52.0 ± 4.0 | |
| 6.70 | 6.66 ± 0.01 | 64.9 ± 2.3 | |
| 6.40 | 6.35 ± 0.02 | 62.3 ± 14.4 |
- 3-day and 7-day HMSCGM and OGM cultures were treated with medium of different pH for 24 h. Cells were harvested, stained with trypan blue to quantitate for viable and non-viable cells, and the pH of the supernatant measured.
- a Original pH of medium.
- b pH of supernatant after 24 h treatment.
- c Percentage of live cells after 24 h period.
Secreted protein levels in the supernatant were assayed by ELISA (Fig. 2). At Day 3 in both HMSCGM and OGM media, CXCL8 secreted protein levels increased as pH decreased from 7.4 to 6.7. The maximum levels achieved were at pH 6.7 (800–900 pg/ml) and decreased slightly as the pH was further reduced to pH 6.4 (200–450 pg/ml). By Day 7, there was no difference in CXCL8 protein in either HMSCGM or OGM medium at pH 7.4, 7.0, or 6.7; although, there was a significant reduction in CXCL8 protein at pH 6.4 in hMSCs cultured in both HMSCGM and OGM.
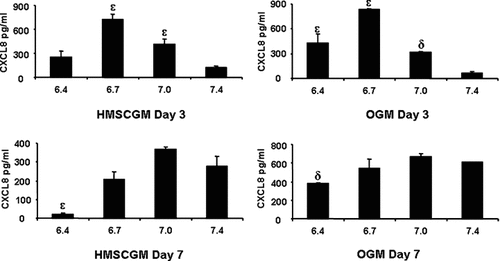
Acidic pH induces secretion of CXCL8 protein. hMSC cells were grown in HMSCGM or OGM medium for 3 or 7 days before treatment with medium of decreasing pH for 24 h. Each experiment was done at least three times in triplicate and CXCL8 protein levels secreted into the medium determined using the human CXCL8/IL-8 DuoSet ELISA Development System from R&D Systems. δP < 0.01, εP < 0.001 indicates values significantly different in comparison to CXCL8 levels at pH 7.4 for each day and condition.
Signal Transduction Pathways
To investigate signal transduction pathways involved in pH-stimulated CXCL8 mRNA expression, various inhibitors were added to the culture media during treatment of the cells with pH 6.7 or 7.4 medium. Treatment of hMSC cells with PTX, an inhibitor of the G-protein inhibitory subunit, Gαi, partially prevented CXCL8 induction in both HMSCGM and OGM media at both pHs (Fig. 3). The suppressive effect was stronger in HMSCGM medium (62% and 50% of the control at pH 6.7 and 7.4, respectively) than in OGM medium (30% and 37% of the control, respectively). In contrast, treatment of the cells with the phospholipase C inhibitor, U73122 did not significantly affect the levels of CXCL8 mRNA in comparison to the negative control U73343 (Fig. 4) suggesting that Gq, G11, and perhaps Gβγ are not involved.
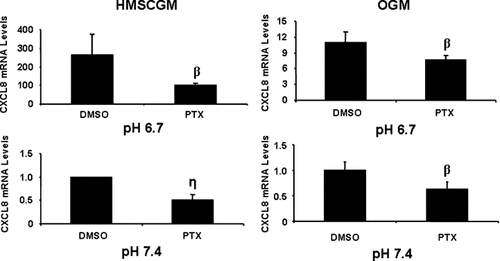
Effect of inhibition of Gαi on acidic pH-induced CXCL8 mRNA levels. hMSC cells were grown in HMSCGM or OGM for 3 days. Gαi signaling was inhibited with PTX (100 ng/ml) for 30 min prior to treatment with pH 6.7 or 7.4 medium containing PTX or the vehicle DMSO for 24 h. CXCL8 levels were determined by real time RT-PCR. βP < 0.02 or ηP < 0.03 indicates significant decreases from mRNA levels in cells treated with DMSO.
