Essential role of PSM/SH2-B variants in insulin receptor catalytic activation and the resulting cellular responses
Abstract
The positive regulatory role of PSM/SH2-B downstream of various mitogenic receptor tyrosine kinases or gene disruption experiments in mice support a role of PSM in the regulation of insulin action. Here, four alternative PSM splice variants and individual functional domains were compared for their role in the regulation of specific metabolic insulin responses. We found that individual PSM variants in 3T3-L1 adipocytes potentiated insulin-mediated glucose and amino acid transport, glycogenesis, lipogenesis, and key components in the metabolic insulin response including p70 S6 kinase, glycogen synthase, glycogen synthase kinase 3 (GSK3), Akt, Cbl, and IRS-1. Highest activity was consistently observed for PSM alpha, followed by beta, delta, and gamma with decreasing activity. In contrast, dominant-negative peptide mimetics of the PSM Pro-rich, pleckstrin homology (PH), or src homology 2 (SH2) domains inhibited any tested insulin response. Potentiation of the insulin response originated at the insulin receptor (IR) kinase level by PSM variant-specific regulation of the Km (ATP) whereas the Vmax remained unaffected. IR catalytic activation was inhibited by peptide mimetics of the PSM SH2 or dimerization domain (DD). Either peptide should disrupt the complex of a PSM dimer linked to IR via SH2 domains as proposed for PSM activation of tyrosine kinase JAK2. Either peptide abolished downstream insulin responses indistinguishable from PSM siRNA knockdown. Our results implicate an essential role of the PSM variants in the activation of the IR kinase and the resulting metabolic insulin response. PSM variants act as internal IR ligands that in addition to potentiating the insulin response stimulate IR catalytic activation even in the absence of insulin. J. Cell. Biochem. 103: 162–181, 2008. © 2007 Wiley-Liss, Inc.
An src homology 2 (SH2) domain-containing sequence termed SH2-B had been identified in the rat [Osborne et al., 1995] and the corresponding mouse and rat proteins, respectively, based on their association with the activated catalytic insulin receptor (IR) domain [Riedel et al., 1997] or as substrates of activated JAK2 [Rui et al., 1997]. Pro-rich putative SH3 domain binding sequences, a pleckstrin homology region (PH) and a SH2 domain implicated a role as a signaling mediator also termed PSM [Riedel et al., 1997] or SH2B1 [Maures et al., 2006]. A total of four alternative splice variants of PSM/SH2-B have been reported in the mouse termed alpha, beta, gamma, and delta [Riedel et al., 1997; Rui et al., 1997; Nelms et al., 1999; Yousaf et al., 2001] and in the human genome [Nishi et al., 2005]. A single SH2-B gene has been mapped to the distal arm of mouse chromosome 7 in a region linked to obesity in mice [Nelms et al., 1999]. In particular, the beta variant was described as a substrate and as a potent cytoplasmic activator of JAK2 in response to growth hormone signaling [Rui et al., 1997, 2000; Rui and Carter-Su, 1999; Carter-Su et al., 2000a,b; Kurzer et al., 2004; Miquet et al., 2005]. Activation of the tyrosine kinase receptors for insulin, insulin-like growth factor-I (IGF-I), platelet-derived growth factor (PDGF), hepatocyte growth factor (HGF), fibroblast growth factor (FGF), or TrkA for nerve growth factor (NGF) resulted in their association with the SH2 domain of PSM/SH2-B, suggesting a putative role in the respective signaling pathways [Wang and Riedel, 1998; Riedel et al., 2000]. Tyrosine phosphorylation of PSM/SH2-B was observed in response to insulin [Kotani et al., 1998], IGF-I, and PDGF [Yousaf et al., 2001]. PDGF stimulated PDGF receptor association with and phosphorylation of PSM/SH2-B on tyrosine, serine, and threonine [Rui and Carter-Su, 1998; Riedel et al., 2000; Yousaf et al., 2001]. cDNA expression of the four PSM/SH2-B variants differentially stimulated the mitogenic response to PDGF, IGF-I, and insulin whereas introduction of cell-permeant, putatively dominant-negative SH2 domain or Pro-rich peptide mimetics interfered with the mitogenic response [Riedel et al., 2000; Yousaf et al., 2001]. Microinjection of an SH2 domain peptide mimetic into transformed fibroblasts partially restored a normal actin stress fiber pattern suggesting a requirement of PSM/SH2-B in phenotypic cell transformation [Riedel et al., 2000]. PSM/SH2-B beta is required for growth hormone-induced actin rearrangement [Herrington et al., 2000] and regulates cellular motility [Diakonova et al., 2002] and membrane ruffling [O'Brien et al., 2003]. PSM/SH2-B interacts with the activation loop of TrkA [Koch et al., 2000] or TrkB [Suzuki et al., 2002] and plays a specific role in TrkA-mediated differentiation [Eggert et al., 2000; Chen and Carter-Su, 2004] with a specific role of PSM/SH2-B shown in the Akt/Forkhead signaling pathway [Wang et al., 2004]. In response to fibroblast growth factor receptor 3 (FGFR3) activation, PSM/SH2-B associates and undergoes tyrosine phosphorylation with a putative role in the activation and nuclear translocation of Stat5 [Kong et al., 2002]. In response to leptin PSM/SH2-B promotes IRS1- and IRS2-mediated activation of the PI 3-kinase pathway [Duan et al., 2004a]. PSM/SH2-B is a key regulator of adipogenesis by regulating the insulin/IGF-I receptor-Akt-Foxo1-PPARγ pathway [Yoshiga et al., 2007]. SH2-B gene polymorphism correlates with serum leptin and body fat [Jamshidi et al., 2007]. Disruption of the SH2-B gene suggests a critical role in the IGF-I-mediated reproductive pathway in mice [Ohtsuka et al., 2002] and as a physiologic enhancer of IR activation as well as a role in the maintenance of normal insulin sensitivity and glucose homeostasis during ageing [Duan et al., 2004b]. PSM/SH2-B shares a high degree of structural similarity with scaffold proteins, Lnk, and APS that participate in B cell receptor and T cell receptor signaling, respectively [Huang et al., 1995; Yokouchi et al., 1997; Iseki et al., 2000; Rudd, 2001; Velazquez et al., 2002; Hu and Hubbard, 2006], and display distinct cellular functions [Kubo-Akashi et al., 2004]. APS and PSM/SH2-B variants engage in cross-regulation in the activation JAK family tyrosine kinases and insulin signaling [O'Brien et al., 2002; Li et al., 2007]. In contrast to the SH2-B gene knockout [Duan et al., 2004b], disruption of the APS gene in mice results in increased insulin sensitivity and hypoinsulinemia [Minami et al., 2003]. In a reported direct comparison in mice, disruption of the APS gene did not display alterations in adiposity, energy balance, glucose metabolism, insulin, or glucose tolerance as opposed to disruption of the SH2-B gene and was interpreted to suggest a specific role of PSM/SH2-B as regulator of energy and glucose metabolism in mice [Li et al., 2006] that is extended to neurons [Ren et al., 2007]. PSM/SH2-B and APS display distinct affinities when compared for binding to IR or JAK2 [Hu and Hubbard, 2006]. In response to insulin both APS [Moodie et al., 1999] and PSM/SH2-B undergo phosphorylation on Tyr, interact with the activation loop of IR, enhance IR autophosphorylation, and enhance (independently of PSM/SH2-B phosphorylation) ERK and Akt activation [Ahmed and Pillay, 2001, 2003]. PSM/SH2-B and APS play a role in neuronal differentiation [Qian et al., 1998; Rui et al., 1999; Koch et al., 2000]. PSM/SH2-B and APS have been reported as homo- or heteropentamers, mediated through the amino terminus in an analogous mechanism and to directly modulate Trk autophosphorylation [Qian and Ginty, 2001]. The activating mechanism has been proposed as homo- or hetero-dimerization via a phenylalanine zipper at the amino terminus which in turn results in the dimerization and activation of associated JAK2 [Dhe-Paganon et al., 2004; Nishi et al., 2005]. In addition, an alternative mechanism to support an active conformation of JAK2 involving only the SH2 domain of PSM/SH2-B and independent of the dimerization domain (DD) has been proposed [Kurzer et al., 2006]. PSM/SH2-B plays a role in the assembly of distinct mitogenic signaling complexes; in cultured normal fibroblasts this involves PI 3-kinase only in response to PDGF when compared to insulin or IGF-I [Deng et al., 2007].
In this study, the four alternative PSM splice variants and individual functional domains were compared for their impact on specific metabolic insulin responses. We found that individual PSM variants in 3T3-L1 adipocytes potentiated insulin-mediated physiologic responses by regulating established key insulin signaling mediators in a signature pattern of variant activities. In contrast, dominant-negative peptide mimetics of the PSM Pro-rich, DD, PH, or SH2 domains inhibited any tested insulin response. Potentiation of the insulin response originated at the IR kinase level by PSM variant-specific regulation of the Km (ATP) whereas the Vmax remained unaffected. Our results implicate an essential role of the PSM variants in the activation of the IR kinase and the resulting metabolic insulin response. PSM variants act as internal IR ligands that in addition to potentiating the insulin response stimulate IR catalytic activation even in the absence of insulin.
