Regulation of arginase-1 expression in macrophages by a protein kinase a type I and histone deacetylase dependent pathway
Abstract
The aim of the current study was to investigate the cAMP-dependent regulation of arginase-1 (ARG1) expression in RAW-macrophages. Basal ARG1 mRNA expression was low and increased upon incubation with the cAMP analogue Br-cAMP. We used selective agonists of protein kinase A type I (PKAI), type II (PKAII) and exchange protein directly activated by cAMP (EPAC) to determine the pathway responsible for ARG1 expression. Activation of PKAI led to a significant up-regulation of ARG1 mRNA expression and arginase enzyme activity. In contrast, neither activation of PKAII nor activation of EPAC affected ARG1 expression. In addition, it has been shown that histone deacetylase (HDAC) activity plays a critical role in cAMP-dependent transcriptional regulation. Incubation with Br-cAMP and the HDAC inhibitor trichostatin A (TSA) led to a concentration-dependent suppression of ARG1 expression. These data indicate that cAMP-dependent activation of ARG1 expression is mediated by PKAI and requires histone deacetylation. J. Cell. Biochem. 103: 520–527, 2008. © 2007 Wiley-Liss, Inc.
Arginase-1 (ARG1) is a key hydrolytic enzyme in the urea cycle, catalyzing the conversion of L-arginine to ornithine and urea, thus eliminating excess nitrogen from the body [Jenkinson et al., 1996]. It is highly expressed in liver, but also found in non-hepatic cells, particularly macrophages [Jenkinson et al., 1996; Yu et al., 2003; Teupser et al., 2006]. In macrophages, ARG1 activation may limit the availability of L-arginine as a substrate for inducible nitric-oxide synthase (iNOS), thereby inhibiting NO-secretion from these cells [Munder et al., 1998, 1999]. In addition, metabolism of ornithine, the primary metabolite of the arginase pathway leads to the formation of polyamines, which display distinct anti-inflammatory effects [Zhang et al., 1997]. Thus, increased arginase activity may have anti-inflammatory properties and might be protective in atherosclerosis and inflammatory disease [Getz and Reardon, 2006; Teupser et al., 2006]. The concept of ARG1 as an anti-inflammatory mediator is supported by the induction of the gene by the Th2 cytokines IL-4, IL-10, and IL-13 [Munder et al., 1998]. In contrast, Th1 cytokines inhibit ARG1 expression [Munder et al., 1998]. Th2-mediated induction of ARG1 is regulated by the assembly of PU.1, IL-4 induced STAT6, and C/EBPβ at an enhancer that lies approximately 3 kb from the mouse basal promoter [Pauleau et al., 2004]. Another potent mechanism of ARG1 induction is via protein kinase A (PKA). Previous studies have shown that the cAMP-analogue Br-cAMP as well as activators of adenylate cyclase significantly up-regulate ARG1 expression and activity [Morris et al., 1998; Hammermann et al., 2000]. In general, PKA-activity may be mediated through two subtypes, PKAI and PKAII as well as exchange protein directly activated by cAMP (EPAC). However, the exact mode of ARG1 induction is presently unknown. In addition, recent studies have demonstrated that histone acetylation plays an important role in cAMP activation of a subset of PKA-dependent genes [Fass et al., 2003]. Thus, the aim of the present study was to elucidate the pathway of PKA-mediated ARG1 induction and the role of histone acetylation in this process.
