CNBP mediates neural crest cell expansion by controlling cell proliferation and cell survival during rostral head development
Abstract
Striking conservation in various organisms suggests that cellular nucleic acid binding protein (CNBP) plays a fundamental biological role across different species. Recently, it was reported that CNBP is required for forebrain formation during chick and mouse embryogenesis. In this study, we have used the zebrafish model system to expand and contextualize the basic understanding of the molecular mechanisms of CNBP activity during vertebrate head development. We show that zebrafish cnbp is expressed in the anterior CNS in a similar fashion as has been observed in early chick and mouse embryos. Using antisense morpholino oligonucleotide knockdown assays, we show that CNBP depletion causes forebrain truncation while trunk development appears normal. A substantial reduction in cell proliferation and an increase in cell death were observed in the anterior regions of cnbp morphant embryos, mainly within the cnbp expression territory. In situ hybridization assays show that CNBP depletion does not affect CNS patterning while it does cause depletion of neural crest derivatives. Our data suggest an essential role for CNBP in mediating neural crest expansion by controlling proliferation and cell survival rather than via a cell fate switch during rostral head development. This possible role of CNBP may not only explain the craniofacial anomalies observed in zebrafish but also those reported for mice and chicken and, moreover, demonstrates that CNBP plays an essential and conserved role during vertebrate head development. J. Cell. Biochem. 102: 1553–1570, 2007. © 2007 Wiley-Liss, Inc.
Abbreviations used:
PCR, polymerase chain reaction; ORF, open reading frame; EGFP, enhanced green fluorescent protein; MO, morpholino; TUNEL, terminal deoxynucleotidyl transferase mediated dUTP nick end labeled; BrdU, bromodeoxyuridine.
The vertebrate head is a composite structure whose formation begins early in development, at neural plate stages. Central to the development of the CNS is the concept of subdivision and segmentation, most overtly manifested in the telencephalon/retina subdivision or the diencephalic prosomeres and hindbrain rhombomeres. These processes begin at gastrulation and afterwards the rostral head regions are determined throughout the three germ layers. Roles for prechordal mesendoderm in rostral brain patterning have been suggested [Ang et al., 1994; Camus et al., 2000]. Cephalic neural crest cells also play essential roles in head development forming different structures such as tendons, which join muscles to bones, the dermis (including the adipose tissue associated with the skin), and most of the skull and all of the facial skeleton [Le Douarin et al., 2004; Creuzet et al., 2005].
The molecular and cellular mechanisms underlying how rostral head structure formation takes place are still largely unknown. Several proteins have been reported as playing roles in head development during organogenesis. These proteins, which show specific anterior expression patterns in the embryo, have been reported to be responsible for cell fate as well as for cell proliferation and cell survival control. The zinc-finger cellular nucleic acid binding protein (CNBP) is one such protein.
CNBP is a single-stranded nucleic acid binding protein that shows striking sequence conservation among vertebrates [Armas et al., 2001]. In the mouse, CNBP is expressed in the forebrain, midbrain, craniofacial structures, limb buds, and somites [Chen et al., 2003; Shimizu et al., 2003]. Homozygous Cnbp-null mutant mice are embryonic lethal and show severe forebrain truncation and facial abnormalities due to a lack of proper morphogenetic movements of the anterior visceral endoderm (AVE) during pre-gastrulation stage. About 40% of heterozygous newborn mutants exhibit multiple defects, including growth retardation and craniofacial malformations (e.g., a smaller mandible and complete lack of eyes), and die shortly after birth [Chen et al., 2003]. Because CNBP upregulates human c-Myc expression [Michelotti et al., 1995] and c-Myc expression is absent in Cnbp−/− mutant mice, it was proposed that during development, CNBP induces the expression of Myc, which in turn stimulates cell proliferation and differentiation required for forebrain induction and specification [Chen et al., 2003]. In chick embryos, Cnbp is expressed in the equivalent tissues compared to the mouse embryo and, furthermore, CNBP siRNA knockdown results in forebrain truncation. Results from the chick indicate that CNBP controls the expression of Six3, Bf-1, and Hesx1 [Abe et al., 2006].
It is noteworthy that CNBP target genes are involved directly or indirectly in cell proliferation control and cell survival. The proto-oncogene Myc has been implicated in the regulation of diverse cellular events such as cell cycle control, proliferation, differentiation, or apoptosis of different cell types in a number of in vitro mammalian cell lines [Grandori et al., 2000; Eisenman, 2001; Liu and Levens, 2006]. In the medaka embryo forebrain, Six3 was shown to facilitate cell proliferation by sequestration of Geminin from Cdt1, a key component in the assembly of the pre-replication complex [Del Bene et al., 2004]. Furthermore, Six3 regulates cell proliferation by the transcriptional control of genes such as Rx1, Bf1, and CyclinD1 [Gestri et al., 2005]. Consequently, it is tempting to speculate that the predominant function for CNBP during forebrain development is to control the balance between cell proliferation and cell survival during craniofacial development.
During the last decade, the zebrafish has emerged as a model system for craniofacial developmental studies. During development, the zebrafish forms essentially all of the same skeletal and muscle tissue types as its higher vertebrate counterparts, but in a simpler pattern, and tissues are composed of smaller numbers of cells. Furthermore, genes identified by random mutational screening have now revealed genetic pathways controlling patterning of the jaw and pharyngeal arches, as well as the midline of the skull, genes that have been found to be conserved between fish and humans [Yelick and Schilling, 2002]. We have taken advantage of these features to investigate further the role of CNBP during craniofacial development.
In this report, we show that cnbp is highly expressed in the head region of zebrafish embryos. Furthermore, we show that morpholino antisense oligonucleotide-mediated knockdown of CNBP protein causes forebrain truncation mainly as a consequence of a reduction in the size in craniofacial structures and neural crest (NC)-derivative depletion. This loss apparently occurs via cell death of the precursor population rather than via a cell fate switch. Our results suggest an essential role for CNBP in mediating NC-derivative expansion by controlling proliferation and cell survival during rostral head development.
MATERIALS AND METHODS
Fish and Embryo Rearing
Adult zebrafish (Danio rerio) were maintained at 28.5°C on a 14-h light/10-h dark cycle as previously described [Westerfield, 1995]. All embryos were staged according to development in hours (hpf) or days (dpf) post-fertilization at 28°C [Kimmel et al., 1995]. In some experiments, 0.03% (w/v) of 1-phenyl-2-thiourea was added to E3 medium to prevent melanin pigment synthesis.
Constructs and Antisense Morpholino
A cnbp-EGFP fusion construct was used as a template for PCR to generate a myc–cnbp-EGFP fusion product. The upstream primer introduced an EcoRI restriction site (5′-GGAATTCCGGGATGGACATGAGTACCAGTGAGTGTTTTGG-3′) and contains the ATG (shown in bold). The downstream primer contains an XhoI restriction site (5′-CCGCTCGAGCGGTTACTTGTACAGCTCGTCCATGCCG-3′). The PCR product was cloned into the pGEM-T Easy vector (Promega, Madison, WI) by TA cloning, digested with EcoRI and XhoI, and subcloned into the pCS2 + MT expression vector. This construct was sequenced to verify the correct ORF of the myc–cnbp-EGFP fusion construct. The resulting plasmid was amplified in E. coli, purified and cut with NotI, and in vitro transcribed with SP6 RNA polymerase. To generate a mutant cnbp-EGFP fusion construct, another upstream primer was designed containing six point mutations (shown in lower case and underlined) that did not modify the CNBP amino acid sequence (5′-GGAATTCCGGGATGGACATGAGcACgAGcGAaTGcTTcGGATGTGG-3′). The cloning procedure was the same as previously described.