MATERIALS AND METHODS
All presented data have been reproduced. Bar graphs are typically shown based on duplicate measurements with the error indicated while for immunoblots one representative experiment is displayed.
Cell-Permeant PSM Fusion Proteins
Complete mouse PSM variant alpha, beta, gamma, or delta protein-coding cDNA was introduced into Escherichia coli expression plasmid pTAT-HA (kindly provided by Steven F. Dowdy, Washington University School of Medicine) under control of the strong T7 phage transcriptional promoter. This plasmid carries amino-terminal coding sequences for a 6-aa His tag peptide to facilitate affinity purification of the recombinant protein as well as an 11-aa HIV TAT protein-derived peptide (YGRKKRRQRRR) that renders the resulting fusion proteins cell-permeant [Wadia and Dowdy, 2005]. The complete protein-coding region of individual mouse PSM variants was assembled starting just upstream of the ATG initiation codon of translation through an Xho I restriction site in the 3'-untranslated region [Yousaf et al., 2001]. A 200-bp cDNA fragment encoding the 5′ end of the mouse PSM variant protein coding sequence was amplified by PCR primers (5′-TGGGGGTACCGGTGGTATGAATGGTGCCCCTTCCCCAGAGG-3′) and (5′-GGAAGAGCTCAGCAAAACGGCCAGAAAAAGCGGCCTCTGCTC-3′) that introduced a 5′ Kpn I site and a 3′ Sac I site into the final PCR product. The remaining protein-coding sequence of the mouse PSM variants was retrieved in a Sac I-Xho I fragment from PSM pIND mammalian expression plasmids [Yousaf et al., 2001]. Both fragments reconstituted the complete PSM variant protein-coding region and were joined with a Kpn I–Xho I fragment of plasmid pTAT-HA to produce cell-permeant TAT-HA-PSM alpha, beta, gamma, or delta fusion protein in E. coli.
Cell-permeant peptides representing the PSM amino-terminal Pro-rich sequence, PH region, or SH2 domain had been prepared by fusion with a sequence of the Drosophila melanogaster antennapedia homeoprotein as described earlier [Riedel et al., 2000; Deng et al., 2007]. The PH region or the SH2 domain had been expressed as a fusion peptide in E. coli and the Pro-rich region was represented by a synthetic peptide mimetic (American Peptide Company) composed of the transduction sequence (RQIKIWFQNRRMKWKK) of the D. melanogaster antennapedia homeodomain followed by the Pro-rich amino terminal PSM sequence FPSPPALPPPPPPSWQ [Riedel et al., 2000]. A cell-permeant peptide mimetic of a fragment FCESHARAAALDFA of the amino terminal PSM DD [Nishi et al., 2005] was created by fusion of the transduction sequence RKKRRQRRR [Wadia and Dowdy, 2005] derived from the HIV TAT protein at the amino terminus via a linker sequence AA (Genscript). A synthetic transduction peptide lacking PSM sequences or a dialyzed column eluate of a control E. coli cell extract served as the respective peptide control.
Recombinant plasmids were introduced into E. coli BL21 (DE3), which carries the lacUV5 repressor to control induction of the T7 transcriptional promoter by isopropyl-β-D-thiogalactopyranoside (IPTG). Cells were grown in Luria-Bertani (LB) medium containing kanamycin (30 µg/ml) at 37°C to OD600 0.2–0.3 followed by 1 mM IPTG induction for 5 h. Cells were sedimented, resuspended in 0.5 M Tris-HCl (pH 6.8), and lysed by French Press (SLM Instruments) treatment. Following centrifugation at 15,000g for 20 min at 4°C the cleared lysate was analyzed by SDS–PAGE (15%). Cell-permeant PSM fusion peptides were purified by nickel column affinity chromatography (Novagen), eluted with 0.4 M imidazole in the presence of 6 M urea, and dialyzed with a 3.5 kDa molecular weight cutoff (Slide-A-Lyzer, Pierce). Precipitated peptides were re-solubilized in 50 mM Tris-HCl (pH 6.8) by addition of DMSO to a final concentration of 0.1%. Protein concentration was determined by Bradford analysis and peptides were stored for up to several months at 4°C and permanently at −80°C. Synthetic peptides were solubilized in PBS at up to 1 mg/ml and kept in aliquots at −80°C for permanent storage.
Antibodies, Peptide Hormones, and Cell Culture
Rabbit polyclonal antibodies against IR, IRS-1, APS, JAK2, and goat polyclonal antibody against PSM/SH2-B were obtained from Santa Cruz Biotechnology, rabbit polyclonal antibodies directed against Cbl, phospho-Ser473 AKT, phospho-Thr308 AKT, and phospho-Ser21/9 GSK-3α/β were from Cell Signaling Technology, mouse monoclonal antibody against phospho-Tyr from BD Biosciences Transduction Laboratories, monoclonal antibody against actin from Sigma, and horseradish peroxidase-coupled immunoglobulin G (IgG) antibody from Kirkegaard and Perry Laboratories. An alternative PSM/SH2-B rabbit antiserum had been custom-produced by Hazelton Research Products, Inc. against a fusion protein composed of glutathione S-transferase (GST) and the SH2 domain of mouse PSM [Riedel et al., 1997]. Human recombinant insulin was obtained from Upstate Cell Signaling Solutions. Mouse 3T3-L1 fibroblast (ATCC) and NIH 3T3 fibroblasts stably expressing human IR were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 1% (v/v) penicillin/streptomycin solution in a 5% CO2 environment at 37°C. 3T3-L1 fibroblasts were differentiated to adipocytes as described earlier [Deng et al., 2003].
Peptide Transduction and cDNA/siRNA Transfection
Differentiated 3T3-L1 adipocytes or NIH 3T3 fibroblasts stably expressing IR were cultured to quiescence for 20 h in serum-free DMEM supplemented with 0.1% BSA. To study cellular responses cultures were typically incubated with 10 µg/ml of cell-permeant peptide for 1 h in serum-free medium followed by addition of 100 nM insulin for 15 min unless indicated otherwise.
Mouse PSM/SH2-B variants were expressed transiently from cDNA plasmids [Yousaf et al., 2001] under control of a constitutively active CMV transcriptional promoter in mouse NIH 3T3 fibroblasts stably expressing IR. Sub-confluent cultures on 10 cm plates were rinsed with antibiotic-free DMEM before 3 ml transfection mix including 5–6 µg of PSM expression plasmid or control plasmid lacking PSM sequences, 30 µl Lipofectamine and 20 µl Plus reagent (Invitrogen) were added according to the instructions of the manufacturer. After 5 h the transfection medium was replaced with DMEM including 10% FBS or, alternatively, for subsequent peptide hormone induction cells were cultured to quiescence for about 20 h in serum-free DMEM.
siRNA pools of three sequences for each mediator were supplied by Santa Cruz Biotechnology: (GUAGGGCAUUGGCUAAUGA, GAAGUCGCUUGGAGUUCUU, CCCGAGCCAUUAAUAAUCA) for PSM/SH2-B (gene accession# NM_011363), (GCAGAAUACAUCCUGGAAA, GGAGAAUCAGUACUCCUUU, GGUCAGCAACACCUAAUGA) for APS (gene accession# NM_018825.2), and (GACAUACUCUACAGGAUAA, CUUCUCGUCUUCAGAACAA, GCAUUAGCAUUACAGUUCU) for JAK2 (gene accession# NM_008413). Mouse NIH 3T3 fibroblasts stably expressing IR at 50–70% confluence in 6-well cell culture plates were transfected with 40–80 µmol siRNA and 5 µl Lipofectamine 200 in 250 µl of Opti-MEM I (Invitrogen).
Cells were subsequently analyzed as described below.
Immunoprecipitation and Immunoblotting
To prepare detergent cell extracts, cultures were rinsed twice with ice-cold PBS and were lysed in buffer containing 1% Triton X-100, 50 mM HEPES, pH 7.4, 10% glycerol, 137 mM NaCl, 10 mM NaF, 100 mM Na3VO4, 10 mM Na4P2O7, 2 mM EDTA, 10 µg/ml leupeptin, and 1 mM PMSF. Cell extract containing 200–500 µg of total protein was incubated at 4°C with the respective specific antibody for 2 h and subsequently with added 25 µl of protein A-Sepharose slurry for 1.5 h. Immunocomplexes were collected by centrifugation and rinsed three times in the same buffer at 4°C. Proteins were re-suspended in 25 µl 2× Laemmli loading buffer, boiled, separated by SDS–PAGE, and transferred to a nitrocellulose membrane. Proteins were identified by immunoblotting with the respective specific antibody, and visualized by enhanced chemiluminescence detection (ECL, Amersham).
