Immediate-early gene regulation by interplay between different post-translational modifications on human histone H3
Abstract
In mammalian cells, induction of immediate-early (IE) gene transcription occurs concomitantly with histone H3 phosphorylation on Ser 10 and is catalyzed by mitogen-activated protein kinases (MAPKs). Histone H3 is an evolutionarily conserved protein located in the core of the nucleosome, along with histones H2A, H2B, and H4. The N-terminal tails of histones protrude outside the chromatin structure and are accessible to various enzymes for post-translational modifications (PTM). Phosphorylation, O-GlcNAc modification, and their interplay often induce functional changes, but it is very difficult to monitor dynamic structural and functional changes in vivo. To get started in this complex task, computer-assisted studies are useful to predict the range in which those dynamic structural and functional changes may occur. As an illustration, we propose blocking of phosphorylation by O-GlcNAc modification on Ser 10, which may result in gene silencing in the presence of methylated Lys 9. Thus, alternate phosphorylation and O-GlcNAc modification on Ser 10 in the histone H3 protein may provide an on/off switch to regulate expression of IE genes. J. Cell. Biochem. 103: 835–851, 2008. © 2007 Wiley-Liss, Inc.
Nucleosomes are the main organizational modules of chromatin and histones are their main protein component. The high conservation of histones throughout evolution attests the basic nature of the nucleosomal design [Tsunaka et al., 2005]. Regulation of gene transcription preferentially occurs by way of post-translational modification (PTM) of the histone in amino terminal tails located outside the compact chromatin structure, as for instance, in the histone 3 (H3) protein [Cheung et al., 2000a]. Several PTMs of histones, namely phosphorylation, acetylation, methylation, and O-GlcNAc modification, regulate the contact of chromatin with DNA [Cheung et al., 2000a]. These PTMs form the basis of a histone code, a specific code that facilitates diverse cellular responses, involving gene expression and orderly completion of the cell cycle [Cheung et al., 2000a; Cosgrove and Wolberger, 2005]. In particular, phosphorylation of H3 and of several transcription factors has been found to closely correlate with immediate-early (IE)-gene transcription under diverse conditions of induction [Thomson et al., 1999; Clayton and Mahadevan, 2003].
The nucleosome response involves alterations in chromatin and nucleosome structure, relies on histone modifications, and is associated with the induction of different genes [Cheung et al., 2000a] including IE-gene transcription [Thomson et al., 1999]. The transcription of IE genes is transiently activated within minutes of cell exposure to a wide range of extracellular stimuli [Thomson et al., 1999]. IE genes encode transcription factors, such as the promoter-specific factor 1 (Sp1) [Chen et al., 1994], activator protein 1 (AP-1) [Angel et al., 1988; Fisch et al., 1989; Herr et al., 1994], and c-AMP-response element-binding protein (CREB) [Gonzales and Bowden, 2002], DNA-binding proteins and proto-oncogene proteins like c-Jun that regulate cell proliferation and apoptosis [Wisdom et al., 1999]. These transcription factors and H3 (on Ser 10 and 28), are phosphorylated by mitogen-activated protein kinases (MAPKs) or their effector kinases such as mitogen- and stress-activated kinases, and the phosphorylated proteins are involved in the induction of several IE genes [Deak et al., 1998; Seassone-Corsi et al., 1999; Clayton et al., 2000; Zhong et al., 2001; Duncan et al., 2006].
Both Ser 10 and 28 are preceded by Lys at −1 position, a residue not found very often in the vicinity of phosphorylated Ser [Iakoucheva et al., 2004; Qazi et al., 2006]. The position of Lys immediately before a phosphorylated Ser appears to be related with its methylation in this particular context. Interestingly, methylated Lys 9 mediates gene silencing and methylated Lys 27, gene repression [Lindroth et al., 2004]. Furthermore, an interplay between methylated and phosphorylated neighboring amino acid residues (Lys 9/Ser 10 and Lys 27/Ser 28) known as “phosphorylation/methylation switching” has been reported in H3 [Wang et al., 2004]. Clearly, the structural motifs consisting of Lys 9 and Ser 10, and Lys 27 and Ser 28 are functionally important.
Amongst the different PTMs, one of the dynamic and regulatory modifications of hydroxyl function of Ser/Thr is the O-GlcNAc modification, which influences protein folding, localization and trafficking, solubility, antigenicity, biological activity, and half-life, as well as cell–cell interactions [Love and Hanover, 2005]. Interplay between O-GlcNAc modification and phosphorylation on the same or neighboring Ser/Thr residues has been observed in several nuclear and cytoplasmic proteins [Comer and Hart, 2000; Wells et al., 2003]. The dynamic O-GlcNAc modification can regulate gene transcription by glycosylating transcription factors like Sp1 [Majumdar et al., 2003] and CREB [Lamarre-Vincent and Hsieh-Wilson, 2003].
Interplays of different PTMs on the same or neighboring residues are known to occur in proteins [Khidekel and Hsieh-Wilson, 2004], and may either facilitate or prevent other modifications, thereby regulating the function of the modified protein. Recently, it has been suggested that an interplay between O-GlcNAc modification and phosphorylation of H3 is involved in the regulation of the cell cycle in mammals [Kaleem et al., 2006], emphasizing the importance of PTMs on proteins that control gene regulation.
The specific combination of different PTMs may provide a basis for H3 to perform multiple functions, and computational methods may help evaluating H3 multifunctionality. Furthermore, these methods have an advantage of being fast, reproducible, and 70–80% accurate [Nielsen et al., 1999]. Several computational methods have been developed to predict glycosylation and phosphorylation sites in proteins. These include NetPhos 2.0 [Blom et al., 1999] and YinOYang 1.2 (unpublished). Most of these prediction methods that compute modification potential are neural network based and recognize specific sequence content through prior learning process. Amino acids involved in maintaining the 3D structure of a protein and hence its functions, have often been found to be highly conserved evolutionarily [Schueler-Furman and Baker, 2003] and interplay of phosphorylation and O-GlcNAc modification on conserved Ser/Thr residues has been proposed to act at key functional sites [Ahmad et al., 2006].