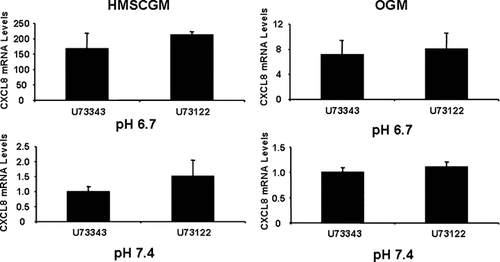
Phospholipase C inhibition has no effect on acidic pH-induced CXCL8 mRNA levels. hMSC cells were grown in basal (HMSCGM) or OGM for 3 days. PLC signaling was suppressed with the specific inhibitor U73122 (100 ng/ml) or the inactive negative control compound U73343 for 30 min prior to treatment with pH 6.7 or 7.4 medium (also containing either U73122 or U73343) for 24 h. CXCL8 levels were determined by real time RT-PCR.
Since Gαi can signal through the MAPK pathway [Gao et al., 2001; Fan et al., 2004] and since pro-inflammatory cytokine-stimulated CXCL8 mRNA expression is known to be mediated by both MAPK and NF-κB [Mukaida et al., 1990; Hwang et al., 2004; Mitsuyama et al., 2004], specific inhibitors of these pathways were also tested. ERK regulation was tested using PD98059, a MEK1 inhibitor, and U0126, a MEK1/MEK2 inhibitor. Both of these MEK inhibitors partially suppressed CXCL8 mRNA expression in comparison to the negative control U0124 (Fig. 5A). For these inhibitors the effect was greatest at pH 6.7 in both HMSCGM and OGM media (70–83% and 94% of the control for HMSCGM and OGM, respectively) than at the normal pH 7.4 (38–45% and 35–57% of the control, respectively).
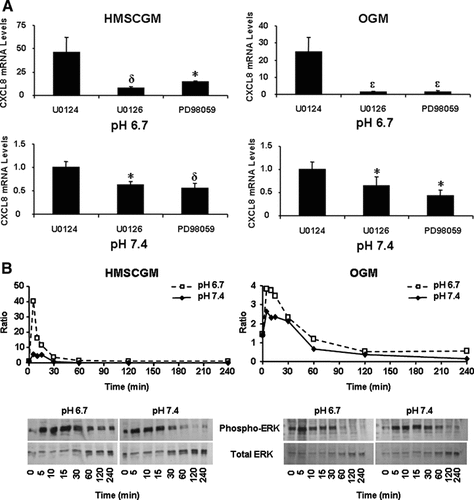
Inhibition of the ERK pathway decreases CXCL8 mRNA levels whereas pH treatment activates ERK phosphorylation. hMSC cells were grown in basal (HMSCGM) or OGM for 3 days. A: ERK signaling was inhibited with the MEK1/MEK2 inhibitor, U0126, or the MEK1 specific inhibitor PD98059 (15 µM) for 30 min prior to treatment with pH 6.7 or 7.4 medium (containing the inhibitors or the negative control U0124) for 24 h. CXCL8 levels were determined by real time RT-PCR. *P < 0.05, δP < 0.01, εP < 0.001 indicates significant decreases in comparison to negative control levels. B: Time course of ERK phosphorylation in response to treatment with pH 6.7 or 7.4 medium. Phospho-ERK1/2 or total ERK was detected by Western blot analysis and then quantitated using the BioQuant software. Values are expressed as a ratio of phospho/total ERK.
Ratios of phosphorylated ERK1/2 to total ERK1/2 were also determined by western blot analysis (Fig. 5B). In HMSCGM, treatment of the cells with pH 6.7 medium resulted in an immediate increase in the level of phosphorylated ERK1/2 (40× that of time 0) peaking 5 min after treatment and then returning to basal levels by 60 min. There is also an increase in pH 7.4-treated cells but to a much smaller level (5×) which may be a consequence of changing the medium at time 0. Cells cultured in OGM also showed increased ERK1/2 phosphorylation upon both pH 6.7 and 7.4 treatment but to a much smaller extent than cells cultured in HMSCGM (4× in OGM vs. 40× in HMSCGM). The peak in ERK1/2 phosphorylation levels in OGM medium was at 5 min and decreases to below basal levels by 120 min. A smaller increase (2.6×) in pH 7.4 medium is also detected by 5 min of treatment.