IR Autophosphorylation
Mouse NIH 3T3 fibroblasts stably (106) expressing IR and transiently transfected with any of the PSM/SH2-B variants or with control expression plasmid, were propagated to quiescence for 20 h in serum-free DMEM supplemented with 0.2% radioimmunoassay-grade BSA (RIA grade, Sigma). Subsequently, cells were stimulated with 100 nM of insulin for various times or were left untreated (“0” control). Cultures were rinsed twice with ice-cold PBS, detergent cell extracts were prepared with lysis buffer, and aliquots containing 500 µg of total protein were immunoprecipitated with IR-specific antibody as described in the preceding section. The level of IR autophosphorylation was identified by immunoblotting with phospho-Tyr antibody.
IR Activation Kinetics
Mouse NIH 3T3 fibroblasts stably expressing IR were cultured to 90% confluence on 10 cm culture plates, washed twice with ice-cold PBS, solubilized with 2 ml lysis buffer and IR was immunoprecipitated as described in the preceding section. Immunocomplexes were rinsed twice with lysis buffer, twice with kinase buffer (25 mM Tris-HCl, pH 7.5, 5 mM β-glycerophosphate, 2 mM DTT, 0.1 mM Na3VO4, 10 mM MgCl2), and were resuspended in 500 µl kinase buffer. In parallel, HEK 293 cells cultured on 10 cm plates were transfected with PSM variant or control expression plasmid, PSM protein was immunoprecipitated from detergent cell extracts with PSM goat polyclonal antibody (Santa Cruz), and immuncomplexes were rinsed and resuspended in 30 µl kinase buffer.
To assay IR catalytic activity in vitro, 90 µl of re-suspended IR immuncomplexes and 30 µl re-suspended PSM immuncomplexes were thoroughly mixed and 20 µl aliquots were transferred to 0.5 ml tubes. Insulin was added to a final concentration of 1 nM and the IR kinase was activated with varying doses of 0, 2.5, 5, 20, 40, 100, and 200 µM ATP in 10 µl kinase buffer at 22°C. After 1 h reactions were stopped by adding 10 µl 4× Laemmli loading buffer, vigorously mixed, immediately heated to 95°C for 5 min and centrifuged. Thirty microliters of supernatant was analyzed by 8% SDS–PAGE, followed by immunoblotting with phospho-Tyr antibody (BD Transduction Laboratories), and catalytic activity was visualized by enhanced chemiluminescence detection (Amersham ECL). IR activation was quantified by scanning densitometry of bands on film in the linear range with UN-SCAN-IT software (Silk Scientific). Densities were normalized to the background detected in the absence of ATP. The Michaelis–Menten activation kinetics of IR was presented with Prizm 4.0 software (GraphPad).
Glycogen Synthesis
A protocol was followed as described below similar to the procedure outlined in [Wada et al., 2001]. Differentiated 3T3-L1 adipocytes were seeded into 12-well plates at densities of 4–5 × 105 cells per well. Cells were cultured for 24 h and subsequently deprived of serum for 18–20 h in DMEM supplemented with 0.1% BSA. Cells were rinsed twice with PBS and incubated for 3 h in 2.5 mM glucose, 0.1% BSA, 25 mM HEPES, pH 7.4, and further for 30 min at 37°C in the presence or absence of 100 nM insulin, cell-permeant peptide mimetics (10 µg/ml), and/or 20 µM PI 3-kinase inhibitor LY 294002. Subsequently, 2.5–5.0 µl of D-[U-14C]glucose (200 µCi/ml, 2–4 mCi/mmol, Amersham Biosciences) was added for 1 h at 37°C (final concentration 0.5–1 µCi/ml). Glycogen synthesis was terminated by rinsing cells three times with ice-cold PBS followed by cell lysis with 0.5 ml of 30% KOH for 1 h at 37°C. Lysates were transferred to 1.5-ml tubes, 50 µl of 20 mg/ml carrier glycogen (final concentration 2 mg/ml) was added, and cell lysates were incubated at 95°C for 30 min. Samples were cooled to room temperature, 0.6 ml of ice-cold ethanol was added, and glycogen was precipitated for 16 h at −20°C. The precipitate was sedimented at 3,000g for 10 min and the supernatant was aspirated. The precipitate was solubilized in 1 ml of H2O and incorporated radioactivity was determined by liquid scintillation spectroscopy.
Glucose Transport
A protocol was followed as described below similar to the procedure outlined in [Wiese et al., 1995]. Differentiated 3T3-L1 adipocytes (1.5 × 105) per well (in 24-well plates) were starved for 3–5 h in serum-free DMEM supplemented with 0.1% BSA and cells were subsequently rinsed twice with KRPH buffer (1% bovine serum albumin, 1 mM MgSO4, 1 mM CaCl2, 136 mM NaCl, 5 mM Na2HPO4, 20 mM HEPES, pH 7.4). Cells were incubated for 15 min at 37°C with 100 nM insulin and/or 10 µg/ml cell-permeant peptide mimetics, 0.5 µl of 2-deoxy-[3H]glucose (1 mCi/ml, 10–20 Ci/mmol, Amersham Biosciences) was added for 4 min (final concentration 0.5 µCi/ml) and nonspecific glucose transport (subtracted for final data presentation) was determined in the presence of 10 µM glucose transport inhibitor cytochalasin B. Cells were subsequently rinsed twice with ice-cold PBS and lysed in 0.5 ml of 0.5 N NaOH, followed by neutralization with 0.5 ml of 2 M Tris-HCl, pH 6.8. Associated radioactivity was determined by liquid scintillation spectroscopy.
Amino Acid Transport
A protocol was followed as described below similar to the procedure outlined in [McDowell et al., 1998]. Differentiated 3T3-L1 adipocytes (1.5 × 105) per well (in 24-well plates) were starved in serum-free DMEM supplemented with 0.1% BSA for 16 h. Cells were incubated for 1 h at 37°C with 100 nM insulin and/or 10 µg/ml cell-permeant peptide mimetics followed by the addition of 1 µl of 2-amino-[1-14C]isobutyric acid (50 µCi/ml, 50–62 mCi/mmol, Amersham Biosciences) for 10 min (final concentration 0.05 µCi/ml). Cells were rinsed three times with ice-cold PBS, lysed in 0.5 ml of 0.2 N NaOH for 30 min at 40°C, and samples were neutralized with 0.5 ml of 0.2 N HCl. Associated radioactivity was determined by liquid scintillation spectroscopy.
Lipogenesis
A protocol was followed as described below similar to the procedure outlined in [Wiese et al., 1995]. Differentiated 3T3-L1 adipocytes (4–5 × 105) per well (in 12-well plates) were starved for 18–20 h in serum-free medium supplemented with 0.1% BSA. Cells were incubated for 1 h at 37°C with 100 nM insulin and/or 10 µg/ml cell-permeant peptide mimetics in the presence of 0.5 µl of 2-deoxy-[3H]D-glucose (1 mCi/ml, 10–20 Ci/mmol, Amersham Biosciences) and 3.5 mM glucose (final concentration 0.5 µCi/ml). Cells were rinsed twice with ice-cold PBS, collected by scraping, and transferred to liquid scintillation vials. Five milliliters of Toluene-Bray scintillation liquid was added and the mixture was incubated for 16 h at 25°C. Four milliliters of the organic phase was removed to quantify incorporated radioactivity by liquid scintillation spectroscopy.
Akt/PKB Kinase Activity
Immunoprecipitation of Akt and analysis of its kinase activity was largely carried out according to the instructions in the employed experimental kit (Upstate Cell Signaling Solutions). Differentiated 3T3-L1 adipocytes (106) in 5-cm culture plates were starved for 20 h in serum-free DMEM. Cells were subsequently incubated with 10 µg/ml cell-permeant peptides for 1 h at 37°C before 100 nM insulin was added for 15 min. Cells were lysed in 1% Triton X-100, 10% glycerol (v/v), 50 mM HEPES, pH 7.6, 150 mM NaCl, 30 mM Na4P2O7, 10 mM NaF, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, 1 mM dithiothreitol, 1 µM microcystin, and 10 µg/ml of each aprotinin, pepstatin, and leupeptin. Twenty-five microliters of protein A-Sepharose pre-equilibrated for 16 h at 4°C and 4 µg of Akt-1/PKB pleckstrin homology domain antibody (Upstate Cell Signaling Solutions) were added to 1 ml of cell lysate and incubated under continuous suspension for 90 min at 4°C. Immunocomplexes were precipitated and rinsed three times with 500 µl lysis buffer containing 0.5 M NaCl, twice with 0.03% (w/v) Brij-35, 50 mM Tris-HCl, pH 7.5, 0.1 mM EGTA, 0.1% (v/v) 2-mercaptoethanol, and twice with 100 µl of AD buffer (100 mM MOPS, pH 7.2, 125 mM β-glycerol phosphate, 25 mM EGTA, 5 mM Na3VO4, 5 mM dithiothreitol). The immunoprecipitate was re-suspended in 10 µl of ice-cold AD buffer followed by the addition of 10 µl of Akt/SGK substrate peptide (RPRRATF) and 10 µl of protein kinase A inhibitor (as supplied in the kit). Phosphorylation was carried out with 0.1 µCi/ml [γ-32P]ATP (5 mCi/ml, 3,000 Ci/mmol, PerkinElmer Life Sciences) for 10 min at 37°C under continuous suspension. The reaction was terminated by the addition of 20 µl of 40% trichloroacetic acid. After 5 min 40 µl of the reaction mixture was spotted on 2-cm2 P81 Whatman phosphocellulose paper, which was rinsed three times for 5 min each with 0.75% phosphoric acid and once with acetone. Associated radioactivity was determined by liquid scintillation spectroscopy.