Available prediction, in silico, data for different PTMs suggest that a complex interplay or a specific combination of these PTMs may regulate repression or induction of different genes, including IE genes. When IE genes are ready for transcription, H3 is phosphorylated on Ser 10 [Thomson et al., 1999], methylated on Lys 4 [Hazzalin and Mahadevan, 2005], and acetylated on Lys 9 [Hazzalin and Mahadevan, 2005] and/or Lys 14 [Cheung et al., 2000b]. We propose that when H3 is O-GlcNAc modified on Ser 10, it may result in deacetylation of Lys 9, which consequently becomes methylated. Thus, a combination of O-GlcNAc modification of Ser 10 and methylation of Lys 9 may result in IE-gene repression.
MATERIALS AND METHODS
The Sequence Data
The sequence data used to predict phosphorylation and O-glycosylation potential of H3 protein in Homo sapiens were retrieved from the Swiss-Prot database [Boeckmann et al., 2003] with primary accession no. P68431. BLAST search was carried out by using NCBI database of non-redundant sequences using all default parameters [Altschul et al., 1997]. The search results were divided into vertebrates and invertebrates. The sequences selected for multiple alignment from different species of vertebrates were from Mus musculus (RefSeq. AAI07286.1), Xenopus laevis (RefSeq. CAA51455.1), Gallus gallus (RefSeq. AAA48795.1), and Xenopus tropicalis (RefSeq. CAJ81662.1). The sequences selected from invertebrates included that of Caenorhabditis elegans (Swiss-Prot P08898), Mytilus chilensis (RefSeq. AAP94665.1), Drosophila melanogaster (RefSeq. CAA32434.1), Lytechinus pictus (RefSeq. AAA30003.1), and Aedes aegypti (RefSeq. EAT45035.1). The chosen sequences were multiple aligned using ClustalW using all default parameters [Thompson et al., 1994].
For comparison of human H3 with human H2A, H2B, and H4, different sequences were retrieved from the Swiss-Prot database [Boeckmann et al., 2003] as follows: H2B1B (Swiss-Prot P33778), H2A1A (Swiss-Prot Q96QV6), and H4 (Swiss-Prot P62805). The four sequences were multiple aligned using ClustalW [Thompson et al., 1994]. BLAST search for human histone H2B was carried out by using NCBI database of non-redundant sequences using all default parameters [Altschul et al., 1997]. The search results were divided into vertebrates and invertebrates. For determination of evolutionary conservation of human H2B, ClustalW [Thompson et al., 1994] was utilized. The sequences chosen from vertebrates included Mus musculus (Swiss-Prot Q64475), Bos taurus (RefSeq. 701196A), Gallus gallus (RefSeq. NP_001026652), Rattus norvegicus (RefSeq. 0506206A), Oncorhynchus mykiss (Swiss-Prot P69069), Rhacophorus schlegelii (Swiss-Prot Q75VN4); and from invertebrates included Drosophila yakuba (Swiss-Prot Q8I1N0), Rhynchosciara americana (RefSeq. AAK58064), Drosophila hydei (Swiss-Prot P17271), Mytilus edulis (RefSeq. CAD37816), Chironomus thummi (Swiss-Prot P21897), Aedes aegypti (RefSeq. EAT45030), Anopheles gambiae (Swiss-Prot Q27442).
Glycosylation and Phosphorylation Prediction Methods
The potential for phosphorylation and O-GlcNAc modification in human histone H3 and H2B was predicted by NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/) [Blom et al., 1999] and YinOYang 1.2 (http://www.cbs.dtu.dk/services/YinOYang/) (unpublished), respectively.
The above two methods are neural networks-based prediction methods. Neural networks are composed of a large number of highly interconnected processing elements (simulated neurons) working in parallel to solve a complex problem. In a neural network-based prediction method, networks are trained by sequence patterns of modified and non-modified proteins so that they become able to recognize and predict a pattern in a new protein for their potential of modification. Artificial neural networks receive many inputs and give one output as a result. NetPhos 2.0 [Blom et al., 1999] was developed by training the neural networks with phosphorylation data from Phosphobase 2.0 [Kreegipuu et al., 1998]. The YinOYang 1.2 server (unpublished) produces neural network predictions for O-GlcNAc attachment sites in eukaryotic protein sequences. This method can also predict phosphorylation potential and thus predicts possible “Yin Yang” sites. A threshold value of 0.5 is used by NetPhos 2.0 to determine possible potential for phosphorylation, while the threshold value used by YinOYang 1.2 is variable, depending upon surface accessibility of the different amino acid residues. False negative sites were also identified, by coupling conservation status and modification potential of the two methods.
Secondary Structure Prediction Methods
The secondary structure (coil, helix, or extended strand) of human H3 and H2B was predicted using GOR IV [Garnier et al., 1996; Combet et al., 2000] to locate and characterize the predicted interplay sites of PTMs in different structural regions, consequently helping in developing structure–function relation for different PTMs. For the purpose of comparing secondary structural characteristics of Ser phosphorylation sites with Lys at −1 position other than human H3, a total of 103 proteins sequence data of the Ser phosphorylation sites with Lys at −1 position was retrieved from Phosphobase 3.0 [Diella et al., 2004] with 124 Ser phosphorylation sites. Similarly, GOR IV [Garnier et al., 1996; Combet et al., 2000] was used to predict the secondary structure of all 124 Ser phosphorylation sites. The secondary structural regions of all these sites were compared with that of human H3.
Kinase Phosphorylating Potential and Methylation Potential Prediction Methods
The kinase phosphorylating potential for 124 known Ser phosphorylated sites was predicted using NetPhosK 1.0 [Blom et al., 2004] to uncover a possible consensus for kinase specificity for Ser with Lys at position −1 along with other neighboring residues.
Similarly, the methylation potential of Lys residues at −1 position of all 124 phosphorylated Ser was predicted using MeMo (a computational method for prediction of protein methylation modifications in proteins) [Chen et al., 2006].
Comparison of the Sequence Motif of O-GlcNAc Modification Sites in Human H3 With Experimentally Known Proteins
The comparison of the sequence motif of O-GlcNAc modification sites, Ser 10 and 28, in human H3 with experimentally known O-GlcNAc-modified proteins was performed. Proteins with experimentally known O-GlcNAc modification sites were manually extracted from the Swiss-Prot database [Boeckmann et al., 2003].
