A chromatin immunoprecipitation screen in mouse keratinocytes reveals Runx1 as a direct transcriptional target of ΔNp63
Abstract
Development of the skin epidermis and appendages such as hair follicles involves coordinated processes of keratinocyte proliferation and differentiation. The transcription factor p63 plays a critical role in these steps as evident by a complete lack of these structures in p63 null mice. The p63 gene encodes for two proteins TAp63 and ΔNp63, the latter being the more prevalent and dominant isoform expressed in keratinocytes. Although numerous p63 target genes have been identified, these studies have been limited to transformed human keratinocyte cell lines. Here, we have employed a genomic screening approach of chromatin immunoprecipitation (ChIP) coupled with an enrichment strategy to identify ΔNp63 response elements in primary mouse keratinocytes. Analysis of p63-ChIP-derived DNA segments has revealed more than 100 potential target genes including novel as well as mouse counterparts of established human p63 targets. Among these is Runx1, a transcription factor important for hair follicle development. We demonstrate that ΔNp63 binds to a p63-response element located within a well-conserved enhancer of the Runx1 gene. Furthermore, siRNA mediated reduction of ΔNp63 in mouse keratinocytes reduces Runx1 expression. Consistent with this, endogenous Runx1 levels are lower in the skin of p63+/− animals as compared to wild type animals. Lastly, we demonstrate that ΔNp63 and Runx1 are co-expressed in specific compartments of the hair follicle in a dynamic fashion. Taken together our data demonstrate that p63 directly regulates Runx1 gene expression through a novel enhancer element and suggests a role for these two transcription factors in dictating skin keratinocyte and appendage development. J. Cell. Biochem. 104: 1204–1219, 2008. © 2008 Wiley-Liss, Inc.
Abbreviations used:
ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; HPRT, hypoxanthine guanine phosphoribosyl transferase; K14, Keratin14; ORS, outer root sheath; Swg, sweat gland; TK, thymidine kinase; PMK, primary mouse keratinocytes.
The p53 family of transcription factors includes p73 and p63 each of which share some degree of structural and functional similarities to the founding member p53 [Yang et al., 2002]. There are, however, unique physiological roles attributed to each factor that do not involve canonical p53-linked pathways such as growth arrest and apoptosis, but rather developmental decisions. This is particularly evident in the case of p63, which is highly and selectively expressed in epithelial cells, where it is thought to play important roles in stem cell self-renewal, morphogenesis and differentiation programs [Koster and Roop, 2004; Barbieri and Pietenpol, 2006; Blanpain and Fuchs, 2007; King and Weinberg, 2007]. The critical role of p63 in regulating various facets of epithelial cell biology is prominently displayed in the striking phenotype of p63 null mice [Mills et al., 1999; Yang et al., 1999]. In the absence of p63, animals do not survive and exhibit a dramatic loss of epithelial structures such as the epidermis and multiple epithelial appendages including teeth, mammary glands, hair follicles, etc.
Our understanding of the molecular mechanisms that underlie p63 function in developmental pathways has been hampered partly by the complexity of the p63 isoforms. The p63 isoforms are derived from multiple transcripts generated from the p63 gene by alternate promoter usage and differential splicing [Yang et al., 1998]. Transcripts (TAp63) that encode for a longer version include a transactivation domain and are generated from a proximal promoter. On the other hand, an internal promoter located within intron 3 directs expression of transcripts that lack this domain (ΔNp63). In addition, alternative splicing results in α, β and γ isoforms of p63 that are unique at their C-terminus. Despite these differences, all six versions of the p63 protein share common DNA-binding and oligomerization domains, which are analogous to that of p53.
Since the ΔNp63 isoforms lack the amino-terminal sequences, it was originally proposed that these proteins might act primarily as dominant negative regulators. However recent studies indicate that ΔNp63 proteins have intrinsic biological activity and can transcriptionally activate or repress target gene expression [King and Weinberg, 2007]. In addition, it is clear that ΔNp63 is the predominant isoform expressed in most epithelial cells. In contrast, the expression profile of TAp63 remains mired in controversy with conflicting results from different laboratories [Mikkola, 2007]. Although both ΔNp63 and TAp63 show similar biochemical properties, in vivo studies have begun to address the unique and independent functions of these proteins [Koster et al., 2004; Suh et al., 2006]. For example, siRNA mediated knockdown of ΔNp63 in the skin of transgenic mice has demonstrated that ΔNp63 proteins are essential for maintaining basement membrane integrity and are required for epidermal morphogenesis and homeostasis [Koster et al., 2007]. Similarly, another study has shown that the skin phenotype of the p63 null animals can be partially rescued by expressing ΔNp63 but not TAp63 [Candi et al., 2006]. Taken together, these data strongly suggest a dominant role for ΔNp63 during the epithelial development and differentiation program.
The primary role of p63 involves regulation of a wide cohort of target genes at the transcriptional level. Despite considerable sequence conservation between the DNA-binding domains of p63 and p53, it has become apparent that there are distinct target genes of p63 independent from those of p53 [Vigano and Mantovani, 2007]. Most of the p63 target genes has been identified by global microarray profiling and by ChIP-Chip experiments [Barbieri et al., 2006; Carroll et al., 2006; Vigano et al., 2006; Yang et al., 2006]. In keeping with the complex biological role of p63, these studies have unearthed a fascinating and diverse range of p63 target genes that are involved in a myriad of cellular processes. The discovery of genomic targets of p63, however, remains far from complete since each search strategy has inherent limitations. What is even more remarkable is the fact that to date none of the p63-target discovery experiments have been performed using mouse models, which have been a steadfast resource for functional studies of p63 over the past several years. This is important and relevant in view of growing evidence for species-specific differences in binding sites for highly conserved transcription factors between human and mouse [Odom et al., 2007]. Thus, continued search for p63 targets is warranted.
We have recently developed and utilized a modified ChIP approach to successfully identify target genes of ΔNp63 in HaCaT cells [Birkaya et al., 2007]. This modified ChIP procedure entails amplification of the immunoprecipitated DNA by ligation-mediated PCR technique followed by selective enrichment. Here, we have taken a similar approach to identify target genes of ΔNp63 in primary mouse keratinocytes. Our screening approach revealed over 100 potential ΔNp63-response elements associated with a variety of target genes. Although some of the genes we identified as potential ΔNp63 targets turned out to be the mouse counterpart of human genes previously identified, several novel candidates also emerged. Finally, as a proof of principle to demonstrate the validity of our approach and to further elucidate the role of ΔNp63 in keratinocytes, we chose to focus our studies on another transcription factor, Runx1, which emerged as a direct target gene of ΔNp63.
Runx1 belongs to a small family of transcriptional regulators (Runx1-3 in mammals), which are characterized by a highly conserved DNA-binding domain named after the Drosophila Runt gene and are evolutionary conserved in all metazoans [Levanon and Groner, 2004]. These factors play roles in proliferation versus differentiation decisions in various tissues, in stem cell maintenance and in organogenesis functioning as activators or repressors of target genes [Levanon and Groner, 2004; Mikhail et al., 2006]. In skin, Runx1 is prominent in several of the epithelial cell layers of the hair follicles and in the bulge, which is the stem cell niche for these appendages [Raveh et al., 2006].