Inhibition of the p38 MAPK pathway with SB202190 or SB203580 also resulted in decreased levels of CXCL8 mRNA in comparison to the negative control, SB202474 (Fig. 6). Similar levels of CXCL8 suppression were detected independent of either pH or medium type (62–85% of the control). JNK inhibition did not significantly affect CXCL8 mRNA expression under any conditions tested (Fig. 7). Simultaneous addition of both ERK and p38 inhibitors (Fig. 8) resulted in an additive effect with even further decreased levels of CXCL8 mRNA (60–83% decrease from control levels in HMSCGM and 77–83% decrease in OGM) than when either pathway was inhibited separately (24–60% decrease in HMSCGM and 51–67% in OGM).
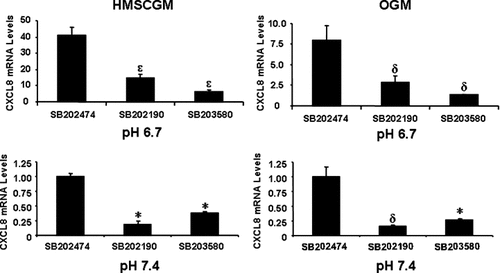
Inhibition of the p38 MAPK pathway suppresses CXCL8 mRNA levels. hMSC cells grown in basal (HMSCGM) or OGM for 3 days were treated with p38 signaling inhibitors SB202190 or SB20358 or the negative control compound SB202474 (15 µM) for 30 min. The cells were then subjected to pH 6.7 or 7.4 medium (also containing the inhibitors or negative control) for 24 h and the CXCL8 levels determined by real time RT-PCR. *P < 0.05, δP < 0.01, εP < 0.001 indicates a significant decrease from levels in the negative control.
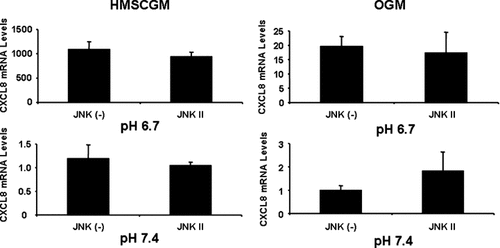
JNK inhibition has no effect on acidic pH-induced CXCL8 mRNA levels. hMSC cells were grown in basal HMSCGM or OGM for 3 days. JNK signaling was inhibited with the JNK II specific inhibitor or the JNK(−) negative control (10 µM) for 30 min prior to treatment with pH 6.7 or 7.4 medium (containing the inhibitor or negative) for 24 h. CXCL8 levels were determined by real time RT-PCR.
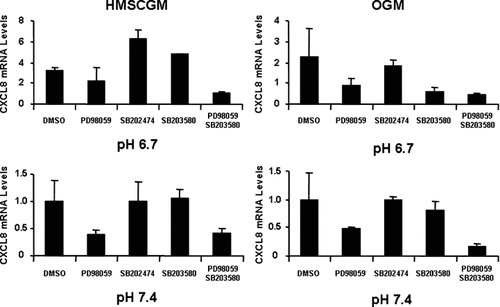
Dual inhibition of the ERK and p38 MAPK pathways further suppresses CXCL8 mRNA levels. hMSC cells grown under basal (HMSCGM) or osteogenic conditions (OGM) for 3 days were treated with the ERK inhibitor PD98059 or the p38 inhibitor SB203580 (15 µM each) either individually or in combination for 30 min prior to pH stimulation (pH 6.7 or 7.4) for 24 h. CXCL8 mRNA levels were determined by real time RT-PCR and compared to levels in the negative controls (DMSO for ERK, SB202474 for p38 both at 15 µM).