Embryos were obtained by natural mating and injected at the one-cell stage into the yolk immediately below the blastomeres using a gas-driven microinjection apparatus (MPPI-2 Pressure Injector, Applied Scientific Instrumentation; Eugene, OR). The antisense morpholino oligonucleotide for cnbp (cnbp-MO) was designed and synthesized by Gene Tools (Philomath, OR). Cnbp-MO was directed against the 5′ region of the mRNA, from nucleotide +6 to +31 relative to the translation start site, with the sequence: 5′-ATCCAAAACACTCACTGGTACTCAT-3′. To inhibit cnbp translation, embryos were injected with 5 nl of 1.75 µg/µl morpholino oligonucleotide solution prepared in Danieau 1× [Nasevicius and Ekker, 2000]. For specificity control experiments, capped mRNA for the wild type or mutant versions of myc–cnbp-EGFP were suspended at 200 ng/µl in distilled water and 5 nl, alone or mixed with cnbp-MO, were injected in one-cell zebrafish embryos. Morpholino specificity control experiments were performed three times independently.
Embryonic Extract Preparation and Western Blot Analysis
Wild type and cnbp-MO-treated zebrafish embryos were homogenized in two volumes of ice-cold extract buffer (20 mM HEPES pH 7.6; 150 mM NaCl; 10 mM MgCl2; 1 mM PMSF; 1 mM DTT; 1 mM EDTA; and 0.5% (v/v) Triton X-100) in a Potter Elvehjem at 0°C. Homogenates were centrifuged twice for 15 min at 21,250 g at 4°C. Supernatants were diluted in Sample Buffer 5× (250 mM Tris-HCl (pH 7.5); 8% (w/v) SDS; 20% (v/v) Glycerol; 0.4 M DTT; and 0.1% (w/v) Bromophenol Blue) and incubated during 10 min at 70°C to allow protein denaturalization. Samples were centrifuged for 2 min at 21,250 g and a proportion of the extracts corresponding to eight embryos were loaded onto a 12% gel for SDS–PAGE. The gel was transferred to Nitrocellulose membrane Hybond™ ECL™ (Amersham Pharmacia Biotech, Freiburg, Germany) and stained for 10 min with PonceauS red solution (0.1% (w/v) PonceauS red in 1% (v/v) acetic acid) at room temperature in shaker, both conditions were used in the following steps. The membrane was washed several times with phosphate saline buffer 1× (PBS) with 0.1% (v/v) Tween-20 (PBT 1×) until complete elimination of the staining. Then the membrane was blocked with several replacements of blocking buffer (PBS supplemented with 5% (w/v) of milk) for 2 h. After five washes of 5 min each with PBT 1×, the membrane was incubated with mouse antibody specific for zebrafish CNBP (obtained by immunization with recombinant purified protein) diluted 1/1,000 in PBT 1× for 2 h. The membrane was washed with PBT 1× five times for 5 min and incubated with anti-mouse Ig HRP-linked antibody (Amersham Life Biosciences) diluted 1/5,000 in PBT 1× for 1 h. The reaction was developed with ECL™ Western Blotting Analysis System (Amersham Biosciences, UK) using Ortho CP-G Plus X-ray films (Agfa-Gevaert, Argentina). Subsequently, the membrane was used to detect β-actin levels. After several washes with PBT 1×, the membrane was incubated for 1 h in blocking buffer. Detection was performed with anti-β-actin antibody (diluted 1/250 in PBT 1×) and anti-rabbit Ig HRP-linked antibody (diluted 1/1,000 in PBT 1×). The enzyme reaction was performed following a similar procedure as previously described.
Whole-Mount mRNA In Situ Hybridization
Embryos were staged and fixed overnight in 4% (w/v) paraformaldehyde (PFA) in PBS 1× at 4°C. After washing, embryos were stored in methanol at −20°C until used. The procedure for whole-mount in situ hybridizations was carried out as previously described [Jowett and Lettice, 1994]. The cnbp probe contained the entire coding region of the cnbp cDNA [Armas et al., 2004]. Digoxigenin-UTP-labeled riboprobes were prepared according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). We used RNA probes prepared from cDNAs corresponding to zebrafish c-myc, emx1, pax2.1, fgf8, krox20, irx3a, col2a1, foxD3, ap2α, and crestin genes.
Cell Proliferation and Cell Death Analysis
For TUNEL analysis, 4% PFA fixed embryos were re-hydrated in PBT 1× and permeabilized by proteinase K digestion. After re-fixing in 4% PFA, embryos were washed three times for 5 min with PBT 1× and incubated with TUNEL reaction mixture (Enzyme reaction + Label solution, In situ Death Detection Kit, AP; Roche) overnight at 37°C in darkness. The reaction was stopped with five washes for 20 min in PBT 1× at room temperature. After 6 h in blocking solution (2% (w/v) blocking reagent (Roche), 20% (v/v) lamb serum in Maleic Acid Buffer (100 mM maleic acid, 150 mM NaCl, 0.1% (v/v) Tween-20, pH 7.5)) at room temperature, embryos were incubated in converter solution (10% (v/v) Converter AP in blocking solution; Roche) overnight at 4°C in darkness. The specimens were washed five times for 20 min in PBT 1× and three times for 5 min in AP Buffer. The label was developed with NBT/BCIP tablets (Roche) diluted in distilled water. Twenty-four hpf live cnbp-MO treated and control embryos were manually dechorionated and stained with the vital dye Acridine Orange at a final concentration of 5 µg/ml in E3 medium, for 30 min at room temperature in darkness. After the incubation, embryos were washed six times for 5 min in E3 medium, and analyzed under fluorescence. The TUNEL assay was repeated four times and the in vivo staining with Acridine Orange two times.
For BrdU treatment, 24 hpf control and treated embryos were manually dechorionated and incubated in a solution of 10 mM BrdU (Sigma) and 10% (v/v) DMSO in E3 medium for 10 min at 28°C. After three washes for 5 min with E3 medium, embryos were fixed overnight in 4% PFA in PBS at 4°C. Then embryos were dehydrated in methanol and stored at −20°C. After re-hydration, embryos were permeabilized with pre-cooled acetone, briefly washed with distilled water, and incubated in 2 N HCl for 1 h at room temperature. After several washes with PBT 1×, preparations were blocked for 2 h in incubation solution (20% (v/v) goat serum, 1% (v/v) DMSO in PBT 1×) at room temperature and then incubated with anti-BrdU antibody, diluted 1/400 for 4 h at 4°C. Finally, embryos were washed with PBT 1× six times for 10 min and incubated overnight at 4°C with anti-mouse Ig Alexa 488 linked antibody diluted 1/200. BrdU experiments were performed three times independently.
