Comparative proteomic analysis of apoptosis induced by sodium selenite in human acute promyelocytic leukemia NB4 cells†
Hua Dong and Tianyi Ying contributed equally to this work.
Abstract
Selenium (Se) is an essential trace element possessing anticarcinogenic properties. Sodium selenite (Na2SeO3) induced apoptosis in human acute promyelocytic leukemia (APL) cell line NB4 with dose and time dependency. In this study, proteomic techniques were used to study the apoptosis of NB4 cells induced by sodium selenite. Twenty-six downregulated and four upregulated proteins were identified, which exhibited a 1.5-fold change or greater. The identified proteins included key regulators of signal transduction such as Rho GDP dissociation inhibitor (Rho GDI) alpha and beta members of the MAPK family, and proteins involved in the regulation of c-fos or c-myc expression. Importantly, the identified proteins, hnRNP D0B and Rho GDI beta, which were related with the regulation of c-myc, c-fos, and c-jun, were determined by reverse transcription-polymerase chain reaction (RT-PCR) to confirm their downregulation in proteomic study. Western blot analysis and RT-PCR were then performed on three associated proteins: c-Myc, c-Fos, and c-Jun, and their expression were observed to be significantly downregulated. Results showed that certain regulation involved in c-myc, c-fos, and c-jun was present in the apoptosis, and the c-Myc dependent-on and Jun N-terminal kinase (JNK) pathway also play roles. J. Cell. Biochem. © 2006 Wiley-Liss, Inc.
Acute promyelocytic leukemia (APL) is a specific type of acute myeloid leukemia (AML). The drug therapy of leukemia has been supported by a great progress in using all-trans retinoic acid (ATRA) and arsenic trioxide (As2O3) to achieve complete remission in patients with APL. But As2O3 has a high toxicity with severe hepatic, cardiac, dermatological, and neural injury. Drug resistance to ATRA and relapse after ATRA treatment are common [Warrell et al., 1994]. So other highly effective and safe drugs are in need.
Selenium is an essential trace element in mammals. Many reports from the numerous studies in animal models and epidemiological investigations show that selenium acts as an antineoplastic agent, and later, it is reported that selenite is able to induce apoptosis in various kinds of human cancer cells, including hepatoma cells, lung cancer cells, and colonic cancer cells [Zhou et al., 2003; Gopee et al., 2004]. But the mechanism involved in the anti-carcinogenic activity of selenium remained to be elucidated.
Apoptosis is a programmed cell death dependent on active participation of cellular regulation. Apoptotic decision of cells is based on the processing of a myriad of impetuses, both external and internal. Our laboratory has been interested in understanding the mechanism that contributes to the apoptosis of human APL NB4 cells induced by sodium selenite [Sun et al., 2002; Zuo et al., 2004]. Quantitation of apoptosis was determined to support that sodium selenite (≥5 µmol/L) suppressed the cell growth, and then induced apoptosis with dose and time dependency [Li et al., 2002a; Zuo et al., 2002]. But the relationship between sodium selenite and signal molecules has not been well elucidated. The application of high-throughput proteomic techniques can systemically identify and characterize the differentially expressed proteins in a time efficient manner, which makes people comprehensively understand the mechanism of life from the indirect gene to the functional executioner protein, and it has been used to study a wide variety of biological processes. So it is necessary to analyze the differentially expressed proteins in the apoptotic NB4 cells following sodium selenite induction by comparative proteomics analysis.
The aim of this study was to further elucidate the mechanisms involved in the anti-carcinogenic activity of selenium by comparative proteomic approach. In this study, two-dimensional gel electrophoresis coupled with mass spectrometry and database-analyzing techniques were used to investigate the global changes in protein expression between the control and the apoptotic NB4 cells induced by sodium selenite. To extend our understanding of the proteome analysis, we analyzed the intracellular signaling pathway involved.