MATERIALS AND METHODS
Cell Culture
The murine macrophage cell line RAW 264.7 was cultivated in Dulbecco's Modified Eagle Medium (containing 10% fetal bovine serum) at 37°C, 5% CO2. For experiments, 106 cells were grown over night in 60 mm dishes and then activated with cAMP analogues. 8-Bromoadenosine-3′,5′-cyclic-monophosphate (Br-cAMP) was obtained from Sigma. The histone deacetylase (HDAC) inhibitors Trichostatin A (TSA) and MS-275 were obtained from Calbiochem and Sigma, respectively. 8-(6-aminohexylamino)adenosine-3′,5′-monophosphate (AHA-cAMP), N6-benzoyladenosine-3′,5′-cyclic monophosphate (Bnz-cAMP), 8-piperidinoadenosine-3′,5′-cyclic monophosphate (PIP-cAMP) and 8-(4-methoxyphenylthio)-2′-O-methyladenosine 3′,5′-cyclic monophosphate (Me-cAMP) were obtained from Biolog, Germany. cAMP-analogues were added to the cells as detailed in the figure legends.
RNA-Isolation and cDNA Synthesis
Total RNA was isolated using the monophasic Trizol reagent according to the manufacturer's instructions (Invitrogen). Reverse transcription was performed with 2 µg of RNA and superscript II enzyme (Invitrogen).
Quantitative TaqMan RT-PCR
Sequences of specific TaqMan primers and probes are provided in Table I in the online-supplement. Probes were obtained from Eurogentec, Belgium and 5′-labeled with FAM and 3′-labeled with TAMRA. TaqMan probes hybridized to two exons to avoid amplification of possible DNA contamination. Cyclophilin A was used as a house-keeping gene [Trogan et al., 2002]. PCR reactions were performed with AmpliTaq Gold Taq polymerase (Applied Biosystems) in 40 two step cycles of 95°C, 15 s and 60°C, 60 s. Serial dilutions of cloned cDNA fragments of the target genes were used for standardization. Expression levels of all genes were normalized to 106 copies cyclophilin A.
| Control | Br-cAMP | PKAI | PKAII | EPAC | |
|---|---|---|---|---|---|
| ABCA1 | 1.0 | 7,360.0* | 4,498.0* | 25.3* | 1.1 |
| c-fos | 1.0 | 79.3* | 228.2* | 134.8* | 78.6* |
| AhR | 1.0 | 24.4* | 17.0* | 2.1* | 0.5 |
| IκBα | 1.0 | 3.4* | 3.1* | 1.5 | 1.1 |
- Br-cAMP (0.5 mM), PKAI activation: Bnz-cAMP and AHA-cAMP (0.1 mM, each), PKAII activation: Bnz-cAMP and PIP-cAMP cAMP (0.1 mM, each), EPAC activation: Me-cAMP (0.1 mM).
- * P < 0.05 compared to control.
Arginase Activity Assay
Cell were washed twice in PBS, lysed in 0.5% Triton-X containing Protease Inhibitor Cocktail Tablets (Roche). Assays were performed as described [Ochoa et al., 2000].
Western Blotting
Cells were lysed with 1% SDS, 1.0 mM sodium ortho-vanadate, 10 mM Tris pH 7.4 and 20 µg protein were size-fractionated on 12% SDS polyacrylamide gels by electrophoresis. Gels were blotted onto a PVDF membrane and blocked (5% BSA in 20 mM Tris, 140 mM NaCl2, 0.1% NP 40) overnight at 4°C. Protein kinase A (PKA) was detected using mouse PKA-RI alpha (PKARI) or mouse PKA-RII alpha (PKARII) antibodies (BD). Alkaline phosphatase-conjugated anti-mouse-immunglobulin (Dako) served as secondary antibody and were visualized using the BCIP/NBT substrate kit (Vector Laboratories).
Statistical Analysis
Statistical evaluation was performed using Student's t-test.