GSK3 Phosphorylation
Differentiated 3T3-L1 adipocytes (106) in 5-cm plates were cultured to quiescence for 20 h in serum-free DMEM. Cells were subsequently incubated with 10 µg/ml cell-permeant peptides for 1 h and stimulated with 100 nM insulin for an additional 10 min at 37°C. Cells were rinsed twice with ice-cold PBS and detergent cell extracts were prepared in ice-cold lysis buffer containing 50 mM HEPES, pH 7.4, 1% Triton X-100, 10% glycerol (v/v), 137 mM NaCl, 2 mM EDTA, 10 mM NaF, 100 mM Na3VO4, 10 mM Na4P2O7, 1 mM PMSF, and 10 µg/ml of each leupeptin and aprotinin (Sigma). About 25 µg of cell protein extract (per lane) was separated by SDS–PAGE (8%), and proteins were transferred to a nitrocellulose membrane. Immunoblots with phospho-specific glycogen synthase kinase 3 (GSK3) antibody (Cell Signaling Technology) were visualized by enhanced chemiluminescence detection (Amersham ECL).
p70 S6 Kinase Activity
p70 S6 kinase activity was largely analyzed according to the instructions in the employed S6 kinase assay kit (Upstate Cell Signaling Solutions). Differentiated 3T3-L1 adipocytes (106) in 5-cm plates were cultured to quiescence for 20 h in serum-free medium. Cells were subsequently incubated with 10 µg/ml cell-permeant peptides for 1 h and stimulated with 100 nM insulin for an additional 15 min at 37°C. Cells were rinsed twice with ice-cold PBS and detergent cell extracts were prepared as described above for the Akt/PKB kinase activity assay. About 200 µg of total cellular protein was immunoprecipitated with p70 S6 kinase antibody (as supplied in the kit) at 4°C for 16 h. Immunocomplexes were rinsed and re-suspended in assay buffer with 10 µl of substrate peptide (AKRRRLSSLRA) and 5 µl of [γ-32P]ATP (5 mCi/ml, 3,000 Ci/mmol, PerkinElmer Life Sciences, final concentration 0.2 µM). Reactions were carried out for 10 min at 30 °C. Twenty-five microliters aliquots were spotted on 2-cm2 P81 Whatman phosphocellulose paper, which was rinsed three times for 5 min each with 0.75% phosphoric acid and once with acetone. Associated radioactivity was determined by liquid scintillation spectroscopy.
Glycogen Synthase Activity
A protocol was followed as described below similar to the procedure outlined in [Sharma et al., 1998]. Differentiated 3T3-L1 adipocytes (4 × 105) per well in 12-well plates were starved for 4 h in serum-free DMEM lacking glucose and supplemented with 20 mM HEPES, pH 7.4, and 1% BSA. Cells were incubated with cell-permeant peptides and insulin in KRPH buffer for 30 min as described above for “Glucose Transport.” Cells were rinsed twice with ice-cold PBS, collected by scraping, sonicated for 30 s on ice in 200 µl of homogenization buffer (50 mM Tris-HCl, pH 7.4, 10 mM EDTA, 10 mM NaF), and centrifuged at 10,000g for 10 min at 4°C. Thirty microliters of homogenate (about 10 µg of protein) was added to 60 µl of glycogen synthase assay buffer containing 50 mM Tris-HCl, pH 7.4, 10 mM EDTA, 10 mM NaF, 10 mg/ml glycogen. Reactions were carried out in the presence of 5 µl of UDP-D-[U-14C]glucose (0.02 mCi/ml, 250–360 mCi/mmol, PerkinElmer Life Sciences) at a final concentration of 0.1 µCi/ml in the absence or presence of 2 mg/ml glucose 6-phosphate for 30 min at 30°C. Forty-five microliters aliquots were spotted on a Whatman no. 4 filter and dried. Filters were washed with ice-cold 70% ethanol once for 20 min at 4°C, twice for 30 min at room temperature, once with acetone, and dried. UDP-[U-14C]glucose incorporation into glycogen was measured as the associated radioactivity by liquid scintillation spectroscopy.
RESULTS
PSM Transduction of 3T3-L1 Adipocytes
In this report we compared four alternative PSM splice variants and individual PSM domains for their role in specific cellular insulin responses. We began by testing the impact of the four PSM variants individually on several of the established physiologic insulin actions in an insulin-responsive cell culture model, differentiated 3T3-L1 adipocytes. Since these cells are not easily transfected by cDNA we produced complete PSM variant proteins as well as peptide mimetics of specific PSM domains in E. coli or synthetically. A transfer sequence had been fused to all peptides to render them cell-permeant and allow their effective transduction into any cell. With this approach we have routinely obtained efficient delivery and physiologic activity of various peptides in any tested cell type including differentiated 3T3-L1 adipocytes [Deng et al., 2003]. We also confirmed in normal NIH 3T3 fibroblasts that PSM transduction when compared to cDNA transfection would produce indistinguishable results [Deng et al., 2007]. This was similarly confirmed for the transduction of dominant-negatively acting peptide mimetics of individual PSM domains that resulted in responses indistinguishable from siRNA strategies for PSM knockdown (see below).
PSM Regulation of Physiologic Insulin Responses
To begin to address the regulatory role of the PSM variants in insulin action each variant was individually transduced into cultured, differentiated 3T3-L1 adipocytes or dominant-negative peptide mimetics of individual PSM domains were alternatively transduced in comparison. Initially, we addressed the impact on insulin metabolic responses, specifically on glucose and amino acid transport, glycogenesis, and lipogenesis (Fig. 1A–D). We observed substantial potentiation of any of the insulin responses by transducing PSM variants. The highest activity was consistently observed for PSM alpha, typically potentiating the regular insulin response two- to threefold, followed by beta, delta, and gamma with decreasing activity. This signature pattern was observed for any of the four tested physiologic responses. In contrast, transduction of any of the dominant-negative PSM domain-specific peptide mimetics strongly inhibited any of the measured insulin responses. The SH2 domain essentially eliminated any response while in case of the PH and Pro-rich regions some insulin stimulation remained with the highest level observed for the PH region in glucose transport, for the Pro-rich region in glycogen synthesis, and essentially similar activities for any domain in amino acid transport (Fig. 1A–C). These results suggest a differential stimulatory role of the four PSM variants in physiologic insulin action. This role appears to involve all three PSM domains tested since each of the specific peptide mimetics inhibited any of the tested insulin response. Our findings are consistent with a critical role of PSM in metabolic insulin action.
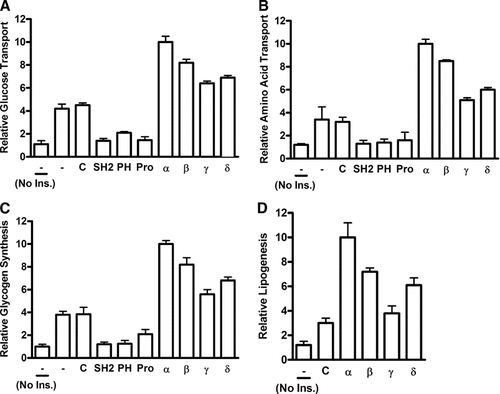
PSM-mediated regulation of metabolic insulin responses in 3T3-L1 adipocytes: (A) glucose and (B) amino acid transport, (C) glycogen synthesis, and (D) lipogenesis. Differentiated 3T3-L1 adipocytes were either treated with cell-permeant complete PSM variant proteins (alpha, beta, gamma, or delta) or cell-permeant peptide mimetics representing the PSM amino terminal Pro-rich sequence (Pro), PH region, SH2 domain, or with peptide control (C), or left untreated (-). Cells were stimulated with insulin or left untreated (No Ins.). The uptake of (A) 2-deoxy-[3H]D-glucose or (B) 2-amino [1-14C] isobutyric acid was quantified by liquid scintillation spectroscopy after cell lysis. C: The incorporation of [U-14C]-D glucose into glycogen was quantified by liquid scintillation spectroscopy after ethanol precipitation. D: The incorporation of 2-deoxy-[3H]D-glucose was quantified in cell extracts by liquid scintillation spectroscopy of the organic phase.
PSM Regulation of Akt and other Key Downstream Insulin Signaling Mediators
To begin to narrow down the specific point of PSM action we tested the regulation of several key enzymes with a role in insulin action specifically PKB/Akt, GSK3, glycogen synthase, and p70 S6 kinase. All are positioned downstream of the central mediator PI 3-kinase and their activation was evaluated with phospho-specific antibodies. The four PSM variants or domain-specific peptide mimetics were transduced as described above. We found that any of the four PSM variants potentiated insulin-mediated Akt, GSK3, glycogen synthase, or p70 S6 kinase activation (Fig. 2A–D) in a signature pattern of activities indistinguishable from that consistently observed for the physiologic responses shown in Figure 1. In addition, cell-permeant, domain-specific PSM peptide mimetics inhibited insulin-stimulated Akt, GSK3, or glycogen synthase function with highest activity consistently observed for the SH2 domain followed by the PH and Pro-rich regions with slightly decreasing activity (Fig. 2A–C). These results demonstrate an essential role of PSM in the regulation of the insulin signal via Akt downstream to GSK3, glycogen synthase, and the resulting synthesis of glycogen. An additional role in a parallel pathway downstream of Akt and directed towards protein synthesis is reflected in the regulation of p70 S6 kinase. In combination, all of our findings are compatible with a point of PSM action upstream of Akt that was subsequently addressed.