MATERIALS AND METHODS
Cell Culture and Sodium Selenite Treatment
The control NB4 cells were cultured in RPMI 1640 medium (Gibco, Paisley, UK) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2. At the same time, the RPMI 1640 medium in which the sodium selenite-induced NB4 cells grew contained 20 µmol/L sodium selenite (Sigma, MO) in addition. Cells were adjusted to a concentration of 5 × 105/ml to maintain a logarithmic growth and were harvested following 36 h [Li et al., 2003]. Sodium selenite treatment was performed in triplicate and repeated three times.
Preparation of Protein Samples
Cells were harvested following 36 h of sodium selenite treatment and then centrifuged for 15 min at 1,500g at 4°C. The pellets were washed three times for 10 min at 1,500g with ice-cold l × PBS. The supernatant was then discarded, and the pellet was resuspended with the extraction solution (7M Urea, 2M Thiourea, 4% CHAPS, 1% DTT) in presence of protease inhibitor cocktail tablet (Roche, Germany). Then the cell lysates were sonicated on ice by ultrasonication (model VC 750, Sonics) for 60 s at 25% power output. After 1 h at room temperature, the resulting homogenate was centrifuged at 40,000g for 30 min at 15°C to sediment the insoluble components. The supernatant was collected, and protein concentration was determined by using PlusOne 2-D Quant Kit (Amersham Pharmacia Biotech, Uppsala, Sweden), and then stored in 500 µg aliquots at −70°C.
Two-Dimensional Gel Electrophoresis
The first-dimensional isoelectric focusing (IEF) was carried out as described previously [Görg et al., 2000]. Precast immobilized pH gradient (IPG) strips (pH 3–10, nonlinear/linear, 24 cm/18 cm long; Amersham Pharmacia Biotech) were used. Equal amount (500 µg) of protein samples were added to the rehydration solution to a final volume of 350 µl (sample loading by rehydration) or 100 µl (sample loading by cup-loading). IEF was conducted at 20°C for 60,000 Vhrs in IPGphor system (Amersham Pharmacia Biotech). After IEF, each strip was equilibrated for 15 min in 10 ml equilibration buffer 1 (6 M urea, 1% DTT, 30% glycerol, 50 mM Tris-Cl pH 8.8) and then in 10 ml equilibration buffer 2 (6 M urea, 2.5% Iodoacetamide, 30% glycerol, 50 mM Tris-Cl pH 8.8) for another 15 min. For the second dimension, vertical slab SDS–PAGE (12.5%) was run with 30 mA/gel in Protean™ II XL system (Bio-Rad, Hercules, CA). The Gels were stained with Colloidal Coomassie Brilliant Blue G-250 (Amresco). High-resolution gel images (300 or 400 dpi) were obtained with scanner (model PowerLook 2100XL, UMAX) and processed with Image-Master 2D Platinum Version 5.0 (Amersham Pharmacia Biotech). The quantity of each spot was normalized by total valid spot intensity. Differentially expressed protein spots were selected for the significant expression variation deviated over 1.5-fold in relative volume (%vol). Each experiment was performed at least three times.
In-Gel Digestion of Proteins and Mass-Spectrometric Analysis
In-gel protein digestion was performed as Liao et al. [2003] described before. The destained gel pieces were completely dried in a Speedvac vacuum concentrator (Savant Instruments) and then were reswollen with 3 µl of 25 mM ammonium bicarbonate containing 10 ng of trypsin at 4°C for 1 h. After 12 h incubation at 37°C, the gel pieces were dried under high vacuum centrifuge to evaporate solvent. Eight microliters of 5% TFA were then added and incubated at 37°C for 1 h. The extract was transfered into another microtube. Eight microliters of 2.5% TFA/50% ACN was used for the second step of extraction at 30°C for 1 h. The extract was transferred to the microtube as described previously. At last another 8 µl 100% ACN was used for the extraction of hydrophobic peptides. All of the extracts were pooled and dried in the Speedvac vacuum concentrator, and resolubilized with 3 µl of 0.5% TFA/30% ACN. Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry analysis was performed on a Bruker Reflex™ III MALDI-TOF mass spectrometer (Bruker-Daltonik, Bremen, Germany) operating in reflectron mode. A saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 0.1% TFA was used as matrix. Totally 1 µl of the matrix solution and sample solution with a 1:1(v/v) ratio were mixed and applied onto the Score384 target well. The MALDI-TOF mass analysis used the following parameters: 20 kV accelerating voltage, 23 kV reflecting voltage.