We show that ΔNp63 binds to a conserved intronic enhancer present in the Runx1 gene and that the expression level of Runx1 is affected by the levels of ΔNp63 in keratinocytes in culture as well as in mouse skin. Our study establishes that ΔNp63 and Runx1 are co-expressed in specific cell types within various skin appendages, such as hair follicles and sweat glands suggesting that these two transcription factors might be interlinked in regulating the expression of downstream genes important for development of these cell types. Collectively our ChIP approach has identified numerous mouse target genes of ΔNp63 and has offered insight into the regulatory pathways controlled by p63 during skin morphogenesis.
MATERIALS AND METHODS
Cell Culture
The mouse keratinocytes utilized in these studies have been well characterized and described before [Romano et al., 2006]. Keratinocytes were maintained in low calcium media supplemented with 10% fetal bovine serum and 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were routinely passaged at 90% confluency. PTK2 (rat kangaroo kidney epithelial) cells were grown in minimal essential medium supplemented with 10% fetal bovine serum, 1% MEM non-essential amino acid solution, 100 U/ml penicillin and 100 µg/ml streptomycin.
Chromatin Immunoprecipitation (ChIP) Assays for Cloning of Immunoprecipitated Products and Confirmation of p63-Binding to Runx1 Enhancer
Chromatin immunoprecipitation (ChIP) experiments were performed with anti-p63 antibodies using mouse keratinocytes. Cells were grown to 80% confluency and then crosslinked with 1% formaldehyde (Sigma) at 37°C for 10 min, rinsed two times with cold PBS, and then harvested in PBS with protease inhibitors by centrifugation for 5 min at 2,000g. The cloning of DNA fragments subsequent to the ChIP procedure was performed as previously described [Birkaya et al., 2007]. For this purpose immunoprecipitated DNA was amplified by PCR after addition of a linker element. Linker DNA was removed by digestion with Hind III restriction enzyme and purified with PCR purification kit (Qiagen) and cloned into pBluescript vector. Plasmid DNAs that contained inserts that were at least 100 bp were chosen for sequencing at the Roswell Park Sequencing Facility. For in vivo ChIP, newborn mouse skin samples were isolated and treated with Dispase II (Roche) overnight at 4°C. The epidermis was separated, finely chopped and washed with PBS followed by crosslinking with 1% formaldehyde in PBS at room temperature for 15 min. To stop crosslinking, 0.125 M glycine was added for 3–5 min followed by washes with PBS and standard ChIP protocols were followed. Binding of ΔNp63 to Runx1 enhancer was evaluated by independent ChIP assay utilizing immunoprecipitated DNA from two different anti-p63 antibodies, H-129 (Santa Cruz) and RR-14. Primers used are listed in Supplemental Table 2. Real time PCR conditions are described for siRNA knockdown of p63.
In Silico Data Analysis
The clones obtained from the ChIPed DNA cloning were sequenced and compared against mouse genome database by BLAST search of various database including UCSC genome browser (http://genome.ucsc.edu/), ENSEMBL (http://www.ensembl.org/index.html), and NCBI (http://www.ncbi.nlm.nih.gov/blast/) to identify potential target genes. Gene Ontology from NCBI was used to identify the function of the identified genes. We utilized the GenomeVISTA (http://pipeline.lbl.gov/cgi-bin/genomevista) program to obtain the ortholog sequences from multiple species and generate the VISTA plot. Multiple sequence alignment was done using CLUSTALW (http:www.ebi.ac.uk/clustalw). The program Transcription element search system (TESS; http://www.cbil.upenn.edu/cgi-bin/tess/tess) was used to identify putative transcription factor binding sites located within the conserved DNA sequence of the Runx1 enhancer.
Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assay (EMSA)
Mouse keratinocytes were grown as described above and nuclear extracts prepared by standard methods as described before [Romano et al., 2006]. EMSAs were performed with 5 µg of nuclear extract and end-labeled double-stranded oligonucleotides. Oligonucleotide sequences are listed in Supplemental Table 2. For competition assays increasing amounts of unlabeled oligonucleotides were incubated with nuclear extract and labeled oligonucleotides. Protein–DNA complexes were resolved by gel electrophoresis on 5% non-denaturing polyacrylamide gels in 1× TBE buffer at room temperature. The gels were dried and visualized by autoradiography after electrophoresis. Anti-p63 antibodies used for supershift experiments have been previously described [Romano et al., 2007].
Plasmids
Recombinant plasmids were constructed using standard molecular biology protocols. ChIP-DNA sequence corresponding to Runx1 enhancer was PCR amplified and cloned into KpnI and NheI sites of the pGL3-basic vector (Promega) containing the thymidine kinase (TK) minimal promoter. Mutations were introduced by PCR amplification. All constructs were verified by sequencing.
Transient Transfections and Reporter Assays
PTK2 cells were seeded in 6-well plates the day before transfection. Transfections were performed using Fugene 6 reagent (Roche) according to the manufacturer's protocol. One microgram of each luciferase reporter construct was transfected per well along with 0.25 µg of pCMVLacZ plasmid to serve as an internal control for transfection efficiency. Twenty-four hours posttransfection cell lysates were prepared by rinsing cells two times in PBS followed by extracting proteins in 200 µl Lysis Buffer (Promega). Each well was scraped and collected into microfuge tubes, vortexed and the debris was pelleted by centrifugation at 4°C. Supernatant solutions were subsequently used in β-galactosidase (Galacto-Light, Applied Biosystem) and luciferase activity assay systems (Promega) following the manufacturer's protocols using a Lumat LB 9501 Luminometer (Berthold). Means and standard deviations were calculated based on data from three independent transfection experiments.
Reverse Transcriptase-PCR
Total RNA from mouse keratinocytes was isolated and purified by Trizol (Invitrogen) according to established protocols. Two micrograms of total RNA from mouse keratinocytes, mouse skin, and spleen was reverse transcribed with the Thermoscript cDNA synthesis kit (Invitrogen). Primer sequences used are listed in Supplemental Table 2. Jump Start Taq Polymerase (Sigma–Aldrich) was used for PCR amplifications.
siRNA Knockdown of p63 in Mouse Keratinocytes
Mouse keratinocytes were seeded 24 h prior to transfection in 6-well plates. Transfections were performed with cells at 30–40% confluency with 10 nM Trp63 siRNA, 10 nM Gapdh siRNA, and 10 nM Negative siRNA (Ambion) using Lipofectamine 2000 (Invitrogen). Cells were collected 72 h after transfection and analyzed for knockdown of p63 by Western blot analysis and quantitative PCR. Anti-p63 antibodies(RR-14) were used for detection of ΔNp63 protein levels and the membrane was stripped and re-probed with antibodies against β-tubulin to confirm equal loading. For estimating transcript levels, RNA was extracted from p63, Gapdh, and negative siRNA transfected mouse keratinocytes and reverse transcribed with the Thermoscript cDNA synthesis kit (Invitrogen). Quantitative PCR was performed with 1 µl of 1:5 diluted cDNA, 10 µM of each primer and the suggested amount of SYBER Green I dye (Bio-Rad) according to the manufacturer's instructions. Cycling parameters were as follows 95°C for 8 min followed by 35 cycles at 95°C for 15 s and 60°C for 1 min. Fluorescent data were normalized to the reference gene β-2-microglobulin. Primer sequences used are listed in Supplemental Table 2. All reactions were repeated at least three times.