NF-κB inhibition by addition of a pyrrolidine derivative of dithiocarbamate (PDTC) resulted in decreased levels of CXCL8 mRNA in HMSCGM medium at pH 6.7 (73% suppression) and at pH 7.4 (44% decrease). In OGM medium, treatment with PDTC resulted in an increase in CXCL8 mRNA levels (4× at pH 6.7 and 2× at pH 7.4) at both pHs (Fig. 9). Several other NF-κB inhibitors were also tested to confirm the differential effects of NF-κB inhibition in HMSCGM versus OGM medium (data not shown). Inhibition of NF-κB with QNZ, MG-132, CAPE, and TMB-8 gave similar results as PDTC with increased levels of CXCL8 mRNA in OGM medium (at both pHs) and decreased levels in HMSCGM at pH 6.7; although, they did not consistently show significant effects on CXCL8 mRNA levels in HMSCGM medium at pH 7.4 (data not shown). Combined use of both the NF-κB inhibitor PDTC and the ERK inhibitor U0126 or the p38 inhibitor SB203580 in hMSCs (Fig. 10) resulted in additional suppression of the CXCL8 mRNA levels from that of the controls or individual inhibitors (84–99% decrease in HMSCGM compared to DMSO vehicle levels in HMSCGM at pH 6.7, 61–98% decrease from PDTC stimulation levels in OGM at either pH 6.7 or 7.4).
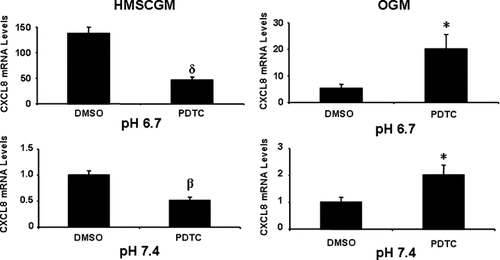
Inhibition of the NF-κB suppresses CXCL8 mRNA levels in multipotent medium (HMSCGM) but increases CXCL8 levels in OGM. hMSC cells grown in basal (HMSCGM) or OGM for 3 days were treated with NF-κB signaling inhibitor pyrrolidine dithiocarbamate (PDTC) at 100 µM or the vehicle (DMSO) for 30 min. The cells were then subjected to pH 6.7 or 7.4 medium (also containing the inhibitor or DMSO) for 24 h and the CXCL8 levels determined by real time RT-PCR. *P < 0.05, βP < 0.02, δP < 0.01 indicates significant difference in comparison to the vehicle.
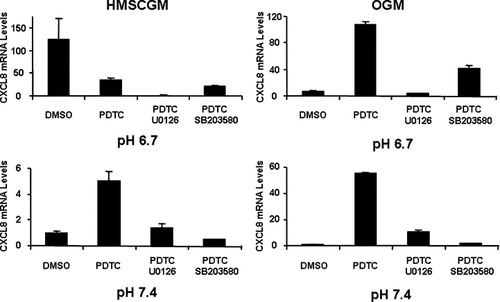
Inhibition of the NF-κB pathway in combination with ERK and p38 MAPK inhibition further suppresses CXCL8 mRNA levels. hMSC cells were grown under basal (HMSCGM) or osteogenic conditions (OGM) for 3 days, treated with the NF-κB inhibitor PDTC in combination with the MAPK ERK inhibitor PD98059 (15 µM) or the p38 inhibitor SB203580 (15 µM) for 30 min, and then subjected to pH stimulation (pH 6.7 or 7.4) for 24 h. CXCL8 mRNA levels were determined by real time RT-PCR and compared to levels in the negative control (DMSO at 15 µM).
DISCUSSION
Acidic pH can be found in tissues that have compromised blood supply when oxygen consumption exceeds its supply or as a result of trauma. Studies using 31P-magnetic resonance spectroscopy have determined that intercellular muscle pH can be decreased to 6.5 within 5–6 min under conditions of high frequency exercise [Roussel et al., 2003]. In rat brain, ischemia trauma resulted in both an intracellular and interstitial pH drop to 6.4 within 5 min that remained at that level for up to 12 min [Kintner et al., 2000]. Therefore, even though systemic pH of 7.0 or below is generally only seen clinically with multi-organ failure with profound hypoperfusion, locally tissue and intracellular pH can reach even lower levels. Hence in bone trauma, it is conceivable that intracellular and interstitial fluid pHs in the immediate area of blood supply disruption could have pHs down to 6.4–6.5. Local pH in bone fractures in a rat crush fracture model was determined in vitro to be approximately 7.2 from fluid aspirated from fracture sites 2 days post-fracture [Newman et al., 1987]. However, it is possible that the pH at the fracture site may be even lower a few hours just post-fracture and the collection of fracture site fluid could have resulted in loss of CO2 thus falsely elevating pH determined by this process.