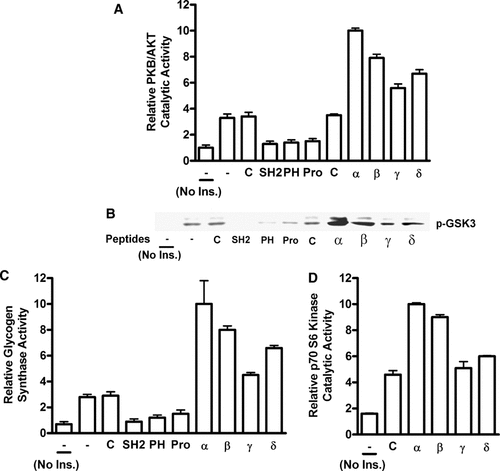
PSM-mediated regulation of PKB/Akt and other established downstream mediators in the insulin signaling pathway. Differentiated 3T3-L1 adipocytes were either treated with cell-permeant complete PSM variant proteins (alpha, beta, gamma, or delta) or cell-permeant peptide mimetics representing the PSM amino terminal Pro-rich sequence (Pro), PH region, SH2 domain, or with peptide control (C), or left untreated (-). Cells were stimulated with insulin or left untreated (No Ins.) and lysed. A: Using [γ-32P] ATP phosphorylation of an Akt-specific peptide substrate served to quantify PKB/Akt catalytic activity by liquid scintillation spectroscopy. B: Immunoblots with a phospho-specific GSK3 antibody from detergent cell extracts separated by SDS–PAGE served to quantify GSK3 catalytic activity (p-GSK3). C: Glycogen synthase activity was determined in the presence and absence of glucose 6-phosphate by incorporation of UDP (uridine-5′-diphospho)-D-[U-14C] glucose into glycogen and quantified by liquid scintillation spectroscopy. D: Phosphorylation of a p70 S6 kinase peptide substrate in the presence of [γ-32P] ATP was quantified by liquid scintillation spectroscopy.
PSM regulation of IR Substrate, Ubiquitin Ligase Cbl
To evaluate PSM regulation of a direct IR substrate we tested the ubiquitin ligase Cbl. PSM super family members, in particular APS link IR to a signaling complex involving Cbl in membrane lipid micro domains that has been implicated in the stimulation of GLUT4 translocation [Katsanakis and Pillay, 2005]. We have investigated the potential PSM regulation of Cbl phosphorylation by IR in analogy to the experiments we presented above. We found that the four PSM variants stimulated insulin-mediated Tyr phosphorylation of Cbl (Fig. 3A) in a signature pattern of activities, consistent with those observed above for the regulation of physiologic insulin responses and key mediator enzymes (Figs. 1, 2). In addition, cell-permeant, domain-specific PSM peptide mimetics inhibited IR-mediated Cbl phosphorylation with the highest activity observed for the SH2 domain followed by the Pro-rich sequence and with the lowest activity found for the PH region (Fig. 3A). When we addressed the potential interaction between PSM and Cbl we were able to demonstrate a complex between cellular Cbl and PSM in response to insulin stimulation. This complex was precipitated from detergent cell extracts with PSM-specific antiserum and demonstrated by immunoblotting with Cbl-specific antibody (Fig. 3B). These results indicate an additional regulatory role of PSM in the insulin signaling pathway towards GLUT4 translocation. The observed PSM regulation of IR-mediated Cbl phosphorylation suggests a point of action for the PSM variants either at or upstream of Cbl that was subsequently addressed.
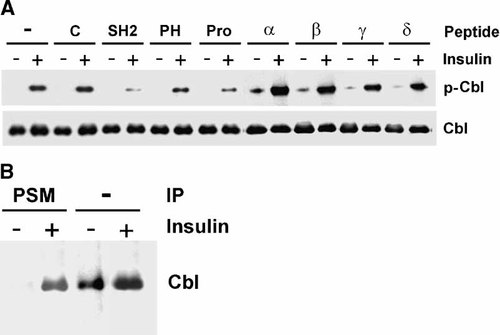
PSM-mediated Cbl regulation in response to insulin. Differentiated 3T3-L1 adipocytes were stimulated with insulin (−/+) and (A) treated with cell-permeant complete PSM variant proteins (alpha, beta, gamma, or delta) or cell-permeant peptide mimetics representing the PSM amino terminal Pro-rich sequence (Pro), PH region, SH2 domain, or with peptide control (C), or left untreated (-). Immunoprecipitate from detergent cell extracts with Cbl antibody was analyzed by immunoblotting with pTyr-specific antibody (p-Cbl) or Cbl antibody (Cbl). B: Cell lysates directly (IP: -) or after immunoprecipitation with PSM antiserum (IP: PSM) were analyzed by immunoblotting with Cbl antibody. The position of Cbl protein is indicated.
PSM Regulation of IR Catalytic Activity Independent of JAK2
The established cross-regulation between PSM and Tyr kinase JAK2 [Rui et al., 1997, 2000; Nishi et al., 2005; Kurzer et al., 2006] suggested the possibility that PSM may activate IR in a mechanism involving JAK2. Such a mechanism should be fundamental and present in most IR expressing cell lines including fibroblasts and not restricted to tissues metabolically responsive to insulin. We tested for PSM-regulation of IR activation and whether JAK2 function was required by siRNA knockdown (Fig. 4A). NIH3T3 fibroblasts stably expressing IR were individually transfected with the four PSM variant cDNAs as well as with JAK2 siRNA. IR was immunoprecipitated from detergent cell extracts and its activation evaluated on immunoblots with pTyr antibody. Significant potentiation of IR autophosphorylation was observed in response to increased levels of the four PSM variants with the signature pattern of the highest activity for alpha, followed by beta, delta, and gamma (Fig. 4B). No detectable impact of JAK2 siRNA was observed when compared to scrambled control siRNA implying that JAK2 function is not required for PSM regulation of IR activity. The function of mouse JAK2 siRNA (Santa Cruz Biotechnology) was confirmed by immunoblotting a fraction of cell extract with JAK2-specific antibody (Fig. 4A). A significant reduction of JAK2 protein was consistently observed in repeated experiments (1, 2) predicted to about 30% of original levels according to the supplier of the siRNA. This result suggested that PSM regulation of insulin action appears to take place at the level of IR kinase through direct association but independent of JAK2. Direct association of PSM with IR and a role of PSM as a physiologic IR enhancer have been described [Kotani et al., 1998; Wang and Riedel, 1998; Nelms et al., 1999; Duan et al., 2004b].
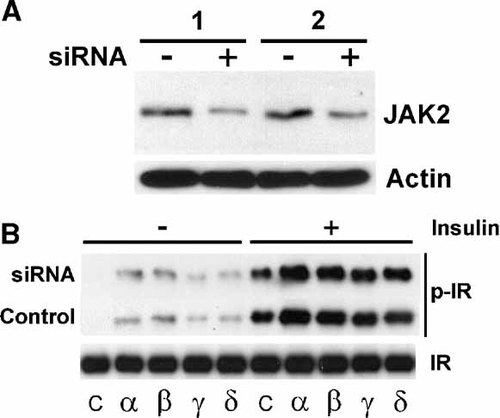
PSM regulation of IR catalytic activity in the presence of JAK2 siRNA. NIH3T3 fibroblasts stably expressing IR were transfected with expression plasmids carrying cDNAs of any of the four mouse PSM alternative splice variants (alpha, beta, gamma, or delta) or control plasmid (C) and either JAK2 siRNA (+) or scrambled siRNA (−/Control). Cells were starved and treated with 100 nM insulin for 30 min. A: The level of JAK2 protein was evaluated in detergent cell extracts by immunoblotting with JAK2-specific antibody followed by ECL (Amersham) detection (shown for two independent experiments: 1, 2). The position of JAK2 has been indicated and the level of actin from an immunoblot with actin-specific antibody is shown as a control (Actin). B: IR was immunoprecipitated from detergent cell extracts with IR-specific antibody and IR catalytic activity (p-IR) was evaluated on immunoblots with pTyr-specific antibody. The level of IR protein from an immunoblot with IR-specific antibody is shown as a control.