RESULTS
Induction of ARG1-Expression is PKAI-Dependent
Incubation of RAW macrophages with Br-cAMP led to a significant, concentration dependent activation of ARG1 mRNA expression (Fig. 1), confirming previously published findings. Br-cAMP is a general activator of the two PKA-isoforms, PKAI and PKAII and exchange protein directly activated by cAMP (EPAC), indicating that induction of ARG1-expression is dependent on activation of either PKA or EPAC. However, it was unknown which of these pathways played a role in this process. We thus used combinations of cAMP-analogues to selectively activate PKAI, PKAII, and EPAC. As shown in Figure 2A, only selective activation of PKAI using a combination of Bnz-cAMP and AHA-cAMP (0.1 mM, each) led to a significant, sevenfold increase of ARG1 mRNA expression (P < 0.01). In contrast, neither activation of PKAII with equimolar concentrations of Bnz-cAMP and PIP-cAMP (Fig. 2B) nor activation of EPAC with Me-cAMP (Fig. 2C) had an effect on ARG1 expression. To determine whether primary or secondary responses to the various stimuli were responsible for ARG1 activation we also performed a time-course experiment and determined ARG1 expression at 1, 2, 4, 8, and 16 h. ARG1 was up-regulated as early as 2 h after activation with Br-cAMP and Bnz-cAMP/AHA-cAMP, suggesting a primary response of ARG1 to these stimuli (Fig. 3).
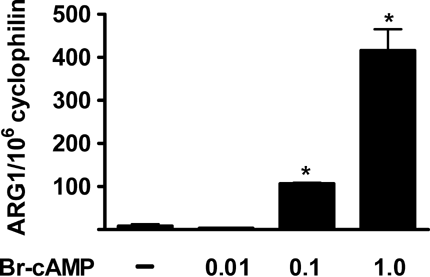
Induction of ARG1 mRNA expression by Br-cAMP. RAW macrophages were incubated with Br-cAMP (0, 0.01, 0.1, 1.0 mM) for 16 h. mRNA was determined by quantitative fluorogenic RT-PCR and expressed as copies ARG1 mRNA normalized to 106 copies cyclophilin. * indicates P < 0.01 compared to control.
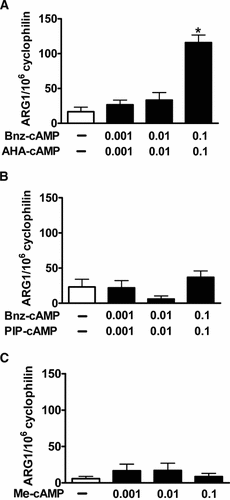
Effect of cAMP-analogues on ARG1 mRNA expression. RAW macrophages were incubated with (A) Bnz-cAMP and AHA-cAMP for induction of PKAI, (B) Bnz-cAMP and PIP-cAMP for induction of PKAII, and (C) Me-cAMP for induction of EPAC at 0, 0.001, 0.01, 0.1 mM for 16 h. mRNA was determined by quantitative fluorogenic RT-PCR and expressed as copies ARG1 mRNA normalized to 106 copies cyclophilin. * indicates P < 0.01 compared to control.
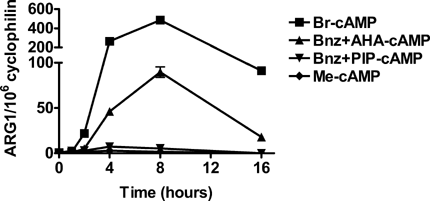
Effect of cAMP-analogues on ARG1 mRNA expression. RAW macrophages were incubated for 1, 2, 4, 8, or 16 h with (0.5 mM) Br-cAMP, Bnz-cAMP, and AHA-cAMP (0.1 mM, each) for induction of PKAI, Bnz-cAMP, and PIP-cAMP (0.1 mM, each) for induction of PKAII and Me-cAMP (0.1 mM) for induction of EPAC. mRNA was determined by quantitative fluorogenic RT-PCR and expressed as copies ARG1 mRNA normalized to 106 copies cyclophilin.
We tested by Western blotting whether PKAI and PKAII were expressed in RAW cells and whether they were influenced by the various stimuli added to the cells. As shown in Figure 4, PKAI and PKAII were indeed expressed in RAW-cells and expression remained constant after incubation with cAMP-analogues.