RNA Isolation From Wild Type and p63+/− Skin
The p63 heterozygous animals were obtained from Jackson Laboratory (strain B6.129S7-Trp63tm1Brd/J) and were mated to generate wild type and heterozygote mutant mice. Animals were genotyped using PCR as described [Mills et al., 1999]. Skin from E18.5 wild type and p63+/− embryos was isolated and processed for RNA isolation using Trizol (Invitrogen) according to established protocols. Two micrograms of total RNA from mouse skin was reversed transcribed with the Thermoscript cDNA synthesis kit (Invitrogen). Real-time PCR was performed as described before.
X-Gal and Immunohistochemical Staining of the Runx1+/lz Animals
The Runx1+/lz mice utilized in the localization studies have been described before [Raveh et al., 2006]. Skin sections (8 µm) were fixed in paraformaldehyde (4% in PBS, 10 min) washed five times in PBS, and incubated in X-gal staining solution at room temperature overnight. After serial washings, sections were treated with 1.5% hydrogen peroxide for blocking endogenous peroxidase activity and incubated for 30 min with blocking solution containing 0.05% Triton X-100, 2.5% normal goat serum (NGS), 1% BSA and 0.25% glycine in PBS prior to 2 h incubation with RR-14 primary antibodies, diluted 1:200 in 50% blocker solution. Biotinylated anti-rabbit secondary antibodies were applied diluted in 1.5% NGS PBS for 1 h and detected by the avidin-biotin peroxidase technique (ABC, Vectastain, Vector laboratories), using DAB reagent (Sigma). Stained sections were observed and photographed using an Olympus BX51 microscope and a Magnafire SP digital camera. Confocal images were taken on a Bio-Rad MRC-1024 confocal microscope.
RESULTS
An Improved ChIP Approach to Identify ΔNp63-Response Elements in Primary Mouse Keratinocytes
Identifying the target genes of ΔNp63 will better elucidate the role of this transcription factor in keratinocyte development and differentiation. One approach to identify these target genes is to locate the specific genomic region to which ΔNp63 is bound in vivo since these regions are likely to be cis regulatory elements controlling the expression of these targets. Such prior studies for p63 have been limited by the fact that these experiments utilized transformed cell lines of human origin and antibodies that recognize both ΔNp63 and TAp63. To overcome such caveats, we utilized a ChIP-based screening strategy with primary mouse keratinocytes, which are known to express high levels of ΔNp63. Furthermore, to specifically probe the ΔNp63 proteins, we used the RR-14 antibody, which has been developed in our laboratory and is directed against an N-terminal peptide unique to this isoform. We have utilized this antibody successfully in prior ChIP experiments in both human and mouse cells [Birkaya et al., 2007; Romano et al., 2007]. Primary mouse keratinocytes were grown in a medium containing low concentration of Ca++ to ensure that the cells do not differentiate and stratify. The keratinocytes were processed for ChIP experiments when they were in active state of proliferation (∼80% confluent), since at this stage these cells contain the highest levels of ΔNp63 (unpublished data). The strategy to enrich, clone and sequence the immunoprecipitated DNA has been described in detail before [Birkaya et al., 2007]. Briefly, we utilized a ligation-mediated PCR technique, to amplify the genomic immunoprecipitated DNA, which was then enriched and purified by incubation with agarose beads containing GST-ΔNp63 protein to reduce the contamination with non-specific DNA fragments. This enrichment procedure allowed us to select for DNA fragments that are more likely to contain binding-sites for ΔNp63. Following immunoprecipitation and purification, the DNA fragments were cloned into the pBluescript vector. The average size of the DNA fragment obtained after the ChIP procedure was ∼250 bp. We sequenced 280 individual clones that represented the library of DNA fragments immunoprecipitated by ΔNp63.
The p63-ChIPed Sequences Correspond to Known and Novel In Vivo Target Genes of ΔNp63 in Primary Mouse Keratinocytes
We analyzed the sequences for every retrieved ChIP fragment by mapping them to the mouse genome database by using a variety of search programs of University of California Santa Cruz genome browser, ENSEMBL, or the BLAST program at NCBI. This allowed us to determine the location of the ΔNp63 immunoprecipitated DNA fragments in relation to known or predicted genes. This study revealed that 109 DNA fragments immunoprecipitated by ΔNp63 antibodies were embedded within or located near known, annotated, or predicted genes. A small number of sequences were of unknown origin since their identity could not be verified in the databases, while a few sequences were of bacterial origin, possibly due to contamination. We chose to assign the DNA fragments to a specific gene if the sequence matched to the intragenic region or a segment within 100 kb upstream or downstream. The 109 gene-associated ΔNp63-binding fragments identified by this approach are listed in Supplemental Table 1 with their precise genomic coordinates. Approximately 16% of the genes we identified in this study were the mouse counterparts of human genes identified by other studies demonstrating the validity of our approach (Table I).