Insulin Dose-Dependent and Independent PSM Regulation of IR Catalytic Activity
A low level of IR kinase activation in the PSM variant-specific pattern even in the absence of insulin stimulation (Fig. 4B) suggested a potential role of PSM as an internal ligand to activate IR even in the absence of insulin rather than only to potentiate insulin action (tested at 100 nM in this experiment). To further address PSM regulation of IR autophosphorylation the insulin dose dependence was determined stepwise down to zero. NIH3T3 fibroblasts stably expressing IR were individually transfected with the four PSM variant cDNAs, starved, and stimulated with varying doses of insulin. IR was immunoprecipitated from detergent cell extracts and its activation evaluated on immunoblots with pTyr antibody. Significant potentiation of IR autophosphorylation was observed by any of the four PSM variants with the signature pattern of highest activity for alpha, followed by beta, delta, and gamma for any insulin dose including zero (Fig. 5A). Increasing the insulin dose to 0.1 nM substantially elevated IR kinase activity in response to PSM variant regulation and successive increases of the insulin dose to 1 and 10 nM proportionally increased the level of PSM stimulation. To test whether PSM variant regulation also extended to a second IR substrate, detergent cell extracts were similarly evaluated after immunoprecipitation with IRS-1 antibody by immunoblotting with pTyr antibody in response to insulin stimulation at 100 nM. IR phosphorylation of IRS-1 was found to be potentiated by the PSM variants in the signature pattern (Fig. 5B) as observed for any other response as shown above; specifically including autophosphorylation of IR (Fig. 5A) or phosphorylation of IR substrate Cbl (Fig. 3A). Efficient transfection of the four PSM variants had been evaluated by immunoblotting of NIH 3T3 detergent cell extracts with PSM antibody. Comparable levels of the four variant proteins were demonstrated (Fig. 5C) at the predicted relative sizes [Yousaf et al., 2001]. Our results suggest a role of the PSM variants as internal IR ligands that differentially activate IR catalytic activity even in the absence of insulin. In addition, the insulin response is potentiated in a variant-specific pattern that extends from IR autophosphorylation via IR substrate phosphorylation to the activation of key enzymes involved in insulin action as well as key physiologic responses. All of these observations can be explained by PSM variant-specific activation of the IR kinase.
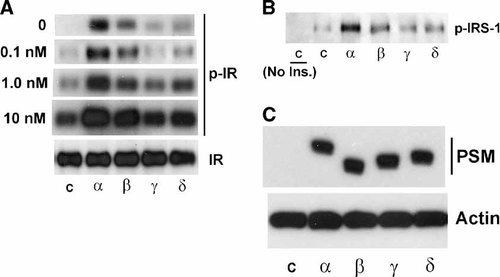
Insulin dose-dependent and -independent PSM regulation of IR catalytic activity. NIH3T3 fibroblasts stably expressing IR were transfected with expression plasmids carrying cDNAs of any of the four mouse PSM alternative splice variants (alpha, beta, gamma, or delta) or control plasmid (C). A: IR or (B) IRS-1 were immunoprecipitated with specific antibodies from detergent cell extracts followed by PAGE or (C) proteins in detergent extracts were directly analyzed by PAGE followed by immunoblotting with PSM-specific antibody and ECL (Amersham) detection. A: Cells propagated to quiescence were treated with insulin for 30 min at doses varying between 0, 0.1, 1.0, and 10 nM as indicated. IR catalytic activity was evaluated on immunoblots with pTyr-specific antibody followed by ECL (Amersham) detection (p-IR). The level of IR from an immunoblot with IR-specific antibody is shown as a control (IR). B: Cells were treated with 100 nM insulin for 30 min. Immunoprecipitates with IRS-1 antibody were evaluated on immunoblots for Tyr phosphorylation of IRS-1 with pTyr-specific antibody followed by ECL (Amersham) detection (p-IRS-1). C: The level of actin from an immunoblot with actin-specific antibody is shown as a control (Actin).
PSM Variant-Specific Regulation of IR Kinase Km for ATP
The impact of the PSM variants on the catalytic characteristics of the IR kinase was addressed by measuring IR autophosphorylation in vitro with partially purified IR and PSM variants at 1 nM insulin and varying concentrations of ATP. IR protein was immunoprecipitated with specific antibody and its activation analyzed by immunoblotting with pTyr antibody and quantified by densitometry (Fig. 6). Whereas the Vmax remained largely unaffected between samples within a range of about 5% the Km for ATP binding was substantially reduced in the same variant-specific signature pattern observed for all other responses (Table I). Compared to control samples at a Km of 34 µM, variant alpha lowered the Km most drastically to 7 µM followed by beta at 8 µM, delta at 13 µM, and gamma with the lowest impact at 24 µM. These changes should explain any of the PSM-mediated potentiation that we measured at various levels in the insulin response.
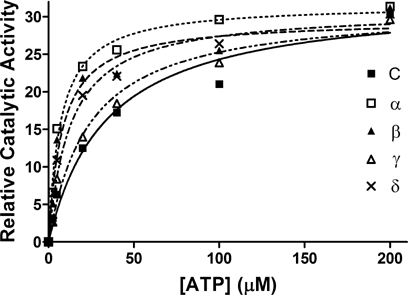
Kinetics of PSM-regulated IR catalytic activity. HEK 293 human embryo kidney cells were transfected with expression plasmids carrying cDNAs of any of the four mouse PSM alternative splice variants (alpha, beta, gamma, or delta) or control plasmid (C) and variant proteins were concentrated by immunoprecipitation with PSM antibody. IR protein was immunoprecipitated from NIH3T3 fibroblasts stably expressing IR and mixed with immunoprecipitated PSM variant protein for 1 h at 25°C to assay IR catalytic activity in the presence of 1 nM insulin and ATP concentrations varying from 0 to 200 µM as indicated. Proteins were separated by PAGE and catalytic activity of IR was detected by immunoblotting with pTyr antibody and quantified by densitometry of the exposed film. The Michaelis–Menten kinetics of the reaction (Prizm 4, GraphPad Software Inc.) has been presented (see Table I for more details).
| Variant | Km (±SD) ATP (µM) | Vmax (±SD) (relative activity) |
|---|---|---|
| C | 34.0 ± 10.2 | 32.6 ± 3.2 |
| α | 7.1 ± 1.1 | 31.7 ± 1.0 |
| β | 8.0 ± 2.2 | 29.6 ± 1.8 |
| γ | 23.8 ± 5.6 | 31.3 ± 2.1 |
| δ | 12.7 ± 2.7 | 30.9 ± 1.6 |
- See the legend to Figure 6 for experimental details.
Inhibition of IR Kinase by Dominant-Negative PSM Peptides or PSM siRNA
PSM dimerization via an amino terminal Phe zipper motif termed DD has been reported crucial for the activation of JAK2 [Nishi et al., 2005]. To address whether an analogous mechanism would apply to IR activation we employed specific peptide mimetics, either a fragment of the DD domain that would disrupt PSM dimerization or a peptide representing the PSM SH2 domain that would disrupt PSM binding to IR. To confirm the selectivity of the peptides we compared their impact on IR activation with PSM siRNA knockdown that would be expected to lead to comparable results. NIH 3T3 fibroblasts stably expressing IR were transfected with mouse PSM siRNA or treated with cell-permeant peptide mimetics of the SH2 domain or of a segment of the DD domain [Nishi et al., 2005]. Cells were starved, stimulated with 100 nM insulin and detergent cell extracts were analyzed in immunoblots with pTyr antibody. Treatment with DD or SH2 domain peptide mimetics individually or in combination essentially eliminated IR activation when compared to control peptide (Fig. 7A). Similarly, insulin stimulation of IR autophosphorylation as well as IRS protein phosphorylation was severely inhibited by PSM siRNA knockdown (Fig. 7B). In contrast, an internal control phosphoprotein of 120 kDa, prominently visible between IRS and IR remained unaffected (Fig. 7B). Inhibition of PSM expression by siRNA transfection had been repeatedly demonstrated in detergent cell extracts by immunoblotting with PSM antibody (Fig. 7C). The impact of the DD or SH2 domain peptide mimetics appears fully equivalent (and not additive) when compared to PSM siRNA knockdown in our experiments. Our results demonstrate an essential role of the PSM variants in insulin action at the level of IR catalytic activation.
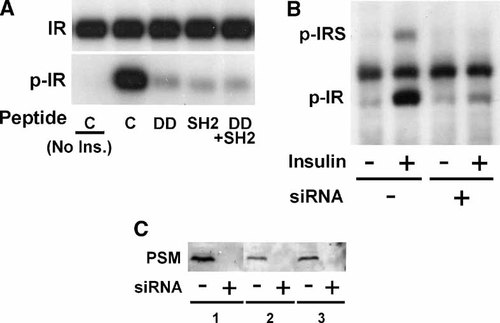
Inhibition of IR catalytic activity by siRNA knockdown of PSM or cell-permeant peptide mimetics of the DD or SH2 domain. NIH3T3 fibroblasts stably expressing IR were (A) treated with cell-permeant peptide mimetics of the PSM DD, and/or SH2 domain or with control peptide (C) for 1 h (Peptide) and with 100 nM insulin for 20 min or control treated (No Ins.) as indicated or were (B,C) transfected with pools of PSM siRNA (+) or scrambled siRNA (−). A: IR was immunoprecipitated from detergent cell extracts with specific antibody and IR catalytic activity was evaluated in immunoblots with pTyr-specific antibody followed by ECL (Amersham) detection (p-IR). The level of IR from an immunoblot with IR-specific antibody is shown as a control (IR). B: After treatment with 100 nM insulin (−/+) as indicated for 20 min proteins from detergent cell extracts were directly separated by SDS–PAGE and phosphoproteins were identified by immonublotting with pTyr-specific antibody. The position of phosphorylated IR or IRS (p-IR, p-IRS) has been indicated. C: Detergent cell extracts were directly analyzed by PAGE and the level of PSM was evaluated with specific antibody in immunoblots (shown for three independent experiments: 1, 2, 3).