Production of CXCL8 by mature or progenitor bone cells has been reported in osteoclasts [Rothe et al., 1998], in subchondral osteoblasts and bone marrow stromal cells in osteoarthritis and rheumatoid arthritis [Lisignoli et al., 1999, 2002], and in mesenchymal stem cells under non-pathologic conditions in culture [Majumdar et al., 2000; Kim et al., 2005]. However, the significance of CXCL8 expression especially by mesenchymal stem cells has not been explored or related to its potential utility in bone repair. CXCL8 expression in breast and prostate tumors that have metastasized to bone has also been described [Bendre et al., 2002; Lehrer et al., 2004; Guise et al., 2005]. Upregulation of CXCL8 expression by acidic pH has been previously described in both solid tumors in vivo [Shi et al., 2000, 2001; Xu and Fidler, 2000; Karashima et al., 2003] and in vitro cerebellar cell cultures [Watanabe et al., 1998]. In this manuscript, we present a novel observation that hMSCs can be stimulated by an acidic pH to express CXCL8 mRNA in both the multipotent state and when differentiated towards the osteogenic lineage. Since osteogenic differentiation results in expression of CXCL8 beginning at 5 days [Bischoff et al., 2007], we looked at early (3 day) and late (7 day) cultures to differentiate between CXCL8 expression due to osteogenic differentiation from that of the pH effects. Our findings demonstrate that there is a pH-dependent increase in CXCL8 mRNA expression in hMSCs cultures with the greatest stimulation at an acidic pH of 6.4.
CXCL8 protein secretion increases as pH decreases from 7.4 to 6.7 for a 24 h period in 3-day cultures and then drops off from pH 6.7 levels as the medium becomes more acidic. It is possible that the protein production or half-life is adversely affected by the acidic pH (acid hydrolysis of proteins) whereas the mRNA expression or stability is not affected at lower pH leading to an overall increased mRNA steady state level but decreased protein secretion into the medium. This has been demonstrated in rat calvarial osteoblasts where protein production of vascular endothelial growth factor (VEGF) is decreased with decreasing pH whereas the rate of hypoxia-stimulated VEGF mRNA decay is not affected by incubation in acidic pH [Spector et al., 2001]. Another possible explanation is that the CXCL8 that is secreted from the cells at the more acidic pH 6.4 is sequestered in the extracellular matrix (ECM) and is therefore not accessible for detection in the supernatant by ELISA. This phenomenon has been suggested for both bFGF and VEGF mRNA induction and bFGF secretion from bovine aortic endothelial cells upon treatment with acidic pH [D'Arcangelo et al., 2000]. Despite the increased expression of these angiogenic growth factors, acidic treatment inhibits endothelial cell proliferation and new blood vessel formation. These growth factors may be sequestered in the ECM at the lower pH and therefore only readily available for increased angiogenesis upon return to more normal pH conditions.
CXCL8 is an ELR+ CXC chemokine that is angiogenic [Strieter et al., 1995]. It binds to both CXC receptor 1 (CXCR1) and CXC receptor 2 (CXCR2); however, CXCL8 angiogenesis has only been correlated with the CXCR2 [Addison et al., 2000; Heidemann et al., 2003]. Knockout models of the CXCR2 equivalent in mice show that wound healing is protracted with decreased cellularity of the granulation tissue and neovascularization [Devalaraja et al., 2000]. Although not yet demonstrated in bone repair, it is possible that the inability of ELR+ CXC chemokines to interact with the CXCR2 could lead to inadequate callus formation during endochondral bone repair due to a lack of sufficient angiogenesis in condensed mesenchyme that would normally differentiate into osteoblasts during intramembranous bone repair. Preliminary experiments in which hMSCs were treated with acidic pH and then switched back to normal pH did not cause differentiation of the cells towards the osteoblastic lineage (data not shown), suggesting that the pH-induction of CXCL8 may have a more prominent role in angiogenesis than differentiation. Although, other factors such as the levels of the CXC receptors (CXCR1 and CXCR2) in cultured hMSCs may directly affect the ability of the expressed CXCL8 to stimulate differentiation of these cells.