RESULTS
Cnbp Expression During Zebrafish Embryonic Development
In a previous report, we had shown that zebrafish cnbp mRNA was maternally inherited and homogeneously distributed during the early embryonic stages [Armas et al., 2004]. In the present work, we further analyzed cnbp mRNA expression pattern by in situ hybridization in whole-mount specimens during embryonic and early larval stages using an antisense cnbp riboprobe (Fig. 1). While cnbp transcripts were ubiquitous in zebrafish embryos up to the 15-somite stage (Fig. 1A), from the 17-somites stage onwards, zebrafish cnbp expression became prominent in myotomes, tectum, hindbrain, and in the eye field (Fig. 1B). By the final segmentation stages, cnbp transcripts appeared mainly in the tectum while it was still clearly detected in the ventral hindbrain, the retina, and myotomes (Fig. 1C). At 36 hpf, cnbp expression was more intense in the retina and at the midbrain–hindbrain border (MHB) while it became almost undetectable in myotomes (Fig. 1D–E). From 40 hpf onwards, MHB expression rapidly refined into a highly specific pattern along the dorsal and posterior edges of the optic tectum (Fig. 1F–G). At this developmental stage, cnbp was also expressed in the pectoral fin buds (Fig. 1G). During hatching period, cnbp expression was localized in the prospective craniofacial structures, the pectoral fins, the retina, and the liver (Fig. 1H–K). It is worth noticing that the zebrafish tectal cnbp expression profile is similar to that of genes involved in cell proliferation, such as some proto-oncogenes and cell cycle regulatory genes [Wullimann and Knipp, 2000; Duffy et al., 2005; Loeb-Hennard et al., 2005]. However, cnbp expression in proliferating brain cells is restricted to the optic tectum and retina and does not localize to other proliferating domains, such as the brain ventricular germinal zones (Fig. 1F–G).
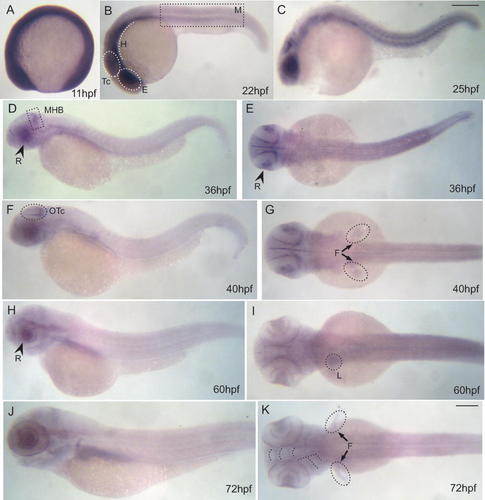
Embryonic cnbp mRNA expression pattern. Whole-mount in situ hybridizations of wild type zebrafish embryos at different developmental stages. (A) 11 hpf; (B) 22 hpf; (C) 25 hpf; (D–E) 36 hpf; (F–G) 40 hpf (H–I) 60 hpf; (J–K) 72 hpf. During early embryonic development until 18 hpf (A), cnbp mRNA is ubiquitously expressed. Since late segmentation to early pharyngula stages (B–C), cnbp gene expression becomes localized in the presumptive retina, tectum, ventral hindbrain, and myotomes. At 36 hpf (D–E), cnbp is expressed in anterior regions and specifically concentrated on the dorsal and lateral optic tectum borders. Expression in retina is also maintained. This gene expression pattern is conserved until late pharyngula period, when expression in the lateral fin buds appeared (G). During hatching period (H–K), a different gene expression pattern is observed in siblings: cnpb is expressed in the future craniofacial structures, the lateral fins and liver. Abbreviations: E, eye field; F, lateral fins; H, hindbrain; L, liver; M, myotomes; MHB, midbrain–hindbrain border; OTc, optic tectum; R, retina; Tc, tectum. All panels: anterior to the left. Panels A–D, F, H, J: lateral views, panels E, G, I, K: dorsal views. Scale bar: in C, 250 µm for A–C; in K, 250 µm for D–K. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
In summary, at the final segmentation stages, cnbp is expressed at the anterior-most end of zebrafish embryos mainly at the border between the midbrain and hindbrain and in the retina. During hatching period, it is expressed in the prospective craniofacial structures, the pectoral fins, and the liver. The expression pattern displayed by cnbp is similar to that of genes involved in cell cycle progression and cell proliferation while its expression coincides with regions that show a high rate of proliferation during development.
CNBP Loss-of-Function With a Specific Morpholino Oligonucleotide
With the goal of studying the role of cnbp during zebrafish development, we inhibited its expression by using a specific antisense morpholino oligonucleotide (cnbp-MO), which binds to the 5′-upstream sequence of the cnbp start codon (see Materials and Methods). In vivo expression analysis and Western Blot experiments were done to test the function and specificity of the cnbp-MO. We constructed a chimera consisting of a fusion between the full-length CNBP and EGFP cDNA sequences. The fusion construct was in vitro transcribed and injected in one-cell stage zebrafish embryos. After microinjection, embryos were incubated at 28°C to allow development to proceed, and EGFP expression was analyzed in vivo at several developmental stages. Most of the mRNA microinjected embryos showed ubiquitous EGFP expression during the first days of development (Fig. 2, Panel I, A–C; 94.5% of injected embryos expressed EGFP; n = 97). However, when embryos were co-injected with CNBP-EGFP mRNA plus cnbp-MO, low (not shown; 27.2% of co-injected embryos expressed low levels of EGFP; n = 92) or undetectable (Fig. 2, Panel II, (D–F); 72.8% of co-injected embryos expressed undetectable levels of EGFP; n = 92) EGFP expression levels were observed during the first 3 days after fertilization. These results indicate that the cnbp-MO is able to inhibit cnbp mRNA translation during the first stages of embryonic development and until 3 dpf. To test the specificity of cnbp-MO, a mutant CNBP–EGFP fusion construct was generated. This chimera has six point mutations in the region where the cnbp-MO binds, without changing the CNBP amino acid sequence. When capped mutant CNBP–EGFP mRNA was co-injected with cnbp-MO in one-cell staged embryos, EGFP expression level was ubiquitously detected (Fig. 2, Panel III, (G–I); 73.7% of injected embryos expressed EGFP; n = 95). This experiment confirmed the specificity of cnbp-MO since it could not inhibit the expression of a CNBP mRNA that lacks a specific target sequence. Furthermore, the development of these treated embryos was completely normal, successfully showing the rescued embryo phenotype (Fig. 2, Panel III, bright-field images).