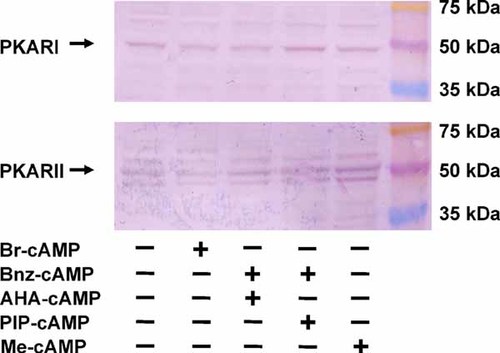
Western blot detection of PKAI and PKAII after incubation with Br-cAMP (0.5 mM) or Bnz-cAMP and AHA-cAMP (0.1 mM, each), Bnz-cAMP and PIP-cAMP (0.1 mM, each), and Me-cAMP (0.1 mM) for 16 h.
We next determined whether induction of ARG1 mRNA was reflected on the protein level by measuring arginase activity in RAW macrophages. As shown in Figure 5, arginase activity increased 18-fold after activation with Br-cAMP, and 12-fold after selective activation of PKAI by co-incubation with Bnz-cAMP and AHA-cAMP (P < 0.05 compared to control). These data show that activation of ARG1-mRNA both by Br-cAMP and by selective activation of PKAI also led to an increase of arginase enzyme activity.
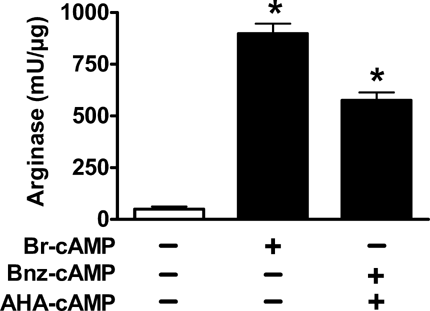
Effect of cAMP-analogues on arginase enzyme activity. RAW macrophages were incubated with 0.5 mM Br-cAMP, 0.1 mM Bnz-cAMP and 0.1 mM AHA-cAMP for 16 h, * indicates P < 0.05 compared to control.
To test whether the mode of transcriptional activation via PKAI was specific for ARG1 or also seen for other target genes containing consensus sites for cAMP-dependent transcription factors we next determined expression of a number of these genes (Table I). mRNA expression of ABCA1, c-fos, aryl-hydrocarbon-receptor (AhR) and NF-kappa-B inhibitor alpha (IκBα) was activated using Br-cAMP. Increased expression levels of ABCA1, AhR and IκBα were mainly due to activation by PKAI (Table I). PKAII clearly mediated mRNA-expression of c-fos, whereas induction of ABCA1 and AhR was low and might have been due to cross-reactivity of Bnz-cAMP and PIP-cAMP with PKAI. In addition, c-fos was also the only gene tested, that was significantly activated by EPAC.
Deacetylase Activity is Required for cAMP Activation of ARG1
Previous studies have shown that histone deacetylases differentially regulate cAMP-inducible target genes [Fass et al., 2003]. To test the role of HDACs in the transcriptional regulation of ARG1, we treated RAW-cells with the HDAC inhibitor trichostatin A (TSA) and simultaneously activated the cells with Br-cAMP. As shown in Figure 6A, TSA suppressed ARG1 activation in a dose-dependent manner. Incubation with TSA at 10 ng/mL led to a partial suppression of ARG1 activation, incubation with TSA at 100 ng/mL led to a complete suppression of ARG1 activation (Fig. 6A). Comparable effects were observed after selective activation of ARG1 via PKAI using Bnz-cAMP and AHA-cAMP (0.1 mM, each) and simultaneously incubating with TSA (1-100 ng/mL) (data not shown). TSA alone without addition of cAMP-analogues had no effect on ARG1 expression (data not shown). In contrast to ARG1 expression, TSA had no influence on c-fos-mRNA expression in cells stimulated with Br-cAMP (Fig. 6B). The effects of TSA on mRNA expression of ARG1 were also reflected on the activity level. As shown in Figure 7, Br-cAMP-activated RAW-cells incubated with TSA (10 ng/mL) significantly reduced arginase activity. In contrast, incubation of non-activated cells with TSA did not alter arginase activity compared to control. Since TSA is an inhibitor of HDACs, these data indicate that deacetylase activity is required for cAMP activation of ARG1.