RESULTS
O-Linked Phosphorylation Sites in Human H3
The results of predictions of phosphorylation sites in human H3 performed by NetPhos 2.0 are given in Table I, and graphically presented in Figure 1. All of the potentially predicted Ser and Thr phosphorylation sites were conserved in vertebrate and in invertebrates as well (Fig. 2). No Tyr residues were predicted to be phosphorylated in human H3.
| Residue no. | Experimental evidence | Prediction of modification potential | |||
|---|---|---|---|---|---|
| Phosphorylation | O-GlcNAc modification | NetPhos 2.0 | YinOYang 1.2 | Yin Yang site | |
| Ser 10 |
Zhang et al. 2003 |
By similaritya | + | + | + |
| Ser 28 |
Zhang et al. 2003 |
By similaritya | + | + | + |
| Ser 57 | − | − | + | + | +/− |
| Ser 86 | − | − | − | + | − |
| Thr 3 | − | − | − | + | − |
| Thr 6 |
Zhang et al. 2003 |
By similaritya | + | + | + |
| Thr 11 |
Zhang et al. 2003 |
By similaritya | + | + | + |
| Thr 22 | − | − | − | + | − |
| Thr 32 | − | − | − | + | − |
| Thr 45 | − | − | + | + | + |
| Thr 80 | − | − | − | + | − |
| Thr 118 |
Zhang et al. 2003 |
By similaritya | + | + | + |
- +, Positive prediction; −, negative prediction; +/− false/negative prediction.
- a Similarity in kinase and OGT recognition of same substrate site.
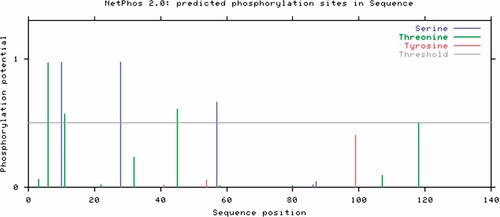
Predicted potential sites for phosphate modification on Ser and Thr residues in human histone 3. The blue vertical lines show the potential phosphorylated Ser residues; the green lines show the potential phosphorylated Thr residues; the red line show the potential phosphorylated Tyr residues. The light gray horizontal line indicates the threshold for modification potential.
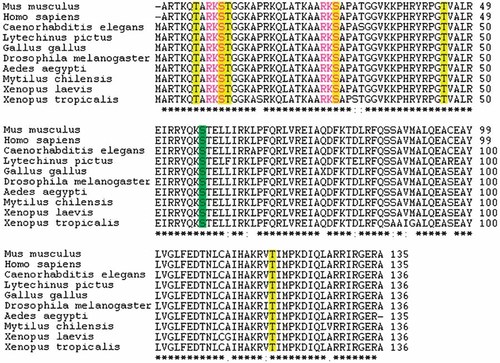
Multiple alignments of five vertebrates sequences (Homo sapiens, Mus musculus, Gallus gallus, Xenopus laevis, Xenopus tropicalis) and five invertebrates (Caenorhabditis elegans, Lytechinus pictus, Drosophila melanogaster, Aedes aegypti, Mytilus chilensis). The consensus sequence is marked by an asterisk, conserved substitution by a double dot, and semiconserved substitution by a single dot. The different sequences are ordered as in aligned results from ClustalW. The positively predicted Yin yang sites are highlighted in yellow, and the negatively predicted Yin yang site is highlighted in green. It is observed that the predicted Ser phosphorylation sites (Ser 10 and 28) have the same sequence motif with Lys on −1 and Arg on −2 positions (highlighted in red).
O-Linked Glycosylation Sites in Human H3
The prediction results of O-GlcNAc modification for human H3 by YinOYang 1.2 have been given in Table I and illustrated in Figure 3. All of the potentially predicted O-GlcNAc modification sites were conserved in vertebrates and in invertebrates (Fig. 2). Furthermore, human H3 showed a higher potential for O-GlcNAc modification compared to phosphorylation.
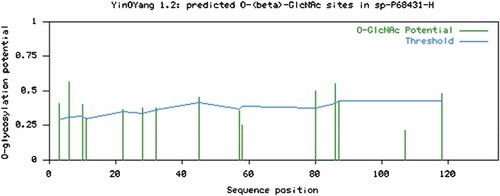
Predicted potential sites for O-GlcNAc modification of Ser and Thr residues in human histone 3. The green vertical lines show the O-GlcNAc modification potential of Ser/Thr residues and the light blue horizontal wavy line indicates the threshold for modification potential. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Yin Yang Sites in Human H3
Yin Yang sites in human H3 were predicted by YinOYang 1.2 and the results have been summarized in Table I and illustrated in Figure 4. All of these sites are of functional importance as these Ser/Thr residues can be modified by kinases as well as by OGT. Only one Ser at position 57 was identified as a false negative Yin Yang site (Table I, Fig. 4). All of the predicted and identified as false negative Yin Yang sites were found to be fully conserved in vertebrates and in invertebrates (Fig. 2).
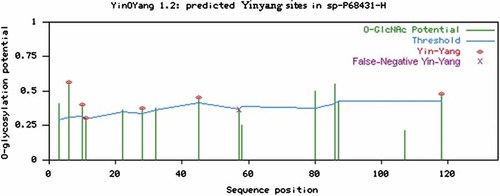
Predicted potential sites for both O-GlcNAc modification and phosphorylation (the Yin Yang sites). The positively predicted Yin Yang sites are shown with red asterisk at the top, and the negative predicted Yin Yang site is shown with purple asterisk on the top, in human H3. The green vertical lines show the O-GlcNAc potential of Ser/Thr residue and the light blue horizontal wavy line indicates the threshold for modification potential. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
It was also observed that the potential Ser phosphorylation sites in the N-terminal of H3 (Ser 10 and 28) contain the same sequence motif with Lys on −1 and Arg on −2 positions (Fig. 2).