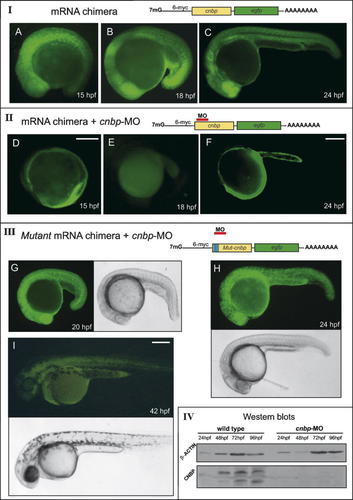
In vivo and in vitro analysis of cnbp morpholino activity and specificity during zebrafish development. Panel I: Representation of the chimera, which contains the fused full-length sequences of CNBP and EGFP. The construction was in vitro transcribed and injected in one-cell zebrafish embryos. Injected embryos were analyzed for green fluorescence (A–C), showing ubiquitous EGFP expression throughout embryonic development. Three different stages are shown, (A) 15 hpf, (B) 18 hpf, and (C) 24 hpf. Panel II: Co-injection of the mRNA chimera and cnbp-MO (D–F). Treated embryos showed reduced or absent levels of EGFP expression. The shown stages are (D) 15 hpf, (E) 18 hpf, and (F) 24 hpf. Panel III: Representation of the mutant chimera, which contains the nucleic acid sequences of the CNBP with six point mutations and the EGFP. Mutant cnbp-EGFP fusion mRNA and cnbp-MO were co-injected in one-cell stage zebrafish embryos. Embryos were analyzed for EGFP green fluorescence detection (green fluorescent images) as well as embryonic phenotypes (bright-field images). Completely rescued embryos at three different stages are shown (G) 20 hpf, (H) 24 hpf, and (I) 42 hpf. In all panels, anterior to the left and lateral views. Scale bars: 250 µm in D for A, D; in F for B, C, E, F; in I for G, H, I. Panel IV: Analysis of CNBP knockdown during zebrafish development by Western Blot. The analyzed developmental stages were 1 dpf, 2 dpf, 3 dpf, and 4 dpf. The membrane was analyzed for CNBP levels with a polyclonal anti-CNBP specific antibody. The level of β-actin was analyzed in order to verify the integrity and amount of protein extracts loaded. CNBP protein levels were not detected in morphant protein extracts compared to the wild type ones, while β-actin protein was detected in all samples. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
We also demonstrated specific inhibition of CNBP protein synthesis in cnbp-MO-treated embryos (morphants) by Western Blot analysis (Fig. 2, Panel IV). Wild type and morphant embryos were collected at different times after fertilization and processed to obtain total protein extracts. Proteins were separated by SDS–PAGE and analyzed by Western Blot using a specific polyclonal antibody made against CNBP. The β-actin protein level was analyzed in order to assure that similar amount of proteins from wild type and morphant embryos were loaded on the SDS–PAGE and the integrity of each embryonic extracts. A band of the correct molecular weight for CNBP was observed in wild-type embryonic extracts whereas morphant extracts did not show detectable levels of CNBP protein. This result confirmed the reduction of CNBP protein levels due to translation inhibition and, furthermore, allowed us to analyze the function of CNBP during embryonic development.
The in vivo effect of CNBP depletion was examined by injecting cnbp-MO into embryos at the one-cell stage. Due to toxicity at high concentrations of the morpholino, we injected different dilutions of cnbp-MO until we obtained a non-lethal concentration. The penetrance of the phenotype was highly dependent on morpholino concentration. When embryos were injected with small amounts of morpholino (less than 7.5 ng per embryo), no effects were observed during embryonic development and larvae developed normally as well. Increasing the concentration of cnbp-MO produced major defects in their development, resulting in a very narrow useful range of concentrations (between 7.5 and 8.1 ng per embryo). These findings suggest that CNBP is required at a threshold amount in order to ensure a normal developmental process in zebrafish embryos. Nonetheless, MO-injected embryos were not visibly distinguishable from wild type embryos until around 20 hpf. After this stage, a mild developmental delay was visible in morphants. Seventy-five percent of cnbp-MO-injected embryos (n = 195) showed alterations in brain and craniofacial structures development. At 24 hpf, some cephalic structures could not be detected, such as the MHB and the hindbrain ventricles (Fig. 3B). At this stage, a reduction of anterior structures such as the eyes and the anterior-most part of the head became evident. Beginning at 30 hpf, apart from the effects on brain development and smaller eyes, the phenotype was more severe: morphants presented fewer pigmented cells, and were smaller than controls (Fig. 3D). At 70 hpf, morphants had hydrocephaly and oedema, their eyes were smaller than controls and their tails were curved, preventing their hatching and/or correct motility (Fig. 3F–G). Moreover, the forebrain–midbrain and MHB areas showed abnormal morphology, and it appeared that the brain ventricles had not been well established. The most seriously affected embryos were developmentally arrested before hatching and they degenerated (Fig. 3G). All cnbp-MO-treated larvae died around 6 to 9 dpf.
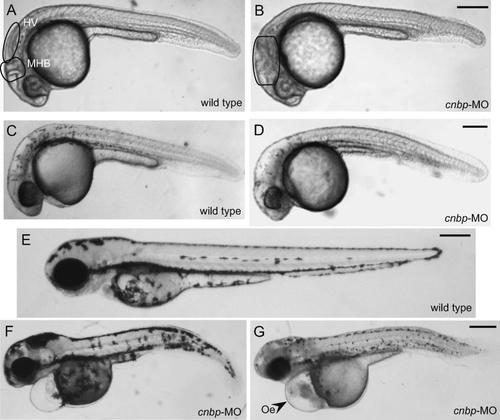
Wild type and cnbp-MO-treated embryos. (A–G) Lateral views of live embryos, anterior to the left; (A, C, E) wild type embryo, (B, D, F, G) cnbp-MO-treated embryo; (A, B) 24 hpf, (C, D) 32 hpf, (E–G) 70 hpf. Morphant phenotype appeared at 20 hpf. At this time, morphant development was slower compared to wild type siblings. Twenty-four hpf morphants (B) showed alterations in the MHB, and midbrain and hindbrain ventricles are affected. At 32 hpf (D), morphants displayed reduced or deformed anterior structures (e.g., retina, midbrain, hindbrain, and brain ventricles). At 70 hpf, larvae were smaller than controls, their eyes are reduced in size, their tails were curved and some larvae had less pigmented cells (F–G). Abbreviations: HV, hindbrain ventricles; MHB, mid-hindbrain border; and Oe, oedema. Scale bars: 250 µm, in B for A and B; in D for C and D; in E for E; and in G for F–G.
Analysis of Cephalic Morphological Defects in CNBP Knockdown Zebrafish Embryos
Analysis of the expression patterns of a number of genes in Cnbp−/− mouse embryos and in cnbp RNAi-treated chicken embryos have been carried out in order to detect the effects of loss of function of this gene [Chen et al., 2003; Abe et al., 2006]. These experiments showed that the expression of several forebrain gene markers, especially those expressed in the telencephalon, was missed in CNBP-depleted embryos. As we detected morphological defects in the brain of zebrafish morphants, we decided to study the expression of anterior neural markers (Fig. S1, Supplementary Material). No differences were observed in the expression patterns for emx1, pax2.1, fgf8, krox20, and irx3a in morphants from late segmentation to early pharyngula stages. These results suggest that CNBP is not necessary for zebrafish early anterior–posterior brain patterning, since all brain structures including telencephalon, forebrain, midbrain, MHB, rhombomeres 3 and 5, and hindbrain are not affected significantly in morphants. Although the morphant phenotype showed severe defects in brain morphology at 24 hpf, we conclude that early anterior–posterior brain gene patterning is normal in CNBP knockdown embryos.
CNBP Is Essential for Cell Proliferation and Cell Survival During Embryogenesis
We observed that zebrafish cnbp morphants grow slower than controls and some embryonic tissues were reduced in size. Both of these phenomena could be attributed either to an increase in cell death or a decrease in cell proliferation rates during development. To elucidate this, we studied cell death levels in cnbp-MO-treated embryos by performing two different assays. Acridine Orange staining permitted the in vivo observation of cells dying by apoptosis. Twenty-four hpf morphants displayed increased cell death in cephalic structures, mainly in the brain (Fig. 4A–B; 89.6% of the treated embryos showed an increment of labeled cells; n = 48). In order to complement this finding, we performed TUNEL analysis. Morphants and wild type embryos were analyzed for TUNEL-positive cells at the 24 hpf stage. The examination of whole-mount-labeled morphant embryos showed an increment of apoptotic cells in different brain structures, such as the eye field, MHB and ventral hindbrain (Fig. 4C–D; 86.6% showed increased TUNEL staining; n = 30). It would thus appear that CNBP is involved in preventing cell death in cephalic structures during embryonic development, since its absence produces death of cells that participate in brain development.