| Chrom | Gene | Description | Location | Accession # | Function | Start | End | References |
|---|---|---|---|---|---|---|---|---|
| 4 C7 | Map1lc3 | Microtubule-associated protein 1 | ND | NG_005200 | Unknown | 105616612 | 105617006 |
Yang et al. 2006 |
| 4 D2.3 | Ibrdc3 | IBR domain containing 3 | 3′ end | XM_204030 | Protein-binding | 128598215 | 128598341 |
Yang et al. 2006 |
| 3 F1 | Ivl | Involucrin | 5′ end | NM_008412 | Structural protein | 92665045 | 92665110 |
Yang et al. 2006 |
| 2 E2 | Fjx1 | Four jointed box 1 (Drosophila) | 3′ end | NM_010218 | Signaling | 102234945 | 102235357 |
Yang et al. 2006 |
| 7 F1 | Plekha7 | Pleckstrin homology domain containing, family mem 7 | 5′ end | NM_172743 | Unknown | 116047718 | 116047842 |
Yang et al. 2006 |
| 5 G2 | Auts2 | Autism susceptibility candidate 2 | Intron 5 | NM_177047 | Unknown | 131860963 | 131861133 |
Yang et al. 2006 |
| 17C | Ddr1 | Discoid domain 1 receptor family 1 | 5′ end | NM_007584 | Cell adhesion | 35380228 | 35380413 |
Osada et al. 2005, Carroll et al. 2006, Yang et al. 2006, Birkaya et al. 2007 |
| 18 A1 | Arhgap12 | RhoGTPase activator protein 12 | 3′ end | NM_02977 | Signal transduction | 5948618 | 5948762 |
Truong et al. 2006 |
| 12 A2-A3 | Dnmt3a | DNA methyltransferase | Intron 1 | NM_007872 | Transcription | 3848079 | 3848465 |
Yang et al. 2006 |
| 11 E1 | Smurf2 | SMAD specific E3 ubiquitin protein ligase 2 | ex 11/in 11 | NM_025481 | Protein folding | 106657541 | 1066557820 |
Vigano et al. 2006 |
| 16 C3.3 | Mrpl32 | Mitochondrial ribosomal protein L39 | 3′ end | NM_017404 | Electron transport | 84577487 | 84577657 |
Yang et al. 2006 |
| 10 A2 | Gpr126 | G protein-coupled receptor 126 | 3′ end | NM_001002268 | Signaling | 14055593 | 14055746 |
Yang et al. 2006 |
| 16 B1 | Trp63 | Transformation related protein 63 | Intron 2 | NM_011641 | Transcription | 25759148 | 25759340 | |
| 12 D3 | Ston2 | Stonin 2 | Intron 1 | NM_175367 | Transport | 921515653 | 92152038 |
Yang et al. 2006 |
| 8 E1 | Cdh13 | Cadherin 13 | Intron 3 | NM_019707 | Cell adhesion | 121597853 | 121597928 |
Carroll et al. 2006 |
| X A2-A3 | G6pdX | Glucose-6-phosphate dehydrogenase X-linked | Intron 2 | NM_008062 | Metabolism | 70676886 | 70676929 |
Yang et al. 2006 |
| 5 E5 | Cds1 | CDP diacylglycerol synthase 1 | Intron 5 | NM_173370 | Metabolism | 102043506 | 102043628 |
Yang et al. 2006 |
| 1 A3 | Xkr9 | X Kell blood group precursor related family member | Intron 1 | NM_001011873 | Membrane protein | 135865082 | 135865154 |
Yang et al. 2006 |
| 3 G1 | Ptbp2 | Polyprimidine tract-binding protein 2 | Intron 6 | NM_019550 | Protein-binding | 119736125 | 119736459 |
Yang et al. 2006 |
| 16 C4 | Runx1 | Runt related transcription factor 1 | Intron 3 | NM_009821 | Transcription | 92515494 | 92515729 | |
| 14 D3 | Ccdc122 | Coiled-coil domain containing 122 | Intron 1 | NM_175369 | Unknown | 75772117 | 75772382 |
Yang et al. 2006 |
| 8 B1.1 | Fat1 | Fat tumor suppressor homolog (Drosophila) | Intron 2 | XM_885736 | Cell adhesion | 46490981 | 46491956 |
Carroll et al. 2006 |
| 13 A4 | Gfod1 | Glucose-fructose oxidoreductase domain containing | Intron 1 | NM_001033399 | Metabolism | 43242969 | 43243250 |
Yang et al. 2006 |
| 16 B2 | Cldn1 | Claudin 1 | 5′ end | NM_016674 | Transport | 26349815 | 26350037 | |
| 16 B2 | Cldn16 | Claudin 16 | 5′ end | NM_053241 | Transport | 26349815 | 26350037 |
Yang et al. 2006 |
| 18 B3 | Centd3 | Centaurin, delta 3 | 5′ end | NM_139206 | Signal transduction | 38123967 | 38124462 |
Yang et al. 2006 |
| 11 B1.3 | Ankrd34 | Ankyrin repeat domain 43 | 5′ end | NM_183173 | Protein-binding | 53333445 | 533333592 |
Yang et al. 2006 |
| 6 A2 | St7 | Suppression of tumorigenicity 7 | 5′ end | NM_022332 | Unknown | 17668908 | 17669384 |
Yang et al. 2006 |
| 12 F1 | Rage | Renal tumor antigen | Intron 9 | NM_011973 | Kinase activity | 111256343 | 111256729 |
Yang et al. 2006 |
| 2 A1 | Gata3 | Gata-binding protein 3 | 3′ end | NM_008091 | Transcription | 9682830 | 9683136 |
Yang et al. 2006 |
| 8 D3 | Cdh3 | Cadherin 3 | 3′ end | NM_001037809 | Cell adhesion | 109461118 | 109461449 | |
| 8 D | Cdh1 | Cadherin 1 | 5′ end | NM_009864 | Cell adhesion | 109461118 | 109461449 | |
| 14 A3 | Arhgef3 | Rho guanine nucleotide exchange factor (GEF) 3 | Intron 3 | NM_027871 | Signal transduction | 2619197589 | 26197814 |
Yang et al. 2006 |
| 17 A2-A3 | Synj2 | Synaptojanin 2 | Intron 2 | NM_011523 | RNA-binding | 5942962 | 5943160 |
Yang et al. 2006 |
In most cases, the ChIPed DNA sequences mapped to a genomic neighborhood that was inhabited by or in close proximity to a single gene making it easy to identify the putative ΔNp63 target. However, in some cases the DNA fragment that was recovered was located in between two genes. For these cases, we have listed both of the genes as ΔNp63 targets. For example, one of the immunoprecipitated DNA fragments mapped to a region that is 16 kb from the 5′ end of Cdh1 (gene encoding for P-Cadherin) and 30 kb from the 3′ end of Cdh3 (gene encoding for E-Cadherin). In view of the known biological role of P-Cadherin and E-Cadherin in epidermal and hair development, it is quite possible that the p63-response element located in the intergenic region may coordinately regulate the expression of both these genes. One of the interesting target sites uncovered by our screen corresponded to a previously described enhancer for the p63 gene itself, located within an intron ∼42 kb downstream from the ΔNp63 transcriptional start site. It has been shown by ChIP experiments that the enhancer is bound by p63 suggesting an auto-regulatory role for p63 [Antonini et al., 2006]. Our data confirms the previous findings and provides further evidence for the efficiency of our approach in uncovering bona-fide p63 bound genomic fragments.
We found that 44% percent of the DNA fragments that were immunoprecipitated mapped within the region spanning 100 kb upstream or downstream of candidate target genes (Fig. 1A). Although many sites were found in the 5′ and 3′ end of the genes, p63-binding sites were also found within a gene, particularly in intronic segments. Indeed, we found that 45% of the DNA fragments immunoprecipitated were located within an intron of known or predicted genes, with an overwhelming preference for the first intron (Supplemental Table 1 and Fig. 1B). This finding is consistent with our previous data of ΔNp63 target genes in HaCaT cells and with the emerging trend of transcription factor-binding sites across the genome. This suggests that ΔNp63 may regulate the gene expression of numerous target genes by associating and binding to enhancer elements embedded within these genes. Intriguingly, only a few binding sites were found in exons corresponding to protein-coding domains, suggesting that potential transcription factor-binding sites in exons may be under negative selection and thus not associated with cis-regulatory functions. Examination of these potential target genes revealed that these genes are involved in a myriad of cellular processes including cell adhesion, transcription, and signal transduction (Supplemental Table 1 and Fig. 1B). Finally, it is important to note that a small percent of the DNA fragments were found to be in gene deserts and thus could not be assigned to any known or predicted gene. Notwithstanding the obvious possibility of false positives, these sites may also reflect enhancer elements located far away or regulatory elements associated with un-annotated genes, non-coding RNAs or novel transcriptional units.