Ser Phosphorylation Sites With Lys at −1 Position in the Secondary Structure of Proteins
The secondary structure prediction results of Ser phosphorylation sites of human H3 and further 103 other proteins suggested that the majority of these Ser residues were located in coiled regions with a small number in the helix region and with a very small fraction in the extended strands (Tables II and III). Sequence motifs with phosphorylated Ser having Lys on −1 position were located. Manual examination of protein sequences resulted in identification of four frequently occurring sequence motifs that is RKS, KKS, PKS, SKS, with K, P, R, and S representing the amino acids lysine, proline, arginine, and serine, respectively, related to phosphorylated Ser with Lys at position −1 (Table IV). Majority of these motifs were in coiled regions (Table IV).
| Total no. of Ser phosphorylation sites in 103 proteins | 124 |
| Phosphorylated Ser residues in coiled structure | 95 (77%) |
| Phosphorylated Ser residues in helix structure | 20 (16%) |
| Phosphorylated Ser residues in extended strands | 9 (7%) |
| Protein ID and amino acid position | Predicted kinases | Predicted methylation sites | Sequence motif | Secondary structure |
|---|---|---|---|---|
| Proteins with RKS motif | ||||
| P01589 [268] | S-268 PKC | QRKS | Extended strand | |
| S-268 PKA | ||||
| P02256 [14;18;22] | S-14 RSK | K13 | PRKS, PRKS, PKKS | Coil, coil |
| S-14 p38MAPK | K17 | Coil | ||
| S-14 PKC | ||||
| S-14 GSK3 | ||||
| S-14 cdk5 | ||||
| S-18 RSK | ||||
| S-18 PKC | ||||
| S-18 GSK3 | ||||
| S-18 cdk5 | ||||
| S-22 RSK | ||||
| S-22 PKC | ||||
| S-22 cdc2 | ||||
| S-22 GSK | ||||
| S-22 cdk5 | ||||
| P04625 [28] | S-28 PKA | KRKS | Coil | |
| P08567 [113] | S-113 PKC | K112 | ARKS | Helix |
| P09543 [9] | S-9 PKA | None | SRKS | Coil |
| P19491 [717] | S-717 RSK | VRKS | Coil | |
| S-717 PKC | ||||
| S-717 PKA | ||||
| S-717 PKG | ||||
| P21730 [314;334] | S-314 PKA | LRKS, ESKS | Coil, coil | |
| S-314 cdc2 | ||||
| S-334 PKC | ||||
| P22613 [8;35;39] | S-8 NP | KLKS, YRKS, SLKS | Coil, coil, coil | |
| S-35 RSK | ||||
| S-35 PKC | ||||
| S-39 PKC | ||||
| P30304 [293] | S-293 RSK | RRKS | Extended strand | |
| S-293 PKA | ||||
| S-293 PKG 0.53 | ||||
| P30443 [336] | S-336 RSK | RRKS | Coil | |
| S-336 PKC | ||||
| S-336 PKA | ||||
| S-336 PKG | ||||
| P38432 [184;202] | S-184 PKC | None | KRKS, NPKS | Coil, coil |
| S-184 GSK3 | ||||
| S-184 cdk5 0.51 | ||||
| S-202 GSK3 | ||||
| P54227 [62] | S-62 RSK | RRKS | Helix | |
| S-62 PKA | ||||
| P68431 [28] H3 | S-28 PKA | K9, K27 | ARKS, ARKS | Coil |
| S-28 PKG | ||||
| P84243 [10;28] | K9, K27 | ARKS, ARKS | Coil, coil | |
| Q14004 [340] | S-340 GSK3 | K339 | SRKS | Coil |
| Q14469 [37] | S-37 NP | HRKS | Coil | |
| Q15172 [28] | S-28 RSK | None | TRKS | Helix |
| S-28 PKC | ||||
| Q15906 [132] | S-132 RSK | K131 | SRKS | Helix |
| S-132 PKC | ||||
| Q9NQU5 [560] | S-560 PKA | None | KRKS | Extended strand |
| Proteins with KKS motif | ||||
| O14920 [705] | S-705 NP | AKKS | Helix | |
| P02256 [14;18;22] | S-14 RSK | PRKS, PRKS, PKKS | Coil, coil, coil | |
| S-14 p38MAPK | ||||
| S-14 PKC | ||||
| S-14 GSK3 | ||||
| S-14 cdk5 | ||||
| S-18 RSK | ||||
| S-18 PKC | ||||
| S-18 GSK3 | ||||
| S-18 cdk5 | ||||
| S-22 RSK | ||||
| S-22 PKC | ||||
| S-22 cdc2 | ||||
| S22 GSK3 | ||||
| S-22 cdk5 | ||||
| P06685 [23] | S-23 PKC | None | DKKS | Helix |
| P11168 [491;503] | S-491 NP | KGKS, QKKS | Coil, coil | |
| S-506 NP | ||||
| P12624 [161] | S-161 PKC | K160 | FKKS | Coil |
| S-161 PKG | ||||
| P16527 [127] | S-127 PKC | K126 | FKKS | Coil |
| S-127 PKG | ||||
| P25107 [467] | S-467 PKC | IKKS | Coil | |
| S-467 PKA | ||||
| P27573 [205;237] | S-205 PKC | None | FHKS, EKKS | Extended strand, helix |
| S-237 NP | ||||
| P29966 [162] | S-162 PKC | K161 | FKKS | Coil |
| S-162 PKG | ||||
| P30009 [155] | S-155 PKC | K154 | FKKS | Coil |
| S-155 PKG | ||||
| P41220 [64] | S-64 PKG | GKKS | Coil | |
| P47736 [484;490] | S-484 p38MAPK | PGKS, RKKS | Coil, coil | |
| S-484 GSK3 | ||||
| S-484 cdk5 | ||||
| S-490 RSK | ||||
| S-490 PKG | ||||
| P61224 [179] | S-179 RSK | RKKS | Coil | |
| S-179 PKA | ||||
| P61586 [188] | S-188 PKA | KKKS | Coil | |
| P62834 [180] | S-180 PKA | KKKS | Coil | |
| Q13002 [697] | S-697 PKC | FKKS | Coil | |
| S-697 PKA | ||||
| S-697 PKG | ||||
| Q13523 [23;277] | S-23 CKII | SEKS, GKKS | Helix, coil | |
| S-277 RSK | ||||
| S-277 PKA | ||||
| S-277 PKG | ||||
| S-277 cdk5 | ||||
| Q16666 [132] | S-132 RSK | K131 | RKKS | Helix |
| S-132 PKC | ||||
| S-132 PKA | ||||
| S-132 PKG | ||||
| Q5T200 [1010] | S-1010 RSK | RKKS | Coil | |
| S-1010 PKA | ||||
| S-1010 PKG | ||||
| Proteins with PKS motif | ||||
| Q01130 [211] | S-211 CKII | K210 | PPKS | Coil |
| S-211 GSK3 | ||||
| S-211 cdk5 | ||||