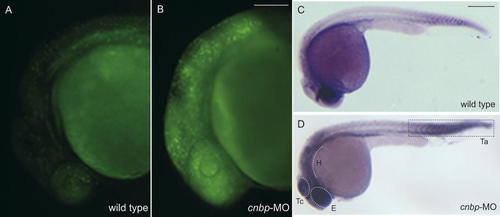
Cell death analysis on morphants and wild type embryos. Acridine Orange (AO) staining in vivo (A–B) and TUNEL analysis (C–D) were performed. Twenty-four hpf wild type (A) and morphant (B) embryos were labeled with AO to observe patterns of cell death. Morphants showed a higher level of AO staining (B), specifically in anterior regions, compared to the wild type embryos (A). TUNEL analysis shows an increased amount of apoptotic cells in morphants (D). The highest levels of apoptosis are detected in the eye field, tectum, hindbrain, and the tip of the tail of morphant embryos. Abbreviations: E, eye field; H, hindbrain; Ta, tail; Tc, tectum. All pictures: lateral views and anterior to the left. Scale bar: in B, 500 µm for A and B; in C, 250 µm for C and D. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
It has been reported that CNBP is involved in cell cycle progression by promoting cell proliferation [Konicek et al., 1998; Shimizu et al., 2003]. We analyzed cell proliferation levels in zebrafish cnbp morphants during embryonic development. Cell proliferation analysis was carried out by in vivo BrdU incorporation experiments. BrdU labeling was performed in morphants and control embryos at two different developmental stages. In order to visualize potential differences in proliferating cell numbers between morphants and wild type embryos, we employed a short pulse of BrdU incorporation. Following BrdU incubation, embryos were fixed and processed to detect labeled nuclei by immunofluorescence (Fig. 5). Morphants exhibited significantly less cells cycling through the S-phase of the cell cycle both at 16 hpf (84% of tested morphants showed less BrdU positive cells; n = 50) and 26 hpf, (91% of tested morphants showed less BrdU positive cells; n = 53) (compare Fig. 5A–C and B–D). It is noteworthy that the regions showing the most evident cell proliferation decrease were the ones most affected in cnbp-MO-treated embryos (i.e., predominantly forebrain, midbrain, and hindbrain), and correspond to the areas where cnbp transcripts are highly expressed. Interestingly, morphants showed normal cellular proliferation levels in the somites and tail. These results suggest that a possible reason for the disorganization of head development may be the excessive cell death or the insufficient proliferation of cells in the anterior–dorsal embryonic region.
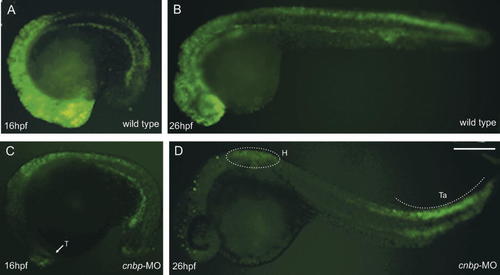
Cell proliferation analysis on morphants and wild type embryos. BrdU-positive cells are detected by immunofluorescence at different developmental stages (A–B 18 hpf, C–D 26 hpf). At 18 hpf, wild type embryos show proliferating cells in the forebrain, midbrain, and hindbrain as well as in the myotomes (A). In contrast, morphant embryos show a highly reduced cell proliferation pattern (B). BrdU-positive cells are exclusively detected in the posterior part of the hindbrain and myotomes. The white arrow shows some proliferating cells in morphant telencephalon. Similarly, 26-hpf wild type embryos show proliferating cells throughout the embryo (C), but in cnbp knockdown embryos, proliferating cells are only detected in the posterior part of hindbrain and at the tip of the tail (D). Abbreviations: H, hindbrain; T, telencephalon; Ta, tail. All embryos are shown in lateral views and the anterior part to the left. Scale bar in D, 250 µm for A–D. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
C-myc Gene Expression Pattern Analysis
One of the reported CNBP targets is c-myc [Michelotti et al., 1995; Chen et al., 2003], a proto-oncogene largely involved in cell proliferation control [Grandori et al., 2000; Eisenman, 2001; Liu and Levens, 2006]. Thus, we wondered whether the phenotypes manifested by cnbp-MO-treated zebrafish embryos may be due to a direct effect on c-myc mRNA synthesis during development. As c-myc gene expression during embryonic development has not been described thus far in zebrafish, we performed whole-mount in situ hybridization with a probe corresponding to the full-length sequence of the gene (Fig. 6). The mRNA was expressed in neural domains during early development. At 15 hpf, c-myc transcripts were localized in the telencephalon, presumptive retina, hindbrain, and neural tube. Expression outside the central nervous system (CNS) was prominent in mesodermal structures, for instance, the somites (Fig. 6A). At 20 hpf, the embryonic midbrain also showed c-myc expression (Fig. 6B). At 26 h of development, c-myc expression was evident in the retina, tectum, ventricular zone, and somites (Fig. 6C). At 44 hpf, c-myc was expressed in neural derivatives such as the retina, the dorsal and posterior limits of the optic tectum and the MHB, and also in pectoral fins and pancreas (Fig. 6E, H–I). At 60 hpf, c-myc expression was detected in the retina and in the dorsal and the posterior optic tectum borders and in the pancreas (Fig. 6F, J). C-myc expression in the retina was maintained after 3 days post-fertilization (Fig. 6G). The neural expression pattern obtained for c-myc appears remarkably similar to that described for n-myc, though n-myc is also transcribed in the cerebellar plate and dorsal rhombomere 2 [Loeb-Hennard et al., 2005]. Interestingly, the c-myc expression pattern is also highly similar to the one observed for cnbp. Indeed, there is a spatial–temporal overlapping of expression, which suggests a possible relationship between both genes.