Nanospray ESI-MS/MS
To confirm the protein marked as spot U10, Electrospray ionization (ESI)-MS/MS was also used. The peptide solution after in-gel protein digestion was collected and dried up and reconstituted with 30 µl of 0.1% TFA in 30% acetonitrile, and then desalted with ZipTip C18™ pipette tips (Millipore). The final solution was 5 µl. ESI-MS/MS was carried out with a hybrid quadrupole orthogonal acceleration tandem mass spectrometer (Q-TOF2) (Micromass, Manchester, UK). Solution (4 µl) from in-gel digestion of Coomassie stained protein spot was loaded into a borosilicate nanoflow sample needle (medium sample needle, PROTANA, Inc.) to run MS and MS/MS experiments. The capillary voltage was set to an average of 900 V and the sample cone to 30 V. Microchannel plate detector (MCP) was applied with 2,200 V. Collision gas was argon with a pressure of 0.1 MPa and collision energy was 50 V. Glufibrinopeptide was used to calibrate the instrument in the MS/MS mode. MS/MS spectra were transformed using MaxEnt3 (MassLynx, Micromass Ltd.), and amino acid sequences were interpreted manually using PepSeq (BioLynx, Micromass Ltd.).
Peptide Mass Fingerprinting
Peptide mass fingerprinting (PMF) search was performed by using the Mascot search engine (http://www.matrixscience.com) (Matrix Science, UK). Monoisotopic peptide masses were used to search the databases, allowing a peptide mass accuracy of 0.3 Da and one partial cleavage. Oxidation of methionine and carbamidomethyl modification of cysteine was considered in the process. For protein identification, peptide masses search against the database of Homo sapiens by Mascot licensed in-house, and the searches against the NCBInr database with free access on the Internet were done.
RNA Isolation and RT-PCR
Selected differentially expressed genes were further evaluated by semi-quantitative RT-PCR analysis. Total RNA from control NB4 cells and treated ones were extracted using the RNeasy kit (Qiagen, Maryland) according to the manufacturer's protocol. All RNA samples were quantified and normalized using spectrophotometric methods. First-strand cDNA was obtained by incubating total RNA samples (2 µg) with the M-MLV Reverse Transcriptase (Promega) in reaction mixture (25 µl) as manufacturer's instructions. The RT product (1 µl) was subjected to semi quantitative PCR using Taq DNA polymerase (Takara, China). Thirty cycles of amplification were carried out for most genes, and 25 cycles for GAPDH and c-jun. The list of gene-specific primers and optimal conditions used in the RT-PCR analysis is detailed in Table I. PCR amplification reactions were performed in a GeneAmp® PCR system 9700 (Applied Biosystems) and the thermo cycling conditions were as follows: 94°C for 5 min followed by 25–30 cycles at 94°C for 1 min, 55–70°C for 1 min, and 72°C for 1 min; and then 1 cycle of 72°C for 5 min was added for completion of the reaction. (Specifically: Rho GDP dissociation inhibitor beta (gene symbol: ARHGDIB): 1 cycle of 94°C for 5 min followed by 30 cycles at 94°C for 1 min and 68°C for 2 min, then 1 cycle of 72°C for 3 min was added). The PCR products were analyzed on 1% agarose gel and visualized by ethidium bromide staining.