| O95684 [160] | S-160 p38MAPK | PPKS | Coil | |
| S-160 GSK3 | ||||
| S-160 cdk5 | ||||
| P10636 [551;712] | S-551 p38MAPK | PPKS, VYKS | Coil, extended strand | |
| S-551 GSK3 | ||||
| S-551 cdk5 | ||||
| S-712 p38MAPK | ||||
| S-712 GSK3 | ||||
| S-712 cdk5 | ||||
| P12839 [502;506;536;603; 608] | S-502 p38MAPK | VEKS, PVKS, GVKS, KAKS, VPKS | Coil, coil, helix | |
| S-502 GSK3 | Coil, coil | |||
| S-502 cdk5 | ||||
| S-506 GSK3 | ||||
| S-536 CKII | ||||
| S-603 GSK3 | ||||
| S-608 GSK3 | ||||
| S-608 cdk5 | ||||
| P23588 [93] | S-93 GSK3 | LPKS | Coil | |
| P33658 [430] | S-430 p38MAPK | QPKS | Coil | |
| S-430 GSK3 | ||||
| S-430 cdk5 | ||||
| P35568 [24;270] | S-24 PKC | KPKS, RSKS | Coil, coil | |
| S-270 RSK | ||||
| S-270 DNAPK | ||||
| S-270 PKB | ||||
| S-270 cdc2 | ||||
| P38432 [184;202] | S-184 PKC | KRKS, NPKS | Coil, coil | |
| S-184 GSK3 | ||||
| S-184 cdk5 | ||||
| S-202 GSK3 | ||||
| Q02224 [2570] | S-2570 p38MAPK | SPKS | Coil | |
| S-2570 GSK3 | ||||
| S-2570 cdk5 | ||||
| Q8N1K5 [584] | S-584 GSK3 | LPKS | Coil | |
| S-584 cdk5 | ||||
| Q15746 [1208] | S-1208 NP | RPKS | Coil | |
| Proteins with SKS motif | ||||
| Q9Y4H2 [306;915] | S-306 RSK | RSKS, EPKS | Coil, coil | |
| S-306 DNAPK | ||||
| S-306 PKB | ||||
| S-915 GSK3 | ||||
| S-915 cdk5 | ||||
| P21730 [314;334] | S-314 PKA | LRKS, ESKS | Coil, coil | |
| S-314 cdc2 | ||||
| S-334 PKC | ||||
| O88809 [306] | S-306 RSK | RSKS | Coil | |
| S-306 GSK3 | ||||
| S-306 cdk5 | ||||
| P33568 [24;270] | S-24 PKC | KPKS, RSKS | Coil, coil | |
| S-270 RSK | ||||
| S-270 DNAPK | ||||
| S-270 PKB | ||||
| S-270 cdc2 | ||||
| P18583 [910] | S-910 NP | GSKS | Coil | |
| P49792 [2280] | S-2280 GSK3 | PSKS | Coil | |
| S-2280 cdk5 | ||||
| P62753 [244] | S-244 RSK | TSKS | Coil | |
| S-244 PKC | ||||
| P70677 [26] | S-26 NP | |||
| Q9JLM8 [307] | S-307 RSK 0.60 | GSKS | Coil | |
| S-307 GSK3 0.50 | ||||
| S-307 cdk5 0.59 | RSKS | Coil | ||
| Q9UKV3 [384;386] | S-384 NP | LKEK, KSKS | Coil, coil | |
| S-386 GSK3 | ||||
| Q9Y4H2 [306;915] | S-306 RSK | RSKS, EPKS | Coil, coil | |
| S-306 DNAPK | ||||
| S-306 PKB | ||||
| S-915 GSK3 | ||||
| S-915 cdk5 | ||||
| Q9Y618 [2261] | S-2261 GSK3 | GSKS | Coil | |
| S-2261 cdk5 | ||||
| Proteins with XKS motif (X = any amino acid except K, R, S, P) | ||||
| O00499 [296] | S-296 GSK3 | GNKS | Coil | |
| O14746 [824] | S-824 PKA | RGKS | Coil | |
| O88498 [109] | S-109 NP | CDKS | Coil | |
| P02671 [576] | S-576 RSK | RGKS | Coil | |
| S-576 PKA | ||||
| S-576 PKG | ||||
| P04083 [26] | S-26 PKC | TVKS | Coil | |
| P06400 [811] | S-811 GSK3 | PLKS | Coil | |
| S-811 cdk5 | ||||
| P06730 [53] | S-53 NP | NDKS | Coil | |
| P07384 [360] | S-360 NP | ALKS | Coil | |
| P08651 [333] | S-333 p38MAPK | MDKS | Coil | |
| S-333 GSK3 | ||||
| S-333 cdk5 | ||||
| P12957 [717] | S-717 p38MAPK | GNKS | Coil | |
| S-717 GSK3 | ||||
| P14164 [624] | S-624 PKC | AHKS | Coil | |
| P14598 [283] | S-283 NP | LQKS | Coil | |
| P17306 [39] | S-39 PKC | SLKS | Coil | |
| P19112 [338] | S-338 PKA | KAKS | Coil | |
| P25090 [236] | S-236 NP | MIKS | Coil | |
| P28749 [749] | S-749 p38MAPK | KVKS | Coil | |
| S-749 GSK3 | ||||
| S-749 cdk5 | ||||
| P35831 [748] | S-748 NP | ITKS | Coil | |
| P51825 [588] | S-588 GSK3 | CQKS | Coil | |
| S-588 cdk5 | ||||
| P52926 [59] | S-59 RSK | KNKS | Coil | |
| S-59 GSK3 | ||||
| S-59 cdk5 | ||||
| P67870 [209] | S-209 cdk5 | NFKS | Coil | |
| Q00987 [186] | S-186 RSK | RHKS | Coil | |
| S-186 PKB | ||||
| S-186 PKA | ||||
| S-186 PKG | ||||
| Q01970 [537] | S-537 NP | PQKS | Coil | |
| Q04726 [245] | S-245 CKII | GDKS | Coil | |
| Q05682 [759] | S-759 p38MAPK | GNKS | Coil | |
| S-759 GSK3 | ||||
| S-759 cdk5 | ||||
| Q12888 [294] | S-294 NP | IQKS | Coil | |
| Q13887 [153] | S-153 ATM | LYKS | Coil | |
| Q15139 [738] | S-738 PKC | GEKS | Coil | |
| Q62736 [491;497] | S-491 p38MAPK | LTKS, GNKS | Coil | |
| S-491 GSK3 | ||||
| S-491 cdk5 | ||||
| S-497 p38MAPK | ||||
| S-497 GSK3 | ||||
| S-497 cdk5 | ||||
| Q92954 [373] | S-373 CKI | TIKS | Coil | |
| S-373 PKG | ||||
| Q99741 [106] | S-106 NP | TIKS | Coil | |
| Q9UNE7 [23] | S-23 GSK3 | PEKS | Coil | |
| S-23 cdk5 | ||||
| Q9UQ35 [901] | S-901 PKA | RVKS | Coil | |
| S-901 PKG | ||||
| Q9Y2W1 [320] | S-320 GSK3 | VGKS | Coil | |
| S-320 cdk5 | ||||
| P30301 [229] | S-229 RSK | RLKS | Coil | |
| S-229 PKA | ||||
| S-229 PKG | ||||
| P33535 [261] | S-261 RSK | RLKS | Helix | |
| S-261 PKC | ||||
| S-261 PKA | ||||
| P46020 [1007] | S-1007 PKC | QLKS | Helix | |
| P78536 [791] | S-791 NP | AAKS | Helix | |
| Q29502 [192] | S-192 NP | HTKS | Helix | |
| Q00960 [383] | S-383 PKA | KDKS | Extended strand | |
| P38398 [988] | S-988 PKC | PIKS | Extended strand | |
| Sequence motif | Coiled structure | Helix structure | Extended strands |
|---|---|---|---|