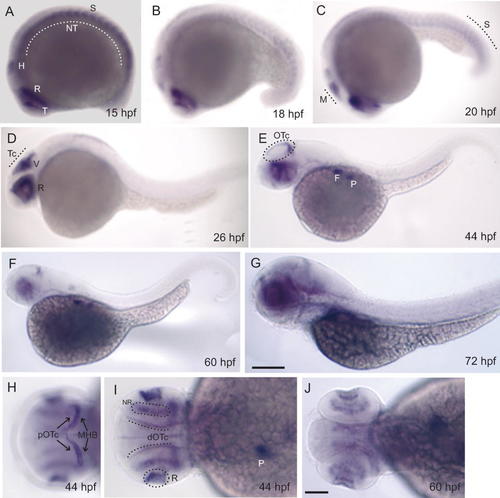
C-myc gene expression pattern during zebrafish development. Whole-mount in situ hybridization of zebrafish wild type embryos at different developmental stages: (A) 15 hpf; (B) 18 hpf; (C) 20 hpf; (D) 26 hpf; (E, H, I) 44 hpf; (F, J) 60 hpf; (G) 72 hpf. At 15 hpf (A), c-myc proto-oncogene expression is observed in the presumptive retina, tectum and myotomes. A very similar gene expression pattern is observed in wild type embryos at 20 hpf (B–C). At 26 hpf (D), c-myc expression becomes reduced in myotomes, while it is maintained in cephalic domains. At 44 hpf (E, H, and I), the proto-oncogene is specifically expressed in the neuroretina and lenses as well as at the dorsal and posterior limits of the optic tectum. In H, there is a double line on the most dorsal part of the embryo. The first line marks the posterior limit of the optic tectum and the second one shows the anterior region of the MHB. In I, the lateral limits could be seen delimiting the dorsal part of the eyes. A similar expression pattern is observed in 60 hpf larvae (F and J). At 72 hpf (G), c-myc gene expression profile is restricted to retinal tissue. Abbreviations: dOTc, dorsal limits of the optic tectum; F, pectoral fins; H, hindbrain; M, midbrain; NR, neuroretina; NT, neural tube; OTc, optic tectum; P, prancreas; pOTc, posterior limit of the optic tectum; R, retina; S, somites; T, telencephalon; Tc, tectum; V, ventricular zone. All panels anterior to the left, A–G lateral views, and H–J dorsal views. Scale bars: in G, 250 µm for A–G; and in J, 100 µm for H–J. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
We studied the expression of c-myc in cnbp knockdown morphants (Fig. 7). After cnbp-MO injection, embryos were fixed and analyzed for c-myc expression by whole-mount in situ hybridization at diverse developmental stages. We did not notice significant differences in c-myc expression level when comparing morphants (Fig. 7D–F) to control (Fig. 7A–C) embryos. However, the in situ hybridization staining allowed us to detect morphological differences between them. At 15 hpf, when embryos were compared in frontal views, the forebrain–midbrain region between the prospective eyes appeared narrower and consequently the presumptive retina fields were closer in morphants (Fig. 7A,D; white dotted circles). When comparing 18 hpf embryos, no significant differences in c-myc gene expression levels were observed between morphants and control embryos (Fig. 7B,E). However, a subtle difference was observed in gene expression pattern of 24 hpf embryos. While morphants showed the prospective eye structure completely stained, wild type embryos had only the dorsal and posterior aspects of the presumptive eye structure labeled (Fig. 7C,F, yellow dotted lines delimitate the expression in the retina while white dotted circles delimitate the lenses, respectively). The changes observed in morphant c-myc gene expression patterns could be due to the different rostral head development, appearing that CNBP is not controlling c-myc mRNA synthesis during zebrafish embryonic development.
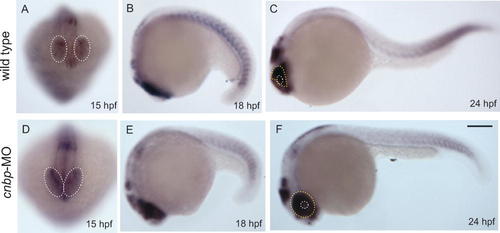
Analysis of c-myc gene expression pattern in cnbp knockdown embryos. Wild type (A–C) and morphant (D–F) embryos of different developmental stages are analyzed for c-myc gene expression pattern. The shown stages are: (A, D) 15 hpf; (B, E) 18 hpf; (C, F) 24 hpf. Morphants express normal levels of c-myc proto-oncogene at all analyzed stages (D–F). At 15 hpf (A, D), morphant embryos show a narrowing of the midline in the anterior brain as the presumptive retina and diencephalon appear closer than in control embryos (compare A to D, white dotted circles delineate the presumptive retina fields, respectively). At 24 hpf, morphants showed the prospective eye structure completely stained, while wild type embryos had only the dorsal and posterior aspects of the retina labeled (C and F, yellow dotted lines delimitate the expression in the retina while white dotted circles delimitate the lenses, respectively). Panels A and D: frontal view; panels B, C, E, and F: lateral views with anterior part to the left. Scale bar in F, 250 µm for A–F. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
CNBP Is Essential for Facial Cartilage Development
Among the diverse alterations in CNBP knockdown zebrafish embryos, rostral head morphology seemed to be the strongest. This malformation could be due to either reduced proliferation of cells that will differentiate and segregate into craniofacial structures or a delay in embryonic development that affects the formation of cartilage and bone. Moreover, by whole-mount in situ hybridization, we have noticed that cnbp is expressed in the future craniofacial structures during hatching period (Fig. 1). In order to detect possible defects in craniofacial structures in CNBP knockdown zebrafish larvae, we studied the expression profile of the col2a1 gene (Fig. 8). This gene encodes a major collagen of cartilage and its expression occurs in differentiating chondrocytes in mice and zebrafish [Vandenberg et al., 1991; Yan et al., 1995]. Embryos at 48 hpf showed col2a1 expression in presumptive craniofacial structures as well as in the notochord. At this stage, cartilage begins to differentiate morphologically in the embryo and it was possible to visualize early pre-cartilage condensations in the developing parachordals, which formed the neurocranium together with the ethmoid plate and trabeculae, the ceratohyal and the notochord (Fig. 8A,B). In CNBP knockdown embryos, the notochord was labeled, but there was a reduction of the parachordals and complete absence of the ceratohyal (Fig. 8C,D). At 72 hpf, col2a1 expression was detected in the presumptive neurocranium, pectoral fins, and pharyngeal skeleton of wild type larvae (Fig. 8E–G). In contrast, expression in morphant neurocranium was restricted only to the developing parachordals, while no label was seen either in the ethmoid plate or the trabeculae. Regarding the presumptive pharyngeal skeleton of morphant larvae, the ceratohyal was the unique structure that appeared labeled (Fig. 8H–I). These results confirmed the abnormal development of craniofacial structures in CNBP knockdown embryos, and allow us to conclude that the formation of facial cartilage is dependent on normal cnbp gene expression during zebrafish embryonic development.