CXCR2 is a seven transmembrane spanning G-protein coupled receptor [Rojo et al., 1999]. Several G-protein α subunits have been described that are coupled to the CXCR2 [Murdoch and Finn, 2000; Mellado et al., 2001; Limatola et al., 2002; Brat et al., 2005]. These include Gαi and Gq/11. Chemokine signaling through Gαi can explain the majority of chemokine responses found traditionally in inflammatory cells that can be inhibited by PTX [Damaj et al., 1996]. However, incomplete blockage by PTX of the calcium mobilization response generally used to test CXC chemokine activity, has led to searches for other G-protein subunits such as Gq/11 capable of eliciting an intracellular calcium response independent of Gαi [Hall et al., 1999]. The pairing of specific G-proteins with CXC receptors in general may be cell type-specific, and it is possible that more than one type of G-protein could be coupled to a specific CXC receptor explaining different physiologic functions induced by specific CXC chemokines in a given cell.
CXCR2 has been shown to be variably expressed in hMSCs [Von Luttichau et al., 2005; Honczarenko et al., 2006]; although, specific α subunits coupled to the CXCR2 in hMSCs have not been determined. Activation of Gαi can lead to downstream stimulation of the MAPK pathway while the Gβγ subunit can activate phospholipase C leading to intracellular calcium signaling and activation of protein kinase C. Additionally, the phosphoinositide-3-kinase (PI3K) pathway can also be activated by CXC chemokines [Knall et al., 1997]. It has been shown that the Gβγ subunit regulates activation of PI3K and that the Gβγ subunits originate from dissociation from either Gαi or Gαq [Krugmann et al., 2002; Brock et al., 2003; Goel et al., 2004]. Stimulation of Gαq can lead to activation of phospholipase Cβ which can activate intracellular calcium and protein kinase C signaling. We have shown that inhibition of Gαi results in a decrease in low pH-stimulated CXCL8 mRNA expression in hMSCs. However, the phospholipase C inhibitor did not alter CXCL8 mRNA expression suggesting that neither Gq/11 nor perhaps the Gβγ subunits may be involved. The involvement of the PI3K pathway in acidic pH-stimulated CXCL8 expression was not tested.
Inhibition of the ERK MAPK pathway suppressed CXCL8 mRNA levels in acidic pH to a greater extent than for p38 MAPK inhibition when the hMSCs are cultured in OGM. This suggests that the dominant pathway for CXCL8 mRNA expression by hMSCs during differentiation with OGM may be via the ERK MAPK pathway. Our results indicate that treatment with pH 6.7 medium results in rapid activation (phosphorylation) of ERK. Involvement of the ERK pathway has also been demonstrated in acid-induced expression of VEGF, an angiogenic stimulator, from glioblastoma cells [Xu et al., 2002]. Comparison of expression levels of CXCL8 in both HMSCGM and OGM may suggest that the basal level of CXCL8 expression at pH 7.4 is controlled more by NF-κB and p38 than by NF-κB and ERK since dual use of NF-κB and p38 inhibitors results in greater inhibition than dual use of NF-kB and ERK inhibitors (Fig. 10). Whereas in acidic pH, dual use of NF-κB and ERK inhibitors suppresses CXCL8 expression to a greater extent suggesting NF-κB and ERK have a more prominent role in CXCL8 expression at acidic pHs (Fig. 10).