| RKS | 15 | 4 | 3 |
| KKS | 15 | 4 | — |
| PKS | 13 | — | — |
| SKS | 11 | — | — |
Phosphorylating Potential of Different Kinases on Ser With Lys at −1 Position in 103 Proteins
Kinases were predicted by NetPhosK 1.0 for the substrates of 124 known phosphorylated sites of 103 proteins retrieved from Phosphobase 3.0 for Ser, with neighboring Lys at −1 position. The predicted kinases that phosphorylate Ser residues with Lys at −1 position in 103 proteins included PKA, PKB, PKC, PKG, RSK, MAPK, cdc2, cdk5, CKI and II, GSK3, and DNAPK. The details of all 103 proteins, their secondary structure prediction results and phosphorylating kinase are given in Table III.
Methylation Potential on Lys at −1 Position of Phosphorylated Ser in 103 Proteins
The MeMo prediction results suggested that methylation of most of Lys at −1 position of phosphorylated Ser is favored by another basic amino acid on −2 position of phosphorylated Ser (Table V). The details of all 103 proteins, their secondary structure prediction results and methylation potential are provided in Table III.
| Sequence motif | Methylated Lys residues preceded by phosphorylated Ser | Functional class of proteins |
|---|---|---|
| RKS proteins, binding | 9 | H1, H3, cell cycle regulator |
| Transcription factors, DNA proteins, PKC | ||
| KKS | 5 | MARCKS family, interferon, actin, synopsin |
| PKS | 1 | Splicing factor |
| SKS | — | — |
Comparison of Human H3 With Human Histones H2A, H2B, and H4
The human histone H3 was aligned with the remainder core human histones to develop a relation between all four core histones. No appreciable sequence similarity was found in other core histones, except for H4, which showed highest sequence similarity as compared to H2A and H2B (Fig. 5).

Multiple alignments of human H2A, H2B, H3, and H4. The conserved site has an asterisk at the bottom, conserved substitution has a double dot and semiconserved substitution has a single dot. The different sequences are ordered as in aligned results for ClustalW. A sequence motif KRS in human H2B, which is similar to proposed sequence motif RKS in H3, at position Ser 33 and 88 (highlighted in blue) is observed. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Then it was investigated if the predicted sequence motif in human H3 (Table IV) also existed in the rest of core human histones. In human H2B, a similar sequence was found (KRS) at position Ser 33 and Ser 88 (Fig. 5). Phosphorylation and O-GlcNAc modification was predicted in H2B. As can be seen in Table VI, Ser 33 is predicted as Yin Yang site and Ser 88 showed potential for O-GlcNAc modification. When H2B was multiple aligned, it was found that Ser 33 is conserved in vertebrates, with a single substitution in Gallus gallus and Ser 88 was fully conserved in invertebrates and vertebrates (Fig. 6). Furthermore, both residues were predicted to be located in coiled regions by GOR IV (Fig. 7).
| Phosphorylated residue | O-Glycosylated residue | Yin Yang sites |
|---|---|---|
| Ser 7, 15, 33, 37, 39, 56, 92, 113, 124 | Ser 5, 7, 33, 88, 113, 124, 125 | Ser 7, 33, 113, 124 |
| Thr 89, 91, 97, 116 | Thr 53, 120, 123 | |
| Tyr 38, 41, 122 |

Multiple alignments of human H2B of six vertebrate sequences and seven invertebrate sequences. The conserved amino acids have an asterisk at the bottom, the conserved substitution is represented by a double dot and semiconserved substitution is represented by a single dot. The different sequences are ordered as in aligned results from ClustalW. It is observed that Ser 33 is fully conserved in vertebrates and Ser 88 is fully conserved in vertebrates and invertebrates. The sequence motif at position Ser 33 and Ser 88 are highlighted in yellow. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]

Secondary structure prediction of core histone H2B. It is observed that the phosphorylated Ser 33 and 88 are found in coiled regions similar to Ser 10 and 28 in human H3. The abbreviation stands for: c, coiled; h, helix; e, extended strand. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The details of sequence alignment of H3 with H2A, H2B, and H4; phosphorylation, O-GlcNAc modification, and Yin Yang sites of H2B; secondary structure prediction in H2B and multiple alignment of H2B in vertebrates and invertebrates are shown in Table VI and Figures 5-7.
Comparison of the Sequence Motif of O-GlcNAc Modification Sites in Human H3 With Experimentally Known Proteins
The sequence motif of the predicted Yin Yang sites, Ser 10 and 28, utilizing YinOYang 1.2 in human H3, was compared with proteins with experimentally known O-GlcNAc modification sites. These results are given in Figure 8. These results showed a similar sequence in experimentally known glycosylated proteins compared to the sequence of human histone H3 at Ser 10 and 28.