| Gene | Primers | Annealing temperature (°C) | Cycle number |
|---|---|---|---|
| c-myc | 5′-TCTGGATCACCTTCTGCTGG-3′ | 58 | 30 |
| 5′-GATTGCTCAGGACATTTCTG-3′ | |||
| GAPDH | 5′-TGGGTGTGAACCATGAGAAG-3′ | 55 | 25 |
| 5′-GCTTGACAAAGTGGTCGTTG-3′ | |||
| ARHGDIB | 5′-GACGTGAAGCACTGAATAAATAGATCAGAATG-3′ | 68 | 30 |
| 5′-GGATGCATTCATTCTGTCCACTCCT-3′ | |||
| hnRNP D0B | 5′-CTATCACAGGGCGATCAA-3′ | 55 | 30 |
| 5′-CTGTTGCTGATATTGTTC-3′ | |||
| c-fos | 5′-AGCTATCTCCTGAAGAGGAA-3′ | 60 | 30 |
| 5′-AGGCTCCCAGTGTGCTGCAT-3′ | |||
| c-jun | 5′-ATGCCCTCAACGCCTCGTTCC-3′ | 59 | 25 |
| 5′-CTGGGCAGCGTGTTCTGGCTGT-3′ |
Western Blot Analysis
Nuclear cell lysates were prepared as described [Sadowski and Gilman, 1993]. The protein concentration was measured by using PlusOne 2-D Quant Kit (Amersham Pharmacia Biotech). The samples of nuclear matrix protein (50 µg) were separated in 12% SDS–PAGE and electro blotted to nitrocellulose membrane with Trans-blot® SD Semi-dry Transfer Cell (Bio-Rad). The membranes were blocked by 5% defatted milk powder/TBST (0.05% Tween 20, 125 mM NaCl, 25 mM Tris-Cl, pH 7.4), and then were incubated with 1:500 dilution of c-Fos (ab2) rabbit polyclonal antibody, c-Jun rabbit polyclonal antibody (Active Motif, CA) and c-Myc mouse monoclonal antibody (Clonetech, Palo Alto, CA) overnight at 4°C, respectively. After washing three times with TBS, the membranes were then incubated with a 1:10,000 dilution of peroxidase-labeled goat anti-rabbit IgGs (KPL) and with a 1:2,000 dilution of peroxidase-labeled goat anti-mouse IgGs (Santa Cruz Biotechonology) at room temperature for 1 h, respectively. The antibody complexes were detected by Enhanced Chemiluminescence (ECL) solution (Amersham Pharmacia Biotech) and exposed to high performance chemiluminescence film for 1–2 min. The visual reaction was scanned with Power-Look 2100XL (UMAX).
Statistical Analysis
Each experiment was performed at least three times. Statistical significance was determined using Student's t-test, and P < 0.05 was considered to indicate statistically significant difference.
RESULTS
Two-Dimensional Gel Analysis of Differentially Expressed Proteins via Sodium Selenite Induction in NB4 Cells
According to the quantitative differences in relative volume (1.5-fold change or greater compared with control group) by image analysis, we addressed the patterns of protein expression of NB4 cells and revealed a total of 121 protein spots altered with the induction of sodium selenite at 36 h, including 31 protein spots that exhibited a 1.5-fold change or greater in response to the induction of 20 µmol/L sodium selenite at 36 h (Fig. 1). All of the 31 protein spots were analyzed by MALDI-TOF mass spectrometry, but no reliable results were obtained for spot U10 and U11. To confirm these two proteins, ESI-MS/MS was used and only spot U10 was identified successfully as Lamin B1. Figure 2 shows the determination of a partial peptide sequence of U10. All the 30 identified proteins (26 downregulated and 4 upregulated proteins) in Table II included key regulators of signal transduction such as Rho GDI beta and alpha, members of the MAP (mitogen activated protein) kinase family, and proteins involved in the regulation of c-fos, c-jun, or c-myc expression; proteins related with metabolism; proteins related with cytoskeleton; and proteins involved in DNA damage. Some of them could be positive modulators to trigger apoptosis.