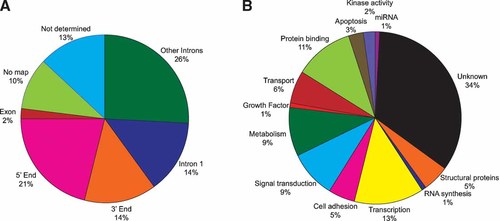
A: Location of the identified p63 ChIPed DNA sites relative to known genes. Examination of 124 potential target genes demonstrates the p63-binding sites are located within introns as well as the 5′ and 3′ end of these target genes. B: Target genes of ΔNp63 identified by ChIP are involved in a myriad of cellular processes.
The Immunoprecipitated DNA Fragment Mapping to the Runx1 Gene Is Highly Conserved
Among the putative p63 target genes that were identified, we chose to focus our attention on the transcription factor Runx1, which is known to play an important role in the development and differentiation of several tissues including the hair follicles. First, we examined the sequence of the DNA fragment that was immunoprecipitated and recovered from the ΔNp63 ChIP screening. Interestingly, this DNA fragment corresponding to an intronic region of the Runx1 gene is highly conserved across species, suggesting that this element may function as a potential enhancer (Fig. 2A,B). Two promoters drive the expression of different isoforms of Runx1 [Ghozi et al., 1996]. While a distal promoter P1 directs expression of Runx1c, the proximal promoter P2 drives expression of Runx1b and Runx1a isoforms. Runx1b is the major isoform in most cell types, and in some cases alternative splicing generates Runx1a (Fig. 2A). The DNA fragment immunoprecipitated in our ChIP approach is embedded within an intron common to both Runx1b and Runx1c making it possible that both isoforms are regulated by ΔNp63. To test this, we designed primers specific for each isoform, indicated by the arrows in Figure 2A. We found that only Runx1b is expressed in mouse keratinocytes grown in culture and in the epidermal compartment of the skin, strongly suggesting that ΔNp63 regulates expression of only Runx1b (Fig. 2C). It is possible that in other cell types, the expression of Runx1c is also under control of this enhancer.
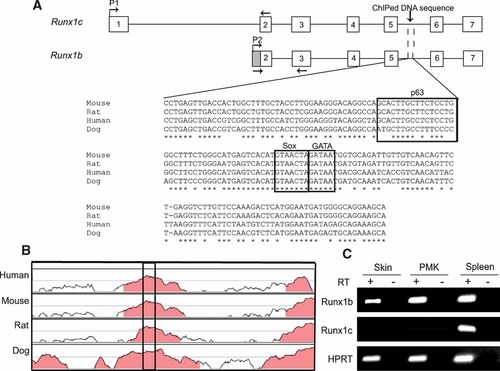
DNA fragment isolated by ChIP is embedded within an intron of the Runx1 gene. A: Shown is the genomic locus of two different isoforms of Runx1 that are generated from two distinct promoters (P1 and P2). The putative enhancer element maps to an intron common to both isoforms and is conserved across species. This enhancer element contains a putative p63-binding site shown in the box. Also shown are predicted response elements for additional transcription factors. B: Genome Vista of 5 kb of the Runx1 intron containing the putative enhancer element. The enhancer segment indicated by the box is highly conserved across several species. C: RT-PCR demonstrates expression of Runx1b in skin, primary mouse keratinocytes (PMK), and spleen, while Runx1c is expressed only in spleen. HPRT was used as a loading control.
Independent ChIP Assay Demonstrates Binding of ΔNp63 to Runx1
To clearly demonstrate that Runx1 is a direct target gene of ΔNp63 we performed an independent ChIP assay (Fig. 3). We used two different anti-p63 antibodies, the ΔNp63 N-terminal specific antibody RR-14, as well as an alpha-specific C-terminal antibody, H-129 to immunoprecipitate crosslinked chromatin from mouse keratinocytes as well as mouse skin tissue. We designed a set of primers to amplify the putative Runx1 enhancer element identified in our screen and as a negative control, we used a set of primers that amplify a genomic segment in the Gapdh gene. As shown by PCR data in Figure 3, specific enrichment of the Runx1 enhancer was observed after immunoprecipitation with both antibodies against p63 as compared to the IgG control. In contrast, there was no localization of ΔNp63 to the Gapdh genomic locus. Three independent ChIP experiments yielded similar results. In addition, we performed quantitative PCR to examine the relative fold enrichment of the Runx1 enhancer when immunoprecipitated with anti-p63 antibodies (Fig. 3B). We determined the relative fold enrichment of Runx1 by comparing the amount of Runx1-specific PCR product amplified from the anti-p63 ChIP to that from the negative control (IgG) ChIP samples. The significant enrichment we observed when we immunoprecipitated with the two anti-p63 antibodies confirms that the Runx1 enhancer is occupied by ΔNp63 protein in mouse keratinocytes grown in culture and more importantly in skin tissue in vivo.
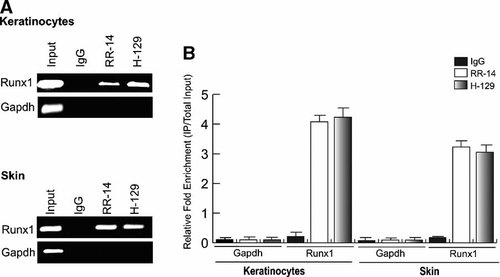
Independent ChIP assay demonstrates binding of ΔNp63 to the Runx1 putative enhancer element. ChIP was performed in mouse keratinocytes and mouse skin with N-terminal RR-14 and C-terminal H-129 anti-p63 antibodies. A: Enrichment of Runx1 enhancer element relative to IgG on an agarose gel. B: Real-time PCR demonstrating the relative enrichment of Runx1 enhancer element with p63 antibodies and IgG.