Inhibition of Akt or GSK3 Activity by Dominant-Negative PSM Peptides or PSM siRNA
To address whether the observed inhibition of insulin action at the level of IR is extended to downstream insulin responses in fibroblasts we evaluated the impact of siRNA knockdown or of PSM peptide mimetics on the insulin activation of mediators Akt or GSK3. NIH 3T3 fibroblasts stably expressing IR were transfected with PSM siRNA or treated with cell-permeant peptide mimetics of the SH2 domain or of a segment of the DD domain [Nishi et al., 2005]. Cells were starved, stimulated with 100 nM insulin and detergent cell extracts were analyzed in immunoblots for the activation of Akt or GSK3 with phospho-specific antibodies. Insulin stimulation of either mediator was readily observed and was substantially inhibited by PSM treatment with either peptide (Fig. 8A) or siRNA knockdown (Fig. 8B). As an internal control an unknown insulin-stimulated pSer protein of 80 kDa (p-80) remained unaffected by either peptide (Fig. 8). The impact of the DD or SH2 domain peptide mimetics appears fully equivalent when compared to PSM siRNA knockdown in our experiments and confirms the essential role of PSM in insulin action.
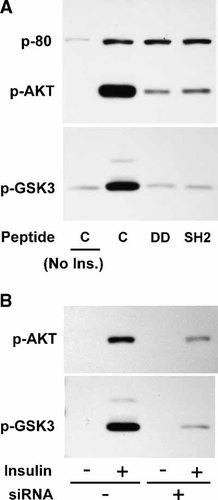
Inhibition of insulin stimulated Akt or GSK3 activation by siRNA knockdown of PSM or cell-permeant peptide mimetics of the DD or SH2 domain. A: NIH3T3 fibroblasts stably expressing IR were propagated to quiescence and treated with cell-permeant peptide mimetics of the PSM DD, or SH2 domains for 1 h (Peptide) and with 100 nM insulin for 20 min or control treated (No Ins.) as indicated. B: NIH3T3 fibroblasts stably expressing IR were transfected with pools of PSM siRNA (+) or scrambled siRNA (−) and stimulated with 100 nM insulin for 20 min (−/+) as indicated. A,B: Detergent cell extracts were analyzed by PAGE and Akt or GSK3 activation was monitored by immunoblotting with antibody specific for phospho-Ser 473 of Akt (p-Akt) or phospho-Ser 21/9 of GSK3 alpha or beta (p-GSK3), respectively, followed by ECL (Amersham) detection.
APS-Independence of IR Catalytic Regulation by PSM
The potential role of the related super family member APS [Katsanakis and Pillay, 2005] in the observed responses was addressed by APS siRNA knockdown. NIH3T3 fibroblasts stably expressing IR were individually transfected with the four PSM variant cDNAs as well as with APS siRNA (Fig. 9A). Cells were starved, stimulated with 100 nM insulin and detergent cell extracts were analyzed in immunoblots with pTyr antibody for IR activity. Basal activation of the IR kinase was significantly reduced by APS siRNA knockdown confirming the role of APS in IR activation (Fig. 9B). Significant potentiation of IR autophosphorylation was observed by the four PSM variants with the signature pattern of highest activity for alpha, followed by beta, delta, and gamma, that was, however, not significantly affected by APS siRNA (Fig. 9B). The function of mouse APS siRNA (Santa Cruz Biotechnology) was confirmed by immunoblotting a fraction of cell extract with APS-specific antibody (Fig. 9A). Substantial inhibition of APS protein expression was consistently observed in repeated experiments. Consequently, whereas APS contributes to IR catalytic activation, it is not required for PSM stimulation of the IR kinase. Both PSM and APS appear to function independently in insulin action possibly in an additive fashion.
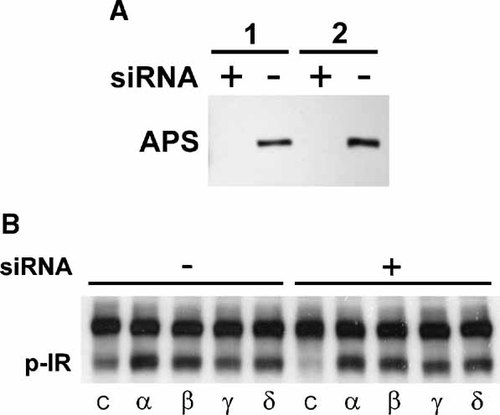
PSM regulation of IR catalytic activity during APS siRNA knockdown. NIH3T3 fibroblasts stably expressing IR were transfected with expression plasmids carrying cDNAs of any of the four mouse PSM alternative splice variants (alpha, beta, gamma, or delta) or control plasmid (C) and either APS siRNA (+) or scrambled siRNA (−) as indicated. A: The level of APS protein was evaluated in detergent cell extracts by immunoblotting with APS-specific antibody followed by ECL (Amersham) detection (shown for two independent experiments: 1, 2). The position of APS has been indicated. B: Cells were propagated to quiescence and treated with 100 nM insulin for 30 min. IR was immunoprecipitated from detergent cell extracts with specific antibody and IR catalytic activity was evaluated on immunoblots with pTyr antibody. The position of IR has been indicated (p-IR).
DISCUSSION
The presence of PSM/SH2-B in the signaling complex of a variety of receptor tyrosine kinases including IR suggested a role in insulin action [Riedel et al., 2000; Ahmed and Pillay, 2003]. Disruption of the SH2-B gene resulted in reduced IR activation and downstream insulin responses consistent with a role as a physiologic enhancer of IR activation and in the maintenance of normal insulin sensitivity and glucose homeostasis during ageing [Duan et al., 2004b]. In this study we have compared the role of the four PSM alternative splice variants in metabolic insulin action by specifically altering PSM function in insulin-responsive cells. We have used a combination of approaches including dominant-negative, cell-permeant peptide mimetics representing individual PSM domains including the amino terminal Pro-rich, DD, PH, and SH2 domain as well as PSM siRNA strategies. Both approaches equally eliminated insulin action at any level confirming the activity and selectivity of the cell-permeant peptides as well as the essential role of PSM in insulin action (Figs. 7, 8). This was complemented by strategies to increase the levels of individual PSM variants by cDNA expression or by transduction of complete cell-permeant proteins. Transduction of complete, individual, cell-permeant PSM variants gave us access to differentiated 3T3-L1 adipocytes that cannot be easily transfected with cDNA. In addition to transfection of complete PSM variant cDNAs, this provided an alternative positive experimental strategy to potentiate insulin action. We have shown cell-permeant PSM domain-specific peptide mimetics to enter cells with high efficiency in a dose-dependent fashion and to selectively interfere with PSM-mediated stimulation of mitogenesis and cell proliferation in response to insulin, IGF-I, or PDGF but not in response to EGF [Riedel et al., 2000; Deng et al., 2007]. In our hands cell-permeant peptide mimetics were shown to impact hormone, mediator, and domain specific physiologic responses [Wang et al., 1999]. In particular the selectivity of Pro-rich peptide mimetics was demonstrated through their distinct hormone and receptor specific functions when comparing respective Pro-rich peptide mimetics of signaling mediator Grb10 [Wang et al., 1999] versus PSM [Riedel et al., 2000]. Our findings are consistent with the specific dominant-negative action routinely observed for isolated SH2 domains including the PSM SH2 domain [Nishi et al., 2005]. A dominant-negative and specific role of PSM peptide mimetics is also supported in a direct comparison with PSM knockdown by siRNA with comparable results (Figs. 7, 8).
We employed positive as well as negative experimental strategies to address the role of PSM variants in various metabolic responses in differentiated 3T3-L1 adipocytes. We found that transduction of individual PSM variant proteins consistently potentiated whereas individual PSM domains essentially eliminated the insulin response. Results were comparable for any specific response tested including glucose or amino acid transport, glycogen synthesis, or lipogenesis (Fig. 1). For the complete PSM variants we observed a differential impact with the highest level of activity consistently observed for alpha, followed by beta, delta, and gamma with decreasing activity. This signature pattern was observed for any of the insulin responses measured in this study. The same pattern was consistently observed for transduced complete variant proteins or variants expressed from transfected cDNA that makes systematic differences between variants in the preparation procedure unlikely. Intriguingly, in the potentiation of mitogenic signaling pathways the four variants consistently displayed a distinct pattern of variant activities led by gamma with the highest level followed by delta, alpha, and beta with decreasing levels [Yousaf et al., 2001]. The molecular basis for the consistent differences in variant activity remains obscure but is likely defined by the distinct carboxyl terminal sequences of the four variant proteins. The distinct pattern of activities of the variants observed in metabolic when compared to mitogenic signaling mechanism is expected to reflect the differences between the pathways and the involved distinct mediators and supports the selectivity of the observed variant responses. By evaluating the activity of individual enzymes in the metabolic insulin response we confirmed the regulation of the pathway towards glycogen synthesis represented by PKB/Akt, GSK3, and glygogen synthase as well as a parallel pathway towards protein synthesis represented by p70 S6 ribosomal kinase (Fig. 2). Activity of all enzymes was consistently potentiated by the PSM variants in the signature pattern and essentially eliminated by individual PSM domains.