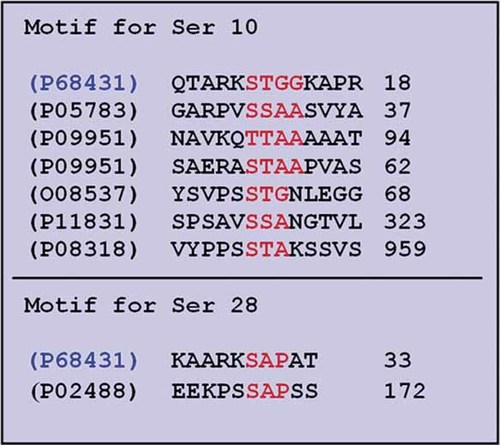
Experimentally known O-GlcNAc-modified protein manually extracted from the Swiss-Prot database [Boeckmann et al., 2003]. P68431 is the accession no. for human histone H3 (highlighted in blue), and the sequence at +1 and +2 positions (highlighted in red) next to Ser 10 and 28 (and +3 position in case of Ser 10, highlighted in red) are compared with experimentally known proteins. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
The different PTMs of H3 result in structural and functional changes. The importance of O-GlcNAc modification in H3 functionality has been put forward and it is suggested in silico that the dynamic intracellular phosphorylation and O-GlcNAc modification of human H3 on Ser 10, together with acetylation and methylation, participate in the control of IE-gene induction.
Phosphorylation sites in bovine H3 have been identified, which include Thr 6 or 11 and 118, Ser 10 and 28 [Zhang et al., 2003]. These are also positive prediction sites for phosphorylation of human H3 (Table I). In addition, these sites have also been predicted positively for the O-GlcNAc modification, i.e., Yin Yang sites (Table I). These Ser/Thr residues of H3 are conserved in all members of vertebrates and invertebrates (Fig. 2), a finding that increases their potential to act as Yin Yang sites, where both phosphorylation and O-GlcNAc modification can occur. Though H3 is almost conserved in all diverse groups of organisms, the Ser/Thr residues, which possess higher potential for O-phosphate and O-GlcNAc modification, can be identified by the 3D structural region of that Ser/Thr.
During mitosis, H3 phosphorylation on Ser 10 is crucial for chromosome condensation and progression of the cell cycle [Prigent and Dimitrov, 2003]. However, this regulation of H3 phosphorylation is affected by other PTMs, such as acetylation and methylation, during interphase [Berger, 2001], result in activation or repression of genes [Berger, 2001]. Phosphorylation of Ser 10 enhances acetylation of Lys 14 [Lo et al., 2000; Cheung et al., 2000b]. In the c-fos promoter, phosphorylated Ser 10 and acetylated Lys 9 can coexist on the same N-terminal of H3 [Edmondson et al., 2002]. Methylation of H3 can mediate transcriptional gene silencing and repression [Bernstein et al., 2002]. Acetylation is rapidly reversible, while methylation is more persistent and can occur even after transcription ceases, providing a memory of a recent transcription. Methylation of different Lys residues of H3 produces different or even opposite gene responses [Strahl et al., 1999; Bernstein et al., 2002; Saccani and Natoli, 2002; Stewart et al., 2005]. In addition to phosphorylation of Ser 10 of H3, a combination of acetylation and methylation on Lys 4, 9, and 14 is important in the induction or repression of IE genes.
Generally, transcriptionally active or silenced genes are associated with distinct combinations of histone PTMs. O-GlcNAc modification, a dynamic modification, has been reported to play a crucial role in chromatin remodeling [Love and Hanover, 2005]. O-GlcNAc transferase (OGT), the enzyme that catalyzes the addition of an O-GlcNAc moiety to the backbone of the protein on Ser and/or Thr residues [Love and Hanover, 2005], is an ubiquitous regulator of transcription, and displays flexibility in recognizing its many substrates [Yang et al., 2002]. The O-GlcNAc modification of same protein may affect different genes differently as for transcription factor Sp1, and may result in different outcomes depending on the type of cell and cellular signaling [Comer and Hart, 1999]. The O-GlcNAc-modified Sp1 induces transcriptional activation in Hela cells, and represses transcription in vascular muscle cells [Comer and Hart, 1999]. Similarly, O-GlcNAc modification of different proteins may result in different gene regulation. For example, O-GlcNAc modification of a transcription factor PDX-1 results in increased DNA binding and hence increased insulin secretion [Gao et al., 2003], whereas, transcriptional inhibition of certain genes is associated with O-GlcNAc modification of transcriptome directly or indirectly through O-GlcNAc modification of the proteasome [Bowe et al., 2006]. This suggests that O-GlcNAc modification plays different and sometimes contrasting roles in the regulation of gene expression through an interplay with phosphorylation. The OGT is recruited to the promoter region by the mSin3A-HDAC1 complex [Yang et al., 2002], where it modifies promoter-bound proteins like histones, RNA-polymerase II, c-Fos, c-Jun, and other transcriptional activators and thus exerts its eukaryotic gene-silencing activity [Lamarre-Vincent and Hsieh-Wilson, 2003; Majumdar et al., 2003; Tai et al., 2004; Toleman et al., 2004] by adding O-GlcNAc moieties on Ser and Thr residues. In some instances, O-GlcNAc modification of proteins induces transcription like in the case of the transcription factor STAT5 [Gewinner et al., 2004]. When STAT5 is O-GlcNAc modified, it interacts with the CREB-binding protein CBP (CBP is a transcriptional co-activator with intrinsic histone acetyltransferase activity) and thereby induces transcription [Gewinner et al., 2004].
OGT [Yang et al., 2002] together with O-GlcNAcase [Toleman et al., 2004] affect gene transcription in mammals. It is well documented that OGT and kinase compete for the same substrate amino acid residue, Ser/Thr [Love and Hanover, 2005] and an interplay of phosphorylation and O-GlcNAc modification on Ser 10 and 28 is therefore most likely to occur. This interplay of O-GlcNAc modification and phosphorylation on Ser 10 and 28 may result in IE-gene regulation.