Depending on cell type and conditions of stimulation, CXCL8 mRNA expression can be regulated by transcriptional or post-transcriptional means [for review see Hoffmann et al., 2002]. For instance, p38 MAPK and ERK activation have been shown to stabilize CXCL8 mRNA in response to tumor necrosis factor-α (TNF-α) stimulation in an intestinal epithelial cell line, HT-29 [Jijon et al., 2002]. However, in Caco-2, another intestinal epithelial cell model, p38 MAPK stimulation by interleukin-1β (IL-1β) directly activated the CXCL8 promoter to increase gene transcription [Parhar et al., 2003]. In human vascular smooth muscle cells CXCL8 mRNA gene transcription was induced by IL-1β in a p38 MAPK-dependent manner [Jung et al., 2002]; whereas, in the human promyelocytic cell line, HL-60, insulin-like growth factor-1 (IGF-1) stimulated CXCL8 mRNA expression via activation of ERK and JNK [Kooijman et al., 2003]. IGF-2 stimulation also increased CXCL8 mRNA in human keratinocytes via the activation of ERK but not p38 MAPK [Kim et al., 2004].
Activation of the transcription factor NF-κB has also been demonstrated to stimulate CXCL8 mRNA expression in HeLa cells [Hoffmann et al., 2002]. In human bronchial epithelial cells, NF-κB regulation of CXCL8 transcription was observed to be dependent on JNK [Li et al., 2002]. However, the lack of inhibition with the JNK inhibitor suggests that this pathway is not involved in pH-stimulated CXCL8 expression in hMSCs. Acidosis has been shown to activate NF-κB leading to CXCL8 expression in several cancer cell lines [Xu and Fidler, 2000; Shi et al., 2001; Karashima et al., 2003]. Dexamethasone is the medium component that is required for CXCL8 expression during differentiation of hMSCs towards the osteoblastic linage [Bischoff et al., 2007]. Stimulation of CXCL8 by the pro-inflammatory cytokines IL-1β and TNF-α has been shown to be regulated in large part through the NF-κB pathway. This pathway is sensitive to inhibition by glucocorticoids [McKay and Cidlowski, 1999; Bosscher et al., 2003], and we have previously shown that the IL-1β/TNF-α-stimulated CXCL8 expression is partially suppressed by DEX in hMSCs [Bischoff et al., 2007]. Thus, there appears to be a DEX-stimulated CXCL8 pathway either independent of the NF-κB signaling pathway involved in pro-inflammatory cytokine stimulation of CXCL8 or overcome when NF-κB-mediated CXCL8 production is induced during hMSC differentiation.
In this report, we have shown that inhibition of NF-κB decreases CXCL8 mRNA expression following acidic pH stimulation when the cells are cultured in HMSCGM. The observation that in OGM medium at both pH 6.7 and 7.4 addition of the NF-κB inhibitor, PDTC, more than doubled the relative expression of CXCL8 mRNA was a unique finding. We tested several NF-κB inhibitors with different mechanisms such as interfering with NF-κB activation (PDTC), inhibition of IκB phosphorylation (BAY11-7082), inhibition of proteasomal degradation (MG132), inhibition of NF-κB translocation but not IκB phosphorylation through an antioxidant mechanism (CAPE), an intracellular Ca2+ antagonist (TMB-8), and one with an unknown mode of action (QNZ). All of these inhibitors lead to increased expression of CXCL8 in the presence of OGM. In general, NF-κB has been demonstrated to bind to the promoter region of the CXCL8 gene to stimulate the transcription of CXCL8 [Hoffmann et al., 2002; Bobrovnikova-Marjon et al., 2004]. In this specific instance of CXCL8 mRNA expression in OGM medium, it can be hypothesized that NF-κB could be repressing either the transcription of CXCL8 mRNA or decreasing the stability of the CXCL8 mRNA directly. Alternatively, NF-κB could be inhibiting the action of ERK or p38 MAPK pathways in OGM. The mechanism of NF-κB inhibition in OGM-stimulated CXCL8 regulation is currently under investigation.
In summary, we have shown that the angiogenic chemokine CXCL8 is induced in both multipotent and OGM-treated hMSCs in response to decreasing extracellular pH through Gαi, ERK, and p38 MAPK signaling pathways. NF-κB activation is also involved as a signaling molecule in CXCL8 stimulation under acidic conditions in mutipotent hMSC cells grown in HMSCGM medium. However, it appears that the NF-κB pathway represses CXCL8 expression when the cells are differentiated towards the osteogenic lineage with OGM treatment.