ΔNp63 Directly Binds to the p63-Response Element Located Within the Intronic Region of the Runx1 Gene
The results of the ChIP assays prompted us to test if ΔNp63 directly binds to the enhancer element. Therefore, we scanned the enhancer element for the presence of a p63 consensus DNA-binding site as defined by several recent studies [Ortt and Sinha, 2006; Perez et al., 2007]. The Runx1 intronic element that was immunoprecipitated by ΔNp63 contains a bona-fide p63 consensus site, with the core sequence being completely conserved among several species (Fig. 2). To determine if ΔNp63 binds to this response element, we designed an oligonucleotide probe (WT) for this site and performed electrophoretic mobility shift assay (EMSA). Upon incubation of the labeled oligonucleotide probe containing the putative Runx1 p63-response element and nuclear extract from mouse keratinocytes, a specific complex was observed (Fig. 4, lane 1). Competition assays with the p63-response element from the K14 enhancer, which has previously been demonstrated to contain a bona-fide p63-binding site, as well as a probe containing a consensus p53-binding site were used to demonstrate specificity [Romano et al., 2007]. When we incubated nuclear extract with increasing amounts of unlabeled oligonucleotide probes containing the K14 p63-binding site, a consensus p53-binding site, or the wild type sequences, the complex was abolished (lanes 2–7). On the other hand, excess amounts of oligonucleotide probes containing two different mutations in the Runx1 p63-binding site (MT1, CTTG > ATTC; MT2, CTTG > ATTG) had no effect (lanes 8–11). Finally, the addition of anti-ΔNp63 specific antibodies (but not preimmune serum) resulted in a distinct supershift of the complex, demonstrating that this complex contains ΔNp63 (lanes 12–13). Our EMSA studies clearly demonstrate that the Runx1 enhancer element contains a p63-binding site and that endogenous ΔNp63 present in mouse keratinocytes can bind to this site.
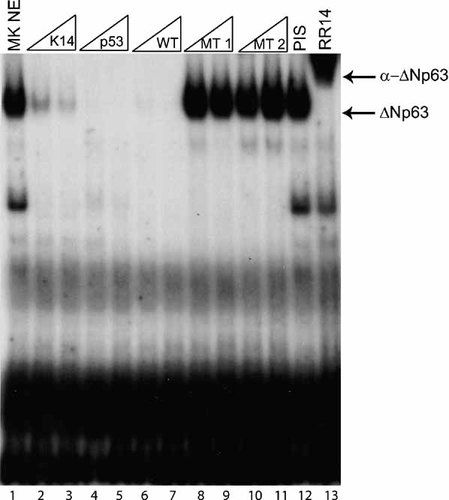
EMSA demonstrates binding of ΔNp63 to the conserved p63-binding site embedded within the Runx1 putative enhancer element. EMSAs were performed with radiolabeled oligonucleotide probe containing the p63-binding site corresponding to the Runx1 enhancer element. Probe was incubated with nuclear extract from mouse keratinocytes. Oligonucleotides containing known p63 response elements, compete while mutant oligonucleotides have no effect at 20- and 100-fold excess amounts. Supershift with anti-p63 antibodies confirms that the complex contains ΔNp63. PIS, preimmune serum
ΔNp63 Transcriptionally Regulates the Runx1 Enhancer Element
Having demonstrated that ΔNp63 can directly bind to the putative Runx1 enhancer, our next goal was to determine if the enhancer element was transcriptionally responsive to ΔNp63 in transient transfection experiments. We utilized PTK2 cells for these experiments since these cells have been shown to lack endogenous p63 expression. The Runx1 enhancer element was cloned upstream of the heterologous minimal TK promoter. The reporter plasmid was co-transfected with either an expression plasmid encoding for HA-tagged ΔNp63 α, β, or γ or an empty HA control vector. Expression of ΔNp63 α, β, or γ resulted in increased levels of reporter activity (four to sixfold) as demonstrated in Figure 5. Interestingly, TAp63γ, the most potent transactivating p63 isoform, also resulted in an increase level of reporter activity. In contrast, when a mutation was introduced in the p63-binding site (MT1, the mutation that abolishes binding of p63 as shown in EMSA experiments), all three ΔNp63 isoforms and TAp63γ failed to activate the reporter construct. Collectively, these data strongly suggest that p63 activates the Runx1 enhancer element through its binding to the p63-binding site.

Runx1 enhancer element is responsive to all isoforms of ΔNp63 and TAp63γ. Luciferase construct containing the Runx1 enhancer element cloned upstream of a heterologous TK promoter demonstrates high levels of activity when co-transfected with expression plasmids encoding ΔNp63 α, β, γ or TAp63γ in PTK2 cells. Luciferase values were determined and normalized against β-galactosidase values. Corrected luciferase values for cells transfected with empty vector pCMV-HA were set at 1. TK, thymidine kinase.
Knockdown of p63 Expression in Mouse Keratinocytes Affects Runx1 Gene Expression
Having shown that ΔNp63 can bind and activate the Runx1 enhancer, we next wanted to demonstrate whether ΔNp63 is directly involved in regulating expression of the Runx1 gene. First we examined the effect of knocking down p63 expression in mouse keratinocytes by employing synthetic dsRNA (siRNA) to reduce p63 expression. We observed a significant reduction in both the mRNA and protein levels of p63 in mouse keratinocytes transfected with the p63 siRNA (Fig. 6A,B). We synthesized cDNA from three independent RNAi experiments and performed quantitative RT-PCR assays to examine Runx1 gene expression in mouse keratinocytes that had reduced p63 levels. As a reference standard, we used the housekeeping gene B2M (beta2 microglobulin) whose expression is not thought to be dependent on p63 or Runx1. Expression of Runx1 was dramatically reduced (∼60%) in keratinocytes transfected with p63 siRNA, whereas cells transfected with control siRNA showed no alteration in Runx1 levels. As demonstrated before, expression of K14, another direct target of ΔNp63 was also reduced in the p63 siRNA treated keratinocytes [Romano et al., 2007].
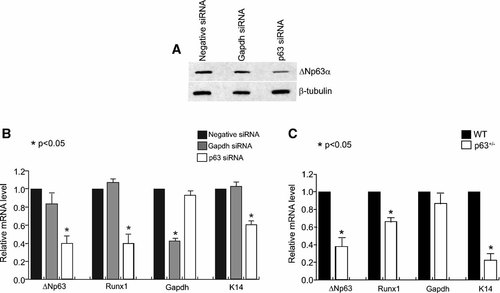
Runx1 levels are reduced when p63 expression is knocked down by siRNA in mouse keratinocytes and in p63 heterozygous skin. Mouse keratinocytes were transfected with negative control siRNA, Gapdh siRNA, and p63 siRNA. A: Western Blot using the RR-14 antibody demonstrates decreased expression of ΔNp63. β-tubulin was used as a loading control. B: Expression of Runx1 was analyzed by real-time PCR. Negative control siRNA-transfected cells were used as a control. C: Skin from wild type and p63+/− embryos was isolated. Expression levels of ΔNp63 and Runx1 in skin were analyzed by real-time PCR. Gapdh and K14 were used as controls. Asterisks indicate statistical significance derived from Student's t-test.