Lys 9 methylation inhibits Lys 4 methylation on H3 in heterochromatic gene silencing [Noma et al., 2001], and methylation of Lys 9 and 27 has been documented to be involved in gene silencing [Lindroth et al., 2004]. It is quite interesting that the basic amino acid, Lys, is preceded (on the left or at −1 position with reference to Ser 10/28) by both Ser 10 and 28, described as Yin Yang sites. According to earlier reports, O-glycosylation of Ser is favored by Pro or a small or neutral amino acid side chain [Christlet and Veluraja, 2001]. Similarly, Ser in close vicinity to Pro is favored for phosphorylation [Iakoucheva et al., 2004; Qazi et al., 2006]. A small fraction of phosphorylated Ser also show basic amino acid residue Lys on −1 position [Qazi et al., 2006]. Furthermore, MAPKs and its effector proteins are known to catalyze phosphorylation of Ser in close vicinity of basic amino acids [Barsyte-Lovejoy et al., 2002]. In H3, both Ser 10 and 28 are preceded by Lys and both these residues are highly conserved in all organisms (Fig. 2). We retrieved 103 phosphorylated protein sequences data from Phosphobase 3.0 [Diella et al., 2004], with 124 phosphorylated Ser and Lys at −1 position. Secondary structure prediction by GOR IV [Garnier et al., 1996; Combet et al., 2000] showed that the phosphorylated Ser residues with Lys at −1 position resides predominantly in coiled structural regions (Table II), whereas, only a fraction was found in the alpha helical region and a very small number of sites were found to be located in extended strands (Table II). Coiled structural regions may provide more space for phosphate modifications in the presence of a bulky Lys residue at −1 position. Both of Ser 10 and 28 of H3 are found in the coiled region, hence attachment of O-GlcNAc on Ser 10 and 28 by OGT can easily result in phosphorylation blockade.
Repeated sequence motifs from secondary structural data of 124 phosphorylation sites were extracted manually. It is striking that four sequence motifs were most frequent: RKS, KKS, PKS, and SKS. The motif RKS sequence is present in all selected sequences of H3 from different species for both Ser 10 and 28 (Fig. 2). Among the other 103 proteins, the most highly repeated pattern was found to be RKS and KKS (Table IV). Table III shows predicted kinases for 124 phosphorylation sites by NetPhosK 1.0 [Blom et al., 2004]. Of the 124 predicted phosphorylation sites, 46 were found in basic amino acid rich motifs (KKS and RKS). From these, 16 phosphorylation sites were predicted by NetPhosK 1.0 [Blom et al., 2004] to be catalyzed by different MAPKs, 11 were found in coiled regions (Table III). This means that 69% of MAPK-catalyzed phosphorylation of Ser residues can be expected in coiled regions. Methylation prediction by MeMo [Chen et al., 2006] showed that, among all the RKS, KKS, PKS, and SKS motifs from all known phosphorylated Ser residues of 103 proteins, only one instance of Lys was found to have potential for methylation in the PKS motifs, no Lys was observed to have potential in the SKS motif, whereas the highest number of Lys with a potential for methylation was found in the RKS and KKS motifs (Table V). Thus, basic residues at position −1 and −2 are preferred for phosphorylation and methylation of adjacent residues. Functional analysis of the 103 phosphorylated proteins on Ser accompanied with Lys at −1 position showed that most of these proteins are nucleus specific (Table V). When human histone H3 was aligned with human core histone H2A, H2B, and H4, they showed very low sequence similarity (Fig. 5), even though the core histones are highly conserved across their entire sequence. When the RKS motif was searched in all core histones, it was found in H3 of all organisms (Fig. 2) but in H2B this motif was found only in one organism Rhacophorus schlegelii (Fig. 6). A similar sequence KRS was identified in human H2B at Ser 33 and Ser 88. Both these sites showed to be conserved in mammals and exhibited a potential for O-GlcNAc modification. Furthermore, Ser 33 also showed potential for both phosphorylation and O-GlcNAc modification (Yin Yang site) (Table VI). Phosphorylation of Ser 33 is essential for transcriptional activation in eukaryotes. It is phosphorylated by the transcription factor TAF1 that is part of the protein complex TFIID in Drosophila [Maile et al., 2004]. Both residues, Ser 33 and Ser 88, were predicted to be located in coiled regions (Fig. 7) as predicted in H3 for Ser 10 and 28 (Table II). This suggests that the sequence motifs containing phosphorylated Ser in the vicinity of basic amino acids, Lys and Arg, at −1 and −2 positions, are important in gene regulation. The phosphorylated Ser 10 of human histone H3 is followed by the amino acids Thr at +1 and Gly and +2 positions (Fig. 2). In case of phosphorylated Ser 28, Ala at +1 and Pro at +2 positions are found (Fig. 2). These sequences, STG and SAP, were compared with experimentally known O-GlcNAc-modified proteins retrieved from the Swiss-Prot database [Boeckmann et al., 2003]. It was observed that several proteins contained a similar though not identical upstream sequence environment like that of Ser 10 (Fig. 8). Together, these results suggest that the sequence motif STG may provide space for OGT to add an O-GlcNAc moiety to the protein. Furthermore, these results indicate that O-GlcNAc modification is most likely to take place at Ser 10 (and Ser 28) of human histone H3.
On the basis of in silico data, we propose that a specific combination of different modifications (phosphorylation, acetylation, methylation, and O-GlcNAc modification) control the activation and repression of genes including the IE genes. It is quite obvious that methylation on Lys 9 results in IE-gene silencing, whereas phosphorylation on Ser 10, acetylation on Lys 9 and Lys 14 might regulate IE-gene induction, along with methylation on Lys 4, suggesting the following sequence of events: when phosphorylation of Ser 10 is blocked by the presence of O-GlcNAc modification, Lys 9 is methylated. Similarly, phosphorylation of Ser 10 and acetylation of Lys 9 and Lys 14 are involved in IE-gene activation. On the contrary, O-GlcNAc modification on Ser 10 and methylation on Lys 9 may lead to gene repression. Thus, a specific combination of different PTMs on Ser/Thr and Lys, involving Ser 10, regulate IE-gene expression and repression. In addition, the interplay of phosphorylation and O-GlcNAc modification emerges to be the regulator of other PTMs.
Acknowledgements
Nasir-ud-Din acknowledges partial financial support from Pakistan Academy of Sciences, and support from Dr. T.A. Khawaja to IMSB.