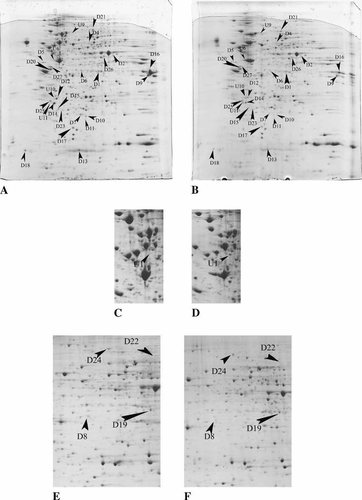
2-DE separation of the total proteins of NB4 cells induced by sodium selenite(Na2SeO3). A, C, E: control group; (B, D, F) Na2SeO3-induced group. Proteins (500 µg) from control NB4 cells and the NB4 cells induced by Na2SeO3 for 36 h, were extracted and then separated on precast IPG strips, followed by 12.5% SDS–PAGE. A, B: sample was loaded by in-gel rehydration and precast IPG strips (pH 3–10, linear, 18 cm long) were used in IEF; (C, D) sample was loaded by cup-loading on the anodic side, and precast IPG strips (pH 3–10, linear, 18 cm long) were used; (E, F) sample was loaded by in-gel rehydration, and precast IPG strips (pH 3–10, nonlinear, 24 cm long) were used. All gels were stained with Colloidal Coomassie Brilliant Blue-G250. C, D: proteins can be separated more clearly in some areas by cup-loading. U1 can only be seen in the map by this kind of sample loading in the first dimensional electrophoresis. The arrows point to proteins exhibit a 1.5-fold change or greater. These labels with “U” and “D” indicate upregulation and downregulation, respectively.
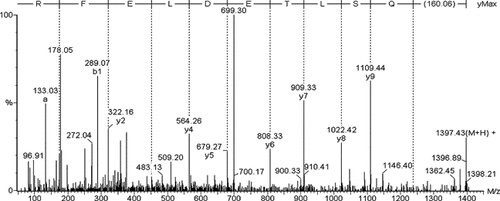
Mass spectra showing the determination of a partial peptide sequence of the protein Lamin B1 (spot U10) showed in Figure 1.
| Spot | Accession No. | Protein name | MW# (kDa) | pI# | MALDI-MS | Function* | ||
|---|---|---|---|---|---|---|---|---|
| Peptides | ||||||||
| Matching | Total | Sequence coverage | ||||||
| D1 | gi|54696696 | Annexin A1 | 8,918 | 6.57 | 15 | 19 | 47% | Signal transduction; cell communication |
| D2 | gi|68293 | Fumarate hydratase (EC 4.2.1.2) precursor, mitochondrial | 54,733 | 8.85 | 13 | 24 | 36% | Catalytic activity |
| D4 | gi|12654583 | Ribosomal protein P0 | 34,424 | 5.42 | 16 | 21 | 56% | Ribosomal unit, protein metabolism |
| D5 | gi|44890755 | Ribosomal protein SA | 32,875 | 4.83 | 14 | 25 | 48% | Ribosomal subunit, signal transdu- ction, cell communication |
| D6 | gi|432376 | Rhodanese;thiosulfate sulfurtransferase | 33,231 | 6.00 | 9 | 13 | 42% | Sulfotranserase activity |
| D7 | gi|56417681 | Splicing factor, arginine/serine-rich 3 | 14,422 | 10.12 | 4 | 9 | 37% | RNA-binding protein |
| D8 | gi|12654583 | Ribosomal protein P0 | 34,424 | 5.42 | 7 | 9 | 30% | Cell envelope, protein metabolism |
| D9 | gi|14043072 | Heterogeneous nuclear ribonucleo protein A2/B1 isoform B1 | 37,464 | 8.97 | 13 | 20 | 47% | mRNA metabolism and transport |
| D10 | gi|56417681 | Splicing factor, arginine/serine-rich 3 | 14,422 | 10.12 | 7 | 9 | 50% | RNA-binding protein |
| D11 | gi|21389379 | Hypothetical protein LOC121355 | 19,682 | 6.04 | 4 | 5 | 24% | Not defined |
| D12 | gi|938026 | Ran-binding protein 1 | 23,396 | 5.19 | 8 | 18 | 30% | Transport activity; signal transduction, cell communication |
| D13 | gi|30582585 | Ubiquitin-conjugating enzyme E2N | 17,184 | 6.13 | 10 | 21 | 56% | Ubiquitin proteasome system protein, protein metabolism |