Since p63 null animals die soon after birth and exhibit a complete dissolution of skin tissue, it is difficult to assess the alterations of putative p63 target genes in these mice. However, we reasoned that examination of skin samples from the p63 heterozygous animals, which appear phenotypically normal, should reveal whether reduction in p63 levels affects Runx1 gene expression. For this purpose, we isolated RNA from skin tissue of newborn p63 heterozygous and control wild type animals. By quantitative RT-PCR, we show that p63 levels are 62% lower in the p63 heterozygous animals as expected (Fig. 6C). Interestingly, in agreement with our p63 knockdown experiments with keratinocytes in culture, p63 heterozygous animals also exhibit lower levels of Runx1 and K14 gene expression as compared to wild type. In light of our ChIP, EMSA and transient transfection experiments, these data argues that Runx1 is a bona-fide target gene of ΔNp63 in mouse skin.
ΔNp63 and Runx1 Are Co-Localized in the Outer Root Sheath of the Hair Follicle
Our biochemical data strongly suggests that Runx1 is a direct target gene of ΔNp63. We next sought to examine the in vivo expression pattern of Runx1 and ΔNp63 to determine if they are co-expressed in skin epithelium and in particular in hair follicles. This is of particular interest since we have recently reported that animals with skin-specific knockout of Runx1 show aberrations in hair follicle morphogenesis associated with specific structural defects. Hence to analyze the distribution patterns of Runx1 and ΔNp63, we investigated their expression in mouse skin during embryogenesis and postnatal development. For this purpose, we utilized mice that have the LacZ reporter gene integrated into the Runx1 genomic locus such that X-gal staining mimics endogenous Runx1 expression. As shown before, at embryonic day 14.5 (E14), the expression of Runx1 is restricted to the dermal compartment (Fig. 7A). This dermal region is distinct from the overlying epidermal layer, which shows strong expression of ΔNp63 as detected by ΔNp63-specific RR-14 antibody. However, at a later stage of development (E18.5), when hair follicles formation has progressed further, nuclear ΔNp63 expression is co-localized with Runx1 along the outer root sheath (ORS) and in the matrix of the hair follicles (Fig. 7A,B). There is also strong ΔNp63 expression in the basal cells of the epidermis, however no blue staining is observed in the interfollicular epidermis suggesting the lack of Runx1 expression in this region of the skin. The pattern of co-localization in the hair follicle is even more apparent in hair follicles at birth (NB) and postnatally (Fig. 7A,B). In view of the intriguing expression profile of ΔNp63 and Runx1 in hair follicle, we decided to investigate their expression in other appendages of the skin. As shown in Figure 7C, in the nail, ΔNp63 is expressed more in basal cells in the matrix and the nail bed compared to Runx1, which is expressed in more differentiated cells, suggesting that there is little overlap of these two transcription factors in this tissue. On the contrary, in the eccrine sweat glands of the toes, ΔNp63 and Runx1 were found to be co-expressed in the proximal loops (Fig. 7C). Taken together, these data suggest that ΔNp63 and Runx1 expression overlap in some regions of the skin tissues, but not all. In particular Runx1 and p63 only partially co-localize in the mouse hair follicle, with p63 mainly detected in the proliferating cells of the ORS and hair bulb matrix, while Runx1 expression is broader and extends into differentiated cells of the hair shaft.
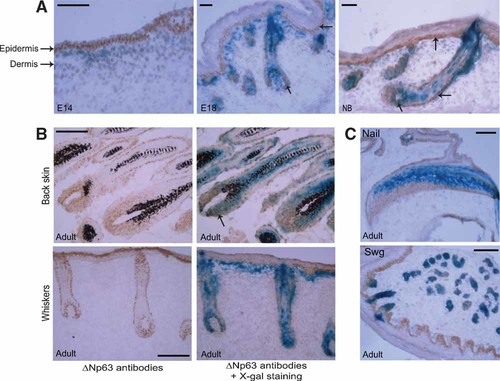
ΔNp63 and Runx1 are co-expressed in the outer root sheath (ORS) of the hair follicle. Expression of Runx1 (X-gal staining, Blue) and ΔNp63 (immunostaining, brown) during hair follicle morphogenesis and in nails and sweat glands. A: At embryonic day 14.5 (E14), Runx1 dermal expression is distinct from the epidermal expression of ΔNp63. At E18.5, nuclear ΔNp63 expression is co-localized with Runx1 along the ORS and in the hair matrix of the hair follicles, while only ΔNp63 is also expressed in the basal cells of the epidermis (indicated by the arrows). This pattern of co-localization is more apparent at birth (NB). B: In adult back skin and whiskers, ΔNp63 is co-localized with Runx1 in the lower portion of the ORS (secondary germ zone; indicated by the arrows). C: In the nail, ΔNp63 is expressed in the matrix and the nail bed in more differentiated cells. In eccrine sweat glands of the toes (Swg), ΔNp63 and Runx1 are co-expressed in the proximal loops. Scale bars = 50 µm (except for Nail and Swg = 100 µm).
DISCUSSION
Identification of binding sites for p63 and the corresponding target genes is essential for deciphering the regulatory circuits through which this transcription factor controls various cellular processes in the epithelium. Although global studies such as microarray profiling assays to probe the transcriptome status in p63 over expressing, knockdown or null cell lines have provided a glimpse into p63-regulated pathways and networks, these types of approaches have inherent limitations. For example, these studies fail to discriminate direct from indirect effects of transcription factors and a priori, do not reveal the location of regulatory response elements that control target gene expression. Recent progress in isolation and mapping of target transcription factor-binding sites by ChIP-based technology have proven to be useful in overcoming some of these shortcomings [Kim and Ren, 2006]. One such strategy, ChIP-chip identifies target-binding regions directly by probing the immunoprecipitated DNA using a DNA microarray that tiles extensive regions of the genome. However, with improved ultrahigh-throughput sequencing capabilities, direct DNA sequencing of ChIP fragments (ChIP-Seq) is gaining momentum and has been successfully performed for some transcription factors.
The network of potential p63 targets has become extensive primarily due to ChIP-chip studies that have been performed recently [Vigano et al., 2006; Yang et al., 2006]. Although these studies have provided an exhaustive list of p63-response elements and putative target genes that are activated and repressed by p63, the database is still evolving and far from completion. The fact that ChIP experiments have only been performed on transformed human cell lines undermines the physiological relevance of the data. In addition, antibodies utilized in these experiments recognize both the TA and ΔN isoforms of p63, further adding to the complexity in data interpretation. It is very likely that the distribution of p63 across the vast array of genomic segments and the consequent transcriptional effects on a battery of target genes is a dynamic event that is dictated by the inherent nature of the cellular environment. The ChIP-based approach can thus provide only a snapshot of the p63-DNA interactions, which depending upon cell type and experimental conditions may vary significantly. Hence, it is important to probe the dynamics of p63 and its target DNA interaction under a variety of experimental conditions to achieve a comprehensive view of its biological function.