In addition to the established insulin signaling pathway centering around PI 3-kinase and Akt, a mechanism involving ubiquitin ligase Cbl, APS, Cbl-associated protein (CAP), adapter Crk II, guanine nuclotide exchange factor C3G, and GTP-binding protein TC10 has been described in insulin-responsive tissues involving flotillin-enriched membrane lipid micro domains and required for GLUT4 translocation in adipocytes [Saltiel and Pessin, 2002]. However, the relevance of this pathway is still under debate [Zhou et al., 2004] and it appears not to be functional in myocytes [JeBailey et al., 2004; Standaert et al., 2004]. A preferred role of APS has been described when compared to PSM in the recruitment of CAP, an early step in this signaling mechanism [Ahmed et al., 2000; Ahn et al., 2004]. We observed Tyr phosphorylation of Cbl to be strongly potentiated by the PSM variants with the described signature pattern and to be substantially inhibited by the PSM SH2 domain or Pro-rich region (Fig. 3A). In combination with the immunocomplex we observed between Cbl and PSM in response to insulin (Fig. 3B) PSM may provide an alternative mechanism when compared to APS in linking IR to the pathway regulating GLUT4 translocation. Using APS siRNA we demonstrated that the presence or absence of cellular APS had no measurable impact on the regulation of IR kinase activity by the PSM variants (Fig. 9) consistent with an alternative and independent mechanism when comparing APS and PSM. These findings also demonstrate the selectivity of the siRNA pools based on the distinct responses we observed when comparing the inhibition of PSM (Fig. 7) versus APS (Fig. 9).
Intrinsic cross-regulation has been reported between Tyr kinase JAK2 and PSM/SH2-B signaling [Rui et al., 1997, 2000; Rui and Carter-Su, 1999; Carter-Su et al., 2000a,b; Kurzer et al., 2004, 2006; Miquet et al., 2005]. This led us to test for a potential role of PSM in combination with JAK2 in the activation of IR. We observed potentiation of IR catalytic activation by the PSM variants in the signature pattern that was indistinguishable in the presence or absence of JAK2 siRNA (Fig. 4) suggesting that PSM-mediated IR activation is independent of JAK2 function. In this experiment a reduction of cellular JAK2 by about 70% was confirmed at the protein level (Fig. 4A) that is consistent with the prediction of the supplier since complete elimination of JAK2 protein is not achievable by this strategy.
To address the impact of the PSM variants directly on the activation of the IR kinase we compared four lower doses of insulin. Substantial activation of IR autophosphorylation was already detected in the absence of insulin with the signature pattern of variant activities (Fig. 5A). The second lowest tested insulin dose of 0.1 nM resulted in a substantial increase in IR kinase activity ensuring that only minimal background levels of insulin could have been present under zero insulin conditions consistent with the use of IRA-grade BSA in the serum-free culture medium. Dose responsive increases in IR catalytic activity were observed with elevated levels of insulin at 1 and 10 nM consistent with the signature pattern of variant activity (Fig. 5A). The same pattern of activation was observed for IR substrates including IRS-1 (Fig. 5B) and Cbl (Fig. 3) demonstrating that the observed IR phosphorylation correlates with substrate phosphorylation. Consistent cDNA expression levels of the variant proteins were monitored by immunoblotting (Fig. 5C). The observation of PSM variant potentiation of IR catalytic activation in the absence of insulin is consistent with PSM dimerization reported as a mechanism for JAK2 catalytic activation [Nishi et al., 2005]. A ligand-induced allosteric mechanism of activation is a key mechanism for receptor tyrosine kinases including IR. It is plausible that a PSM dimer can mimic this mechanism as an internal ligand by interacting with the cytoplasmic receptor domain rather than the extracellular domain.
To further characterize the impact of the PSM variants on the catalytic characteristics of the IR kinase, the Michaelis–Menten kinetics was determined with partially purified PSM variants and IR in vitro in an ATP dose response (Fig. 6). The Km for ATP binding was substantially reduced in the signature pattern of variant activity whereas the Vmax remained largely unaffected between samples (Table I). These changes should explain any of the PSM variant-regulated responses that we measured at various levels in the insulin signaling pathway. Our measurements are consistent with the reported catalytic characteristics of the IR kinase [White et al., 1984]. A major decrease in the IR Km for ATP has been reported in response to insulin for IR partially purified from skeletal muscle [Bak et al., 1990] and in response to iodoacetamide treatment during receptor purification [Li et al., 1991] which in parallel both resulted in increased IR catalytic activity including increased Vmax. In addition, an increase in the Km for ATP has been reported for a soluble IR kinase domain through replacement of the two carboxyl terminal Tyr phosphorylation sites [Tennagels et al., 2000] similarly correlating with IR catalytic activation. The decrease in the Km for ATP from 35 to 8 µM reported in response to insulin [Bak et al., 1990] approximates the decrease we observed in response to transfection with the most active PSM variants alpha or beta (Table I).
The SH2-B gene is fairly ubiquitously expressed in most tissues tested as shown by northern analysis or RT-PCR including significant levels in insulin responsive tissues such as adipocytes and muscle except for liver where expression is consistently detectable but low [Yousaf et al., 2001; Nishi et al., 2005]. In most tested tissues pairs of variants were expressed at elevated levels typically either alpha and delta, or beta and gamma, possibly due to shared alternative splice mechanisms, however, the underlying mechanism remains unclear [Yousaf et al., 2001]. Increasing the cellular levels of the PSM variants either by transduction of cell-permeant proteins or cDNA expression consistently stimulated any of the measured insulin responses in our experiments indicating that the level of cellular PSM may typically be limiting the insulin response under normal physiologic conditions. All other mediators of insulin action and the resulting signaling pathways appear to be in place to sensitively respond to altered levels of PSM activity supporting the perspective of a critical regulatory role of the PSM variants in insulin action. An essential role is also supported by our observation that interference with PSM function eliminates IR activation and any resulting insulin response (Figs. 7, 8).
PSM associates via its SH2 domain with various receptor tyrosine kinases including IR [Wang and Riedel, 1998; Riedel et al., 2000]. PSM dimerization via an amino terminal Phe zipper motif termed DD has been proposed as a model for the activation of JAK2 by PSM binding via the SH2 domain [Nishi et al., 2005] that may be extended to other receptor tyrosine kinases. This model would hypothesize that either a fragment of the DD domain that would disrupt PSM dimerization or a peptide representing the PSM SH2 domain that would disrupt PSM binding to IR or a combination of both would equally inhibit IR kinase activation in response to insulin. Our experiments support this model since either peptide essentially eliminated IR kinase activation in response to insulin and as predicted the combination of both peptides did not appear to result in an additive effect (Fig. 7A). Either peptide individually would be expected to disrupt the signaling complex predicted between IR and a PSM dimer indistinguishably from the simultaneous presence of both peptides. Either peptide was found at least as active as treatment with PSM siRNA that similarly inhibited IR catalytic activation (Fig. 7B). PSM regulation of IR activity is expected to be propagated to IR-dependent downstream responses including the regulation of Akt and GSK3 activity that we specifically demonstrated as examples. Either PSM DD or SH2 domain peptide mimetic inhibited the insulin response indistinguishable from PSM siRNA (Fig. 8). Like siRNA knockdown, in particular the DD peptide mimetic is expected to display the highest level of selectivity since this domain is not shared with other proteins except for the related mediator APS [Dhe-Paganon et al., 2004; Nishi et al., 2005]. Consistent with our findings yeast tri-hybrid experiments had suggested the formation of heterotetramers composed of PSM dimers linking two insulin receptor sequences [Nishi et al., 2005]. An alternative mechanism to support an active conformation of JAK2 involving only the SH2 domain of SH2-B and independent of the DD domain has also been proposed [Kurzer et al., 2006]. The model proposing a critical role of PSM dimerization in Tyr kinase activation [Nishi et al., 2005] is supported by our findings in the case of IR activation.
Our results implicate an essential role of the PSM variants in the activation of the IR kinase and the resulting metabolic insulin response. PSM variants act as internal IR ligands that in addition to potentiating the insulin response stimulate IR catalytic activation even in the absence of insulin.
Acknowledgements
We are grateful to Dr. Nora Riedel for the critical discussion of experimental strategies, data, and the manuscript. This work was supported (to H. R.) in part by National Institutes of Health Grant R01 CA77873, by National Science Foundation Grant MCB-9808795, by American Diabetes Association Grant 7-02-RA-76, and Juvenile Diabetes Foundation International Grant 197048.