| D14 | gi|56676393 | Rho GDP dissociation inhibitor (GDI) beta | 22,988 | 5.1 | 6 | 13 | 36% | Signal transduction; cell communication |
| D15 | gi|182385 | Long-chain acyl-CoA synthetase | 78,297 | 7.99 | 6 | 8 | 8% | Activates long chain fatty acids |
| D16 | gi|35053 | Uracil DNA glycosylase | 35,698 | 8.22 | 14 | 20 | 45% | DNA repair protein |
| D17 | gi|61364276 | dUTP pyrophosphatase | 17,908 | 6.15 | 8 | 12 | 48 | Metabolix;energy pathway |
| D18 | gi|38383133 | Ribosomal protein P2 | 11,658 | 4.42 | 4 | 7 | 69% | Ribosomal subunit; protein metabolism |
| D19 | gi|47124405 | Poly(rC)-binding protein 1 | 37,987 | 6.66 | 17 | 28 | 48% | Signal transduction |
| D20 | gi|58476967 | HNRPC protein | 27,861 | 4.55 | 15 | 21 | 40% | RNA-binding. metabolism |
| D21 | gi|4505257 | Moesin | 67,778 | 6.08 | 16 | 28 | 25% | Structural constituent |
| D22 | gi|37078490 | Far upstream element-binding protein 1 | 67,431 | 7.18 | 16 | 26 | 33% | Transtiption regulatory activity |
| D23 | gi|20072835 | ACSL1 protein | 59,945 | 7.18 | 6 | 8 | 11% | Metabolism |
| D24 | gi|46249758 | Cytovillin | 69,313 | 5.94 | 20 | 36 | 30% | Cytoskeletal anchoring activity |
| D25 | gi|30582607 | Rho GDP dissociation inhibitor (GDI) alpha | 23,250 | 5.02 | 7 | 11 | 28% | Signal transduction; cell communication |
| D26 | gi|2773158 | Heterogeneous nuclear ribonucleoprotein D0B | 32,120 | 8.73 | 8 | 26 | 26% | Regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolism |
| D27 | gi|55667133 | Serine/threonine kinase receptor-associated protein | 38,756 | 4.98 | 7 | 14 | 27% | Regulator of cell cycle, signal transduction;cell communication |
| U1 | gi:14389309 | Tubuliln alpha 6 | 49,895 | 4.96 | 12 | 23 | 28% | Cell structure and transduction |
| U6 | gi|15277503 | ACTB protein | 40,542 | 5.55 | 14 | 22 | 44% | Cell structure and transduction |
| U9 | gi|61358117 | Mitogen-activated protein kinase kinase 6 | 37,468 | 7.01 | 6 | 13 | 28% | Signal transduction |
| U10§ | gi|576840 | Lamin B1 | 66,412 | 5.20 | / | / | / | Structural molecule activity, cell growth, and/or maintance |
- These labels with “U”and “D” indicate upregulation and downregulation, respectively.
- # Theoretical.
- * All the information about function come from HPRD, SWISS-PROT, NCBI database.
- § ESI-MS/MS was used to confirm the protein marked as spot U10.
RT-PCR Demonstrates the Complex Regulation of c-fos, c-myc, and c-jun
Among our identified proteins, the hnRNP D0B (Fig. 1, D26), Rho GDP dissociation inhibitor (GDI) beta (gene symbol: ARHGDIB) (Fig. 1, D14) and Rho GDI alpha (GDIR) (Fig. 1, D25), members of the Rho GDI family, were all downregulated in this apoptotic process. The results achieved by the semi-quantitative RT-PCR analysis of hnRNP D0B and ARHGDIB shown in Figure 3A illustrated that there was essentially downregulation in the expression of the two genes as a function of sodium selenite treatment.