With that in mind, in this study we have utilized ChIP in combination with sequencing and mapping of isolated genomic DNAs to locate binding sites for ΔNp63 and its potential target genes in primary mouse keratinocytes in vivo. Although sequencing ChIPed DNA fragments (ChIP-Seq) is potentially straight forward, limiting amounts of DNA obtained during the procedure and the overwhelming excess of non-specifically precipitated DNA is a complicating issue. Hence, there is a need to explore experimental parameters important for enhancing the performance of ChIP-seq strategy particularly for complex genomes such as those of mammalian cells. We have previously demonstrated a modified and improved ChIP approach to successfully identify target genes of ΔNp63 in HaCaT cells, which are immortalized human keratinocytes [Birkaya et al., 2007]. Since the typical ChIP procedure generates only minute amounts of immunoprecipitated genomic DNA, which is often associated with non-specific DNA contamination, direct sequencing and identification of the genomic DNA presents a number of technical challenges. We have overcome these limitations by incorporating a PCR-based amplification and GST-purification step to enrich the immunoprecipitated genomic DNA. As we demonstrate in this report, this strategy allowed us to identify numerous target genes of ΔNp63 in primary mouse keratinocytes.
A careful evaluation of the ∼100 ΔNp63-reponse elements and their corresponding genes lead to numerous interesting observations. A number of the genes identified in our study were the mouse counterparts of human genes identified in previous studies. There are also a significant number of genes that have thus far not been reported to be direct targets of p63. These might represent a subset of p63 targets that are unique to ΔNp63 since many of the prior studies did not discriminate between the ΔN and TA isoforms. Alternatively, differences in cell type, growth conditions, and antibodies may account for the lack of complete overlap in p63 targets obtained from various studies. We believe that most of the ChIPed DNA fragments represent true p63-response targets since these DNA segments contain one or more p63-binding sites as identified by sequence comparison with the p63 consensus binding site. This coupled with the fact that many of the target genes are linked to pathways (such as the Notch pathway) that have been implicated to be under the control of p63 or have appeared in multiple other screens for p63 targets (such as DDR1), strongly suggests the functional relevance of these segments. Interestingly, most of the response elements were found to be located in intragenic regions or in segments that are located far away from the promoters demonstrating the propensity for ΔNp63 to regulate gene expression through the utilization of distal enhancer elements. This seems to be a recurring theme for a vast majority of transcription factors and should be an important consideration when trying to decipher locations of transcription factor-binding sites.
It is important to identify the ΔNp63 target genes to better understand its role in the development of stratified epidermis and associated appendages. We have focused our study on Runx1 to illustrate the validity of our ChIP based search approach for ΔNp63 target genes and to evaluate its role as a potential target of ΔNp63 in keratinocytes. Runx1 is a member of a family of three transcription factors in mammals that are involved in the control of proliferation and differentiation in various tissues. The role of Runx1 as a regulator of hematopoiesis is well established, particularly for megakaryocytic maturation and lymphocyte differentiation [Mikhail et al., 2006]. However, it is becoming increasingly clear that Runx1 can also play a role in the homeostasis of other tissues such as the nervous system and skeletal muscles [Wang et al., 2005; Kramer et al., 2006; Marmigere et al., 2006]. Of the three Runx proteins, Runx1 is prominently expressed in mouse skin, exhibiting a dynamic expression pattern particularly in the hair follicles. Using epidermal-targeted Runx1 knockout mice we have shown that Runx1 affects hair structure and morphogenesis through its involvement in the differentiation program of the hair shaft [Raveh et al., 2006].
Here we identify a putative enhancer element within the Runx1 gene that is evolutionarily conserved. We show that this putative enhancer element is occupied by ΔNp63 in vivo and can be transcriptionally activated by all isoforms of ΔNp63 in transient transfection assays. Importantly, ΔNp63 is a key transcriptional mediator of Runx1, since expression of Runx1 is decreased in mouse keratinocytes that have reduced p63 levels. The Runx1 gene gives rise to two major mRNA products, Runx1c and Runx1b that result from transcriptional initiation at either a distal P1 or proximal P2 promoter. The novel enhancer described in our study is embedded within the Runx1 gene in an intronic region, which is common to both Runx1b and Runx1c. However, since we observe that Runx1b is the predominant isoform expressed in mouse skin and keratinocytes in culture, we speculate that this enhancer is likely to influence the activity of the proximal P2 promoter, which is relatively closer as compared to the distal P1 promoter. Prior studies evaluating the transcriptional regulation of Runx1 have been focused primarily on B cells and have implicated Ets sites, CCAAT boxes and Runx1 elements present in the proximal promoter [Ghozi et al., 1996]. Interestingly we find that there are no p63-response elements in the promoter regions of Runx1 (unpublished data). It will be interesting to test if the enhancer reported here also plays a role in other cell types and if p63 or any of its closely related family members influence the enhancer activity in other cell types as it does in keratinocytes.
One interesting observation from our studies is the fact that Runx1 and p63 do not completely overlap in the various keratinocyte cell types in the mouse skin. This is clearly evident by the fact that whereas ΔNp63 is present primarily in both the proliferating cells of the interfollicular epidermis and the ORS of the hair follicles, Runx1 expression is restricted to follicular epithelial cells that also include the differentiated cells of the inner root sheath. In hair follicle development, there is evidence for a role for GATA-3 and Sox9, both of which are not only direct transcriptional targets of p63 but also have been functionally linked with Runx1 [Kaufman et al., 2003; Vidal et al., 2005; Zhou et al., 2006; Naoe et al., 2007]. In this regard, it is important to note that the Runx1 intronic enhancer identified in this study contains binding sites for both GATA and Sox family of transcription factors. Thus an interesting possibility is that the development and differentiation of hair follicle keratinocytes involves extensive crosstalk and feedback among these critical transcription factors.
There are a number of skin diseases and syndromes involving Ectrodactyly, Ectodermal Dysplasia and alopecia in human patients that are associated with mutations in the p63 gene [van Bokhoven and McKeon, 2002]. In view of our findings, it is tempting to speculate that the hair follicle phenotype in these patients could be a consequence of de-regulated Runx1 expression. Our identification of a wide range of in vivo target genes of ΔNp63 provides only a glimpse of the function of p63 during epithelial development and maintenance. The target genes identified are involved in numerous cellular pathways further illustrating the complexity and importance of this transcription factor. The role of p63 in hair follicle morphogenesis is an interesting aspect and unearthing the target genes that may be involved in this process, such as Runx1, is one step into revealing the molecular mechanisms by which p63 masterfully controls this intricate process. Finally, the success of our improved ChIP strategy in identifying bona-fide target genes of ΔNp63 suggests its usefulness as a genomic tool for other transcription factors.
Acknowledgements
We are especially grateful to Dr. Lee Ann Garrett-Sinha and Rose-Anne Romano for helpful discussion and advice. We also thank Kirsten W. Smalley for excellent technical assistance in mouse husbandry and other members of our laboratory for useful comments on this study. We would like to thank Nancy Speck for providing the Runx1+/lz mice. This work was supported by a grant from NIH R01AR049238 (S.S.) and in part by a grant support from United States-Israel Binational Science Foundation grant 2005197 (U.G. and S.S.).