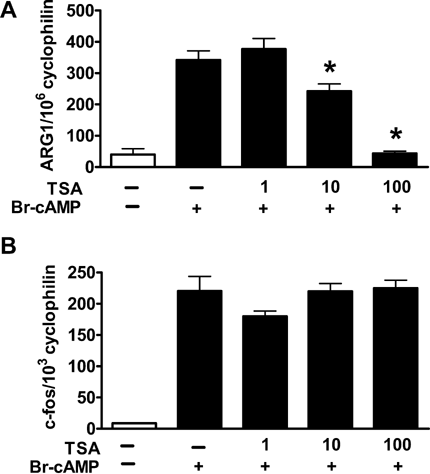
Effect of the HDAC-inhibitor TSA on ARG1 (A) and c-fos (B) mRNA expression. RAW macrophages were incubated with Br-cAMP (0.5 mM) and increasing concentrations of TSA (0, 1, 10, 100 ng/mL) for 16 h. * indicates P < 0.01 compared to cells incubated with 0.5 mM Br-cAMP only.
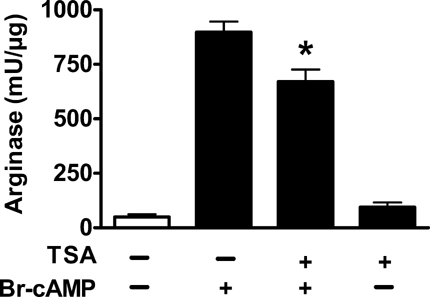
Effect of the HDAC-inhibitor TSA on arginase enzyme activity. RAW macrophages were incubated with 0.5 mM Br-cAMP for 16 h, * indicates P < 0.05 compared to control.
To validate these findings, we repeated our experiments using MS-275 as an alternative HDAC-inhibitor. We found that ARG1-mRNA expression of Br-cAMP-activated cells was significantly reduced by 84% (10 µM MS-275), confirming our finding that ARG1 mRNA expression was dependent on HDAC-activity.
We then tested the effects of HDAC inhibition on expression levels of other genes. TSA alone without addition of cAMP-analogues led to a significant activation of expression of ABCA1, c-fos, and AhR, while expression of IκBα was down-regulated (Table II).
| TSA | Br-cAMP + TSA | PKAI + TSA | PKAII + TSA | EPAC + TSA | |
|---|---|---|---|---|---|
| ABCA1 | 33.0* | 3,236.2* | 1,631.5* | 347.3* | 259.8* |
| c-fos | 40.1* | 58.0 | 203.8 | 97.5 | 168.3* |
| AhR | 3.0* | 70.4* | 64.1* | 17.1* | 14.5* |
| IκBα | 0.6* | 5.2 | 6.8 | 2.5* | 2.5* |
- Br-cAMP (0.5 mM), PKAI activation: Bnz-cAMP and AHA-cAMP (0.1 mM, each), PKAII activation: Bnz-cAMP and PIP-cAMP cAMP (0.1 mM, each), EPAC activation: Me-cAMP (0.1 mM).
- * P < 0.05 compared to incubation without TSA (see Table I).
When TSA was added to activators of PKAI, expression of ABCA1 was inhibited in a similar way as expression of ARG1. In contrast AhR expression was increased in response to TSA, whereas TSA had no effect on the activation pattern of c-fos and IκBα.
DISCUSSION
The question addressed in the present study was to elucidate the pathway of cAMP-dependent ARG1 induction in macrophages and the role of histone acetylation in this process. We found that ARG1 was induced by a PKAI dependent pathway. In addition, histone deacetylase activity was required for PKAI-mediated activation of ARG1 expression.