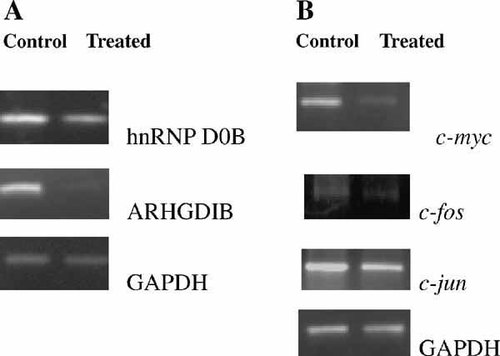
Confirmation of differentially expressed proteins by semi-quantitative RT-PCR analysis in control and sodium selenite-induced NB4 cells. The expression of GAPDH was used as an internal control for equal loading.
The hnRNP D0B (Fig. 1, D26), play certain roles in the complex regulation of c-fos or c-myc expression. The cleavage of Rho GDI alpha could lead to activation of Jun N-terminal kinase (JNK), which phosphorylate Jun within its N-terminal transactivation domain (residues Ser63 and Ser73) and thereby enhance its transactivation potential.
So, we further investigated the mRNA level of c-myc, c-fos, and c-jun. The results of RT-PCR (Fig. 3B) analysis showed that their expressions were significantly decreased after 36 h culture with 20 µmol/L sodium selenite.
Western Blot Analysis of c-Fos, c-Jun, and c-Myc
To confirm the results of mass spectrometry and RT-PCR, three proteins, c-Fos, c-Jun, and c-Myc, under the regulation of certain identified proteins such as the hnRNP D0B, Rho GDP dissociation inhibitor (GDI) beta (gene symbol: ARHGDIB), and Rho GDI alpha (GDIR), were measured by Western blot analysis. The results showed that there is also significant downregulation in expressions of c-Fos, c-Jun, and c-Myc in the apoptosis of NB4 cells (Fig. 4).
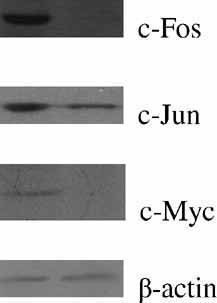
Western blot analysis of c-Fos and c-Jun in response to sodium selenite for 36 h. The expression of β-actin was used as an internal control for equal loading.
DISCUSSION
Among the identified proteins, some proteins of interest may play certain roles in the complex regulation of c-Myc, c-Fos, and c-Jun expression.
The hnRNP D0B (Fig. 1, D26), belongs to hnRNP D0, which is one of the five mCRD (the c-fos major protein coding-region determinant)-associated proteins. Overexpression of the five proteins stabilized mCRD-containing mRNA by impeding deadenylation. The decay of mRNA is mediated by the instability of c-fos major protein coding-region determinant [Grosset et al., 2000; Chang et al., 2004]. The hnRNP D0 binds with high affinity to RNA molecules that contain AREs (AU-rich elements) from myc, fos mRNAs, and other proto-oncogenes and cytokine mRNAs. The hnRNP D0 acts as a potential mediator of ARE-directed mRNA degradation [Brewer, 1991; Zhang et al., 1993].
Expression of FBP 1 (far upstream element (FUSE)-binding protein 1) (Fig. 1, D22) in human leukemia cells stimulates the activity of the myc promoter in a FUSE-dependent manner. The c-myc regulatory region includes binding sites for a large set of transcription factors. Certain study demonstrated that in the absence of FBP, the remainder of the set fails to sustain endogenous c-myc expression [He et al., 2000]. Deregulated expression of c-Myc can either induce or sensitize cells to apoptosis. Apoptosis associated with inappropriate Myc expression limits the tumorigenic effect of the c-myc proto-oncogene [Hoffman and Liebermann, 1998]. Reversal of aberrant gene expression that is induced by the proto-oncogene c-myc is likely to be effective for treating a variety of tumors that rely on this pathway for growth. It may be theoretical targets for small-molecule therapy [Huth et al., 2004]. Fos-mediated apoptosis was accelerated by deregulated c-Myc. Fos/Jun transcription factor complexes play a role in modulating both myeloid cell survival and differentiation, and suggest that genetic lesions that alter Fos expression may cooperate with deregulated c-Myc in leukemogenesis [Shafarenko et al., 2004].