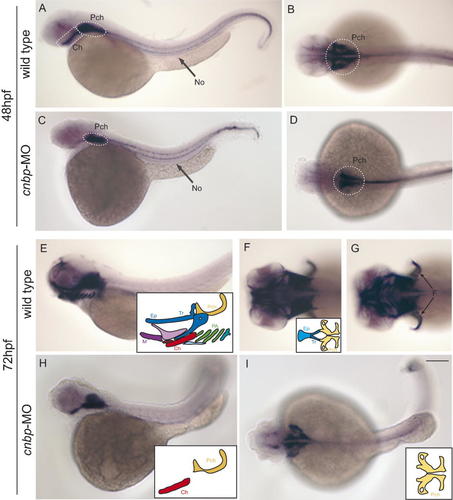
Craniofacial abnormalities were detected in cnbp-MO-treated larvae. The expression pattern of col2a1 is studied during the pharyngula stages in wild type (A–B and E–G) and morphant (C–D and H–I) larvae. At 48 hpf, wild type col2a1 expression is detected in the notochord, and in the presumptive mandibular and hyoid arches (A–B). Morphants show highly reduced col2a1 expression in the prospective craniofacial structures, such as the parachordals and presumptive ceratohyal, while expression in the notochord is completely normal (C–D). Three dpf morphant larvae display strongly reduced and aberrant facial cartilage structures (H–I), compared to control embryos (E–G). Colored inboxes representing the different craniofacial structures that appeared in morphant and control embryos were added in E, F, H, and I for better understanding the col2a1 gene expression pattern. Abbreviations: Ch, ceratohyal; Ep, ethmoid plate; F, pectoral fins; M, Meckel's cartilage; No, notochord; PA, pharyngeal arches; Pch, parachordals; Tr, trabeculae. In all pictures: anterior to the left. Pictures A, C, E, and H: lateral views; Pictures B, D, F, G, and I: dorsal views. Scale bar in I, 250 µm for A–I. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To further analyze the relationship between cranial structure establishment and cnbp gene expression during embryonic development, we studied the expression profiles of NC markers. The NC develops at the border between the neural plate and the epidermis, and following closure of the neural tube these cells delaminate from the dorsal neural tube to migrate along different pathways. On reaching their destination in the embryo, they differentiate into a wide variety of derivatives, including neurons and glia of the peripheral nervous system, pigment cells, and craniofacial cartilage and bone. Different studies performed in avian and amphibian embryos showed that cranial NC cells are involved in development of the neurocranium, specifically in formation of the trabeculae, while the pharyngeal skeleton derives from paraxial mesoderm [Kimmel et al., 2001]. In our analysis, we had initially noted that morphant larvae displayed lower amounts of pigmentation and that the trabeculae was not formed when col2a1 gene expression pattern was analyzed (see Figs. 3 and 8), suggesting that there is an effect of CNBP loss of function on NC development. By whole-mount in situ hybridization, we analyzed the gene expression patterns of NC markers (Fig. 9). FoxD3 is one of the earliest NC genes to be expressed in mice, zebrafish, Xenopus, and chick embryos [Steventon et al., 2005]. When the expression pattern of foxD3 was analyzed in CNBP knockdown embryos, differences were found when comparing to wild types. At the 1–4 somites stage, expression of foxD3 was detected at normal levels in the posterior hindbrain and spinal cord (Fig. 9E–F). At 14 hpf/10–13 somites, foxD3 expression in cnbp-MO-treated embryos was normal in the somites (Fig. 9H), but it was not detected in the dorsal anterior brain regions (Fig. 9G, black arrowhead; compare to 9C). Ap2α, another early NC marker, appears to be also involved in late NC development [Steventon et al., 2005]. Its expression pattern was analyzed at early as well as late NC development in wild type (Fig. 9I–L) and cnbp morphant embryos (Fig. 9M–P). Expression of ap2α in 10–13 somites stage morphants was lower than in controls in the cephalic domain (Fig. 9, compare I to M, black arrow, and J to N). At 26 hpf, ap2α is transcribed in a highly restricted manner in several anterior structures derived from the NC (Fig. 9K–L). Lower levels of ap2α gene expression were observed in some cephalic structures in morphant embryos (Fig. 9O–P, black arrow), most notably in the pharyngeal arches (Fig. 9L, P, white brackets). Finally, we analyzed the expression profile of the post-migratory NC cell marker crestin (Fig. 9Q–X). Although at 14 hpf no high differences were observed in the expression of this marker between wild type and morphant embryos (Fig. 9Q–R and U–V), 26 hpf morphants showed diminished expression in anterior structures (Fig. 9W–X, red arrowheads).
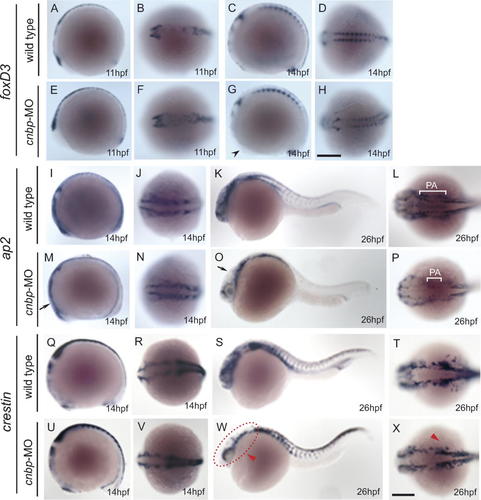
Gene expression pattern of neural crest markers by whole-mount in situ hybridization. (A–H) foxD3, (I–P) ap2α, and (Q–X) crestin gene expression patterns in wild type (A–D, I–L, Q–T) and morphant (E–H, M–P, S–X) embryos during zebrafish development. In all panels, the stages of development are indicated in the right-inferior corners, anterior to the left. Differences in NC expression patterns are detected in morphant compared to wild type embryos. FoxD3 expression appears normal in morphants during the 1–4 somites stage (compare A–B with E–F). At 10–13 somites stage, cnbp-MO-treated embryos showed normal foxD3 expression in the somites but no expression was detected in the dorsal brain regions (compare C, D with G, H). Ap2α displayed lower levels of expression in anterior domains at both analyzed developmental stages (M, O, black arrow). Ap2α expression in 26 hpf embryos labels pharyngeal arches in wild type embryos (L, white bracket), while in morphants, some pharyngeal arches were not detected (P, white bracket). CNBP depleted (Q, R) and wild type (U, V) embryos display similar expression of crestin at 10–13 somites stage. However, lower levels of crestin are detected in anterior regions of 26 hpf morphant embryos (W, X, red arrowhead) when compared to control embryos (S, T). Abbreviation: PA, pharyngeal arches. Panels B, D, F, H, J, L, N, P, R, T, V, and X the embryos are shown dorsally, while in the others laterally. Scale bars represent 250 µm, in H for A–H; in X for I–X. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Given these results, there appears to be a function for CNBP in specification, establishment, and/or migration of NC cells. This role could involve the control of the correct amount of proliferating NC cells while they are delaminating from the neural tube, migrating and colonizing the different anterior domains.
Taken together, our results provide evidence for a critical role for CNBP in the preservation of the balance between cell death and proliferation in anterior tissues during embryonic development. A defect in this process can account for the changes in the expression of markers we have observed, such as those of the NC, and for the loss of craniofacial structures.
DISCUSSION
CNBP expression has been reported in amphibians [Flink et al., 1998; Calcaterra et al., 1999], chicken [van Heumen et al., 1997; Ruble and Foster, 1998], fish [Armas et al., 2004; Liu and Gui, 2005], and mammals [Palis and Kingsley, 1995; Shimizu et al., 2003]. CNBP homologs are widespread throughout the animal kingdom; gene organization as well as nucleic acid and amino acid sequences are highly conserved, suggesting a fundamental role in different species throughout evolution. Two previous investigations have pointed out the role of CNBP in the development of the forebrain of vertebrates [Chen et al., 2003; Abe et al., 2006]. The results presented here bring about new facts on the expression of the CNBP gene in craniofacial structures and its effect on NC-derived cell proliferation and survival control.
To further understand CNBP function during vertebrate embryogenesis, we analyzed the expression pattern of cnbp in zebrafish embryos using whole-mount RNA in situ hybridization. According to our previous results, which reported ubiquitous expression during early zebrafish embryonic development [Armas et al., 2004], it would seem that cnbp expression is not conserved among zebrafish, mammals, and the chicken. A possible reason for the discrepancy might be the existence of high amounts of maternally inherited cnbp transcripts in early zebrafish embryos [Armas et al., 2004], which might preclude cnbp detection in specific structures. However, the present study demonstrates that cnbp expression in late zebrafish embryos is highest in the forebrain, midbrain, retina, and craniofacial structures, as has been reported for mouse and chick embryos (Fig. 1). This finding reinforces the importance of CNBP for the development of specific anterior tissues during vertebrate antero-posterior axis formation and for face morphogenesis. We have also found that cnbp expression is sharply delimited to the rostral head and it is upregulated in structures that show high rates of cell proliferation, such as the retina, tectum, and pectoral fins.