Induction of ARG1 by cAMP has first been demonstrated in liver [Nebes and Morris, 1988; Morris et al., 1989]. Later studies have shown that the cAMP-analogue Br-cAMP as well as activators of adenylate cyclase significantly up-regulate ARG1 expression and activity in macrophages [Morris et al., 1998; Hammermann et al., 2000]. Consistent with these findings, analysis of the ARG1 promoter region had revealed a putative cAMP-responsive element (CRE) approximately 280 base-pairs upstream of the transcription start site [Takiguchi et al., 1988]. Interestingly, also cytokine-mediated induction of ARG1 by IL-13 was preceded by a transient increase of intracellular cAMP, suggesting that the underlying mechanism of ARG1-induction is cAMP dependent while other pathways of ARG1-induction appear to be cAMP-independent [Chang et al., 2000]. We confirmed in our experimental system that ARG1 could be activated by incubation of RAW-macrophages with Br-cAMP (Fig. 1). PKA-activity is mediated by two subtypes, designated PKAI and PKAII as well as EPAC. In its inactive state, PKA exists as a tetrameric holoenzyme composed of two regulatory and two catalytic subunits. The two classes of PKA isoenzymes, types I and II differ in their subunits, designated RI and RII, respectively [Kopperud et al., 2003]. cAMP binds cooperatively to two sites termed A and B on each regulatory subunit. Binding of four cAMP molecules, two at each subunit, leads to a conformational change and release of the catalytically active subunits. The catalytic subunits released from the holoenzyme subsequently enter the nucleus by passive diffusion, activating transcription of cAMP-dependent genes by phosphorylation of the transcription factor cAMP response element binding protein (CREB) [Kopperud et al., 2003; Tasken and Aandahl, 2004]. Striking differences in the binding affinity of cAMP analogues to cAMP binding sites A and B between the regulatory subunits RI and RII have been reported [Christensen et al., 2003]. We used this effect for selective activation of PKAI, PKAII and EPAC to identify the mechanism leading to ARG1 expression. PKAI was activated by combining Bnz-cAMP with AHA-cAMP. Bnz-cAMP is an activator of the A-site of both regulatory subunits RI and RII (Ki 4.0 and 3.8, respectively), while AHA-cAMP has a selective affinity for the B-site of the regulatory subunit RI (Ki 4.1) but not for the B-site of the regulatory subunit RII (Ki 0.39), thus predominantly activating PKAI [Christensen et al., 2003]. Activation of PKAI using these agents at equimolar concentrations led to a significant increase of ARG1 expression (Fig. 2A). Induction of ARG1 mRNA expression was also reflected on the level of arginase activity (Figs. 1, 2 and 5). Incubation with Br-cAMP led to a 30-fold increase of ARG1 mRNA and a 18-fold increase of arginase activity, while incubation with Bnz-cAMP/AHA-cAMP led to 7-fold increase of ARG1 mRNA and 12-fold increase of arginase activity. This discrepancy in fold-induction of ARG1 mRNA expression and arginase activity might be explained by low basal expression of ARG1, where minor differences in the measurement of basal expression have a major impact on relative expression levels.
In a time-course experiment, ARG1 was up-regulated as early as 2 h after activation with Br-cAMP and a combination of Bnz-cAMP and AHA-cAMP, suggesting a primary response of ARG1 expression to these stimuli (Fig. 3).