With the aim of further understanding the role of CNBP during vertebrate embryonic development, we knocked down CNBP by specific morpholino-oligonuclotide depletion assays. It was observed that injected embryos exhibited reproducible morphological defects in cephalic structures (Fig. 3). The diencephalic, mesencephalic, and rhombencephalic ventricles were not well formed in cnbp-MO-injected embryos. In addition, the constriction normally present at the midbrain–hindbrain boundary was absent. Although loss of cnbp expression leads to defects in forebrain and midbrain development, the most posterior CNS looked normal. While most cnbp-MO-injected embryos showed defects in the head region, tissues in the trunk such as the notochord, somites and spinal cord appeared normal, as has been reported for both mice and chick CNBP-depleted embryos [Chen et al., 2003; Abe et al., 2006]. It is noteworthy that narrow differences in morpholino concentration were enough to produce either no effect or lethal phenotypes in zebrafish development. This finding suggests that CNBP must be expressed above a threshold level to ensure normal development of anterior tissues in zebrafish embryos, in agreement with previous observations made in both mice and chick CNBP depleted embryos [Chen et al., 2003; Abe et al., 2006]. Thus, CNBP appears to play a conserved and essential role in vertebrate development.
Several mechanisms could account for the abnormal development of anterior structures in CNBP-depleted embryos. One possibility is that the lack of CNBP drives neural precursors into another lineage. Alternatively, CNBP may play a role in promoting proliferation and/or survival of a specific cell precursor pool. Our results from BrdU incorporation assays showed a significant decrease in the number of proliferating cells in the anterior head of morphant embryos, mainly within the cnbp expression territory (Fig. 5). A decrease in cell proliferation was also reported in Cnbp−/− mutant mice, which was attributed to the absence of c-Myc expression [Chen et al., 2003]. The zebrafish c-myc developmental pattern expression is coincident with the cnbp expression profile during pharyngula period (Figs. 2 and 6), suggesting either that both genes are co-regulated or that there is a regulatory interaction between them. However, we found near normal levels of c-myc transcripts in the anterior region of cnbp-MO-injected embryos (Fig. 7). This finding suggests that the deformities observed in the anterior-most structures of zebrafish embryo were not due to changes in c-myc transcription. Moreover, a survey of the zebrafish genomic sequence surrounding the c-myc gene analyzed showed that its promoter region does not contain the CT regulatory element found in human c-Myc, a possible reason for the normal c-myc expression profile observed in cnbp-MO-injected embryos. Interestingly, it was reported that c-Myc is not essential for forebrain development in the mouse, given the low penetrance of anterior neural fold truncation in c-Myc mutant mice [Davis et al., 1993]. It seems likely that CNBP regulates anterior cell proliferation and tissue development by controlling the expression of cell proliferation genes other than c-Myc.
In addition to cell cycle progression, a possible explanation for our observations is that depletion of CNBP may cause cell death. To test this possibility, we performed TUNEL and Acridine Orange assays to examine whether CNBP knockdown led to increased apoptosis in the affected region of the embryos. Examination of the CNBP-depleted embryos revealed that there was, indeed, an increase in the number of dying cells, relative to the control embryos (Fig. 4) possibly due to a blockade of the cell cycle and subsequent cell death. CNBP is not a typical transcription factor considering that it binds to single-stranded nucleic acids [Armas et al., 2004] modifying the conformational torsion of a DNA sequence [Michelotti et al., 1996]. A possible regulatory mechanism could involve the binding of CNBP to single-stranded regions in the DNA preventing the binding of a transcription factor that is required for activating the expression of cell cycle inhibitors, facilitating cell cycle progression and cell survival. In a similar way, CNBP may upregulate the expression of factors required for cell proliferation. What factors control cell proliferation and survival and how are they related to regulators of cell fate determination and differentiation? The shortened head observed in mouse, chick, and zebrafish embryos when CNBP is downregulated may be due to abnormal development of the CNS or the face cartilages and bones. In cnbp-MO-injected embryos, the expression pattern of a number of neural gene markers (e.g., fgf8, krox20, emx1, pax2.1, and irx3a) did not change, suggesting that CNS patterning was not affected by CNBP depletion. In contrast, comparison of the expression pattern of the chondrogenic marker gene col2α1 in normal and treated embryos made evident that CNBP downregulation causes a number of craniofacial abnormalities in the pharyngeal skeleton such as the Meckel's cartilage and the pharyngeal arches (Fig. 8). According to fate mapping studies in avian [Le Lievre, 1978] and amphibian [Chibon, 1967] embryos, and extirpation experiments in lamprey [Langille and Hall, 1986, 1988], paraxial mesoderm forms the parachordals and cranial NC forms the trabeculae [Kimmel et al., 2001]. The absence of trabaculae development in CNBP-depleted zebrafish embryos, strongly suggests that CNBP may be involved in NC derivative development. In agreement with this finding, the expression of col2α1 in cnbp-MO-injected embryos was retained in the parachordals and the presumptive ceratohyal while was abolished in the rest of the neurocranium, the branchial arches and the others ventrolateral craniofacial structures (Fig. 8). These results, in addition to the fact that CNBP-depleted embryos showed lower pigmentation than controls (Fig. 3), allowed us to hypothesize that CNBP might control the proliferation or survival of NC-derived cells. We noted that the expression patterns of the NC markers foxD3, ap2α, and crestin were modified by cnbp-MO injection (Fig. 9); for example, ap2α whole-mount in situ hybridization displayed abnormalities in shape, size, and number of pharyngeal arches (Fig. 9). It is well known that in the developing head, the cephalic NC cells migrate from posterior midbrain and hindbrain regions to the branchial arch system and execute specialized programs of migration, patterning, proliferation, and differentiation [Le Douarin et al., 2004; Creuzet et al., 2005]. Moreover, recent experiments have revealed that the cephalic NC is required for the development of the forebrain and midbrain. Indeed, the NC forms the meninges of the forebrain (while the meninges of the other parts of the CNS are of mesodermal origin) [Etchevers et al., 1999; Creuzet et al., 2004]. Thus, if CNBP were essential for cell proliferation and survival of cells responsible for craniofacial skeleton development, that is, the NC cells, CNBP depletion may cause a significant deficit in the number of cells available to form the anterior-most region of the embryo and, consequently, neurocranial truncation, disorganization, and anterior–posterior axis disruptions of pharyngeal cartilages would ensue. It is worth mentioning that this possible role of CNBP may not only explain the craniofacial anomalies observed in zebrafish but also those ones reported for mice and chicken. Indeed, CNBP knockdown in chick embryos showed severe craniofacial defects [Abe et al., 2006], which may be attributed to abnormal cartilage development. Similarly, morphological analysis of heterozygous Cnbp+/− newborn mutants revealed growth retardation and craniofacial defects (e.g., a smaller mandible and complete lack of eyes) or had mild eye and skeleton defects [Chen et al., 2003] probably due to abnormal facial cartilage development. Therefore, our data expand and contextualize the basic understanding of the molecular mechanisms of CNBP function.
Our results suggest a critical role for CNBP in development of head structures, mainly through the control of proliferation and survival of cell precursor populations. Future works that address the functional role of CNBP should tie together and clarify these intriguing observations. The morpholino methodology, though widely used, does not allow loss-of-function assays at specific times during embryonic development. We assume that to better understand CNBP function as well as to learn about its molecular targets during embryonic development, it will be necessary to use methods that allow greater temporal and spatial control of the manipulation of CNBP expression. An approach to solve this difficulty is being currently developed in our laboratory.
Acknowledgements
We thank Dr A. Reyes for the generous gift of crestin, ap2α, and foxD3 probes. Work reported in this paper was undertaken during Research Training Programs awarded by Coimbra Group – University of Bergen and AMSUD-Pasteur - MNDB to AMJ Weiner.