In contrast, neither activation of PKAII nor activation of EPAC led to a significant increase of ARG1 expression. PKAII was activated by combining Bnz-cAMP with PIP-cAMP. The latter has a selective affinity for the B-site of the regulatory subunit RII (Ki 2.7) but not for the B-site of the regulatory subunit RI (Ki 0.06), thus only activating PKAII [Christensen et al., 2003]. Me-cAMP was used for specific activation of EPAC (Ki 7.1). Since PKAI and PKAII were expressed in RAW macrophages (Fig. 4), these data suggest that cAMP-dependent activation of ARG1 was mediated specifically by PKAI. As a limitation of our study and others [Skalhegg et al., 1992; Braun et al., 1998; van Bemmelen et al., 2005; Tan et al., 2007] we should mention that selective stimulation of EPAC and PKA-subtypes with combinations of cAMP-analogues in cells has not been directly proven and is an extrapolation from the purified system [Christensen et al., 2003].
Surprisingly little is known about the specific role of the two subtypes of PKA and EPAC in cAMP-dependent gene activation. We thus compared the mode of activation of ARG1 with activation of a number other known cAMP target genes. To this end, we confirmed that mRNA expression of ABCA1 [Smith et al., 2004], c-fos [Fass et al., 2003], AhR [Oesch-Bartlomowicz et al., 2005] and IκBα [Kamthong and Wu, 2001] was activated in a cAMP-dependent manner. However, we showed that the specific response to activation via PKAI, PKAII or EPAC was quite different for these genes. The pattern of ARG1-activation most closely resembled the pattern of activation of ABCA1, AhR and to a lesser extent IκBα, which were all predominantly induced via PKAI (Table I). In contrast, c-fos was equally inducible via PKAI, PKAII, and EPAC. These differences might be due to differences in cAMP-responsive elements in the promoter regions of these genes. It is of interest, that the major-pathway of PKA-dependent gene activation in four of the five genes tested was mediated by PKAI.
Previous studies have shown that histone deacetylation plays a role in regulating cAMP-dependent genes. Phosphorylated CREB does not only bind to cAMP-response elements (CREs) in promoters of cAMP-regulated genes but also promotes the recruitment of the transcriptional co-activator CREB binding protein (CBP) [Chrivia et al., 1993]. After its recruitment, CBP is proposed to mediate activation of target genes through its association with RNA polymerase II complexes and through its intrinsic histone acetlytransferase activity that acetylates promoter bound histones [Mayr and Montminy, 2001]. Other studies have shown that HDAC-1 blocks phosphorylation of CREB, thus attenuating cAMP-response, while HDAC inhibitors potentiate CREB activity [Canettieri et al., 2003].
To test the role of HDACs in the transcriptional regulation of ARG1, we treated RAW-cells with the HDAC inhibitor TSA and simultaneously activated the cells with Br-cAMP. As shown in Figure 6A, TSA suppressed ARG1 activation in a dose-dependent manner. This was unexpected, since TSA has been shown to enhance activation of cAMP-dependent genes [Canettieri et al., 2003; Lu et al., 2003]. However, suppression of ARG1 activation could be confirmed with a different HDAC-inhibitor, MS-275. We thus compared the effect of TSA on ARG1-expression to other cAMP-dependent genes and found that some where activated while others were suppressed (Table II). Comparable results have been obtained by Fass et al. 2003 systematically testing the role of HDAC-activity in a microarray study. These authors showed that HDACs differentially regulated CREB target genes by contributing to either activation of cessation of transcription. They speculate that the promoters of genes activated by HDAC-inhibition are in a state of readiness to bind the preinitiation complex and rapidly begin transcription. In the case of genes suppressed by HDAC-inhibition, initiation factor TFIIB and RNA polymerase II are not bound to the promoter under basal conditions and TSA does not promote the recruitment of these two proteins [Fass et al., 2003]. The latter might be the reason for suppression of ARG1 gene expression by TSA.
In conclusion, we found that ARG1 was induced by a protein kinase A type I dependent pathway. In addition, histone deacetylase activity was required for PKA-mediated activation of ARG1 expression.
Acknowledgements
We thank Franziska Jeromin, Claudia Weise, and Wolfgang Wilfert for expert technical assistance. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) grant Th 374/2-3 to Dr. Joachim Thiery.