The sodium selenite induced suppression of Poly (rC)-binding protein 1(Fig. 1, D19) (HNRPE1) and HNRPC protein (Fig. 1, D20). Both of them have been shown to bind and enhance translation at the internal ribosomal entry site element (IRES) of c-myc [Kim et al., 2003; Evans et al., 2003]. The ATRA treatment of APL demonstrated the suppression of Poly (rC)-binding protein 1(HNRPE1) and HNRPC protein involved in the complex regulation of c-myc expression is sharply downregulated [Dimberg et al., 2002; Harris et al., 2004].
Evidence for further confirmation came from the RT-PCR results of the mRNA that encodes suppressed hnRNP D0B and c-myc (Fig. 3). In Figure 3, it was noticed that the expression of both hnRNP D0B and c-myc declined by 36 h of induction. This pattern is consistent with their known way we have just discussed. In addition, the RT-PCR analysis also showed that the expression of c-fos was suppressed by the induction of sodium selenite (Fig. 3). The Western blot analysis showed that the expression level of c-Fos and c-Myc was decreased (Fig. 4). So these mechanistic studies provide vital information regarding the role of c-myc and c-fos in the apoptotic process induced by sodium selenite.
Rho GDI beta (Fig. 1, D14) and alpha (Fig. 1, D25), are members of the Rho GDI family. As its name implies, Rho GDI inhibits the dissociation of GDP from the GDP-bound form and sequesters Rac, Rho, and Cdc42 in the inactive form. Rho GDI alpha was identified and characterized as a cell death mediator and as a potential anticancer target [Mackeigan et al., 2003]. Implicating Rho GDI alpha in cell death suggests the importance of the traditional role Rho GDI alpha plays in controlling cellular responses through the small GTPases Rac, Rho, and Cdc42. And Rho GDI alpha has been identified as downregulated during ATRA and the cyclin-dependent kinase inhibitor bohemine treatment [Kovarova et al., 2000; Wan et al., 2001]. These results add a novel role for Rho GDI alpha as an important event in the apoptotic response in cancer cells. The downregulation of Rho GDI beta and alpha in this study may play active roles in the apoptosis induced by sodium selenite.
The cleavage of Rho GDI alpha could lead to activation of JNK, which had been implicated as an upstream regulator of apoptosis in some systems [Krieser et al., 2002]. The activation of JNK, phosphorylates Jun within its N-terminal transactivation domain and thereby enhances its transactivation potential. In this study, the decreased mRNA level of c-jun was investigated by RT-PCR analysis (Fig. 3B). And the Western blot analysis confirmed a decreased amount of c-Jun at protein level (Fig. 4). Thus, the decrease of c-Jun in the apoptotic NB4 cells may be important.
JNKs are members of the mitogen-activated protein kinase(MAPK) superfamily. Activated by MAPK cascades, the JNKs translocate to the nucleus, where they phosphorylate Jun [Hess et al., 2004]. There are other identified proteins related with the MAPK family in this study. All of these show that the MAPK pathway may be involved in the apoptosis induced by sodium selenite.
Several of the differentially expressed proteins identified in this study, which are related with the MAPK family will be discussed below.
Serine threonine kinase receptor-associated protein (UNRIP) (Fig. 1, D27), which interacts with UNR, was also found. UNR is the other one of the five mCRD-associated proteins. Matsuda et al. [2000] isolated cDNAs encoding UNRIP, which they termed as MAPK activator with WD repeats, or MAWD. They reported overexpression of MAWD induces expression of activated MAPK and correlates with anchorage-independent growth in vitro. They detected expression of MAWD as a 39-kD protein in breast cancer but not normal tissue and proposed that the MAWD gene may be a frequent target for alteration in breast cancer. So UNRIP (also named as MAWD) may be also a target in the apoptosis of NB4 cells.
Cytovillin (ezrin) (Fig. 1, D24) is a microvillar cytoplasmic peripheral membrane protein that is expressed strongly in placental syncytiotrophoblast and in certain human tumors. It is the same as ezrin, a component of the microvilli of intestinal epithelial cells that serves as a major cytoplasmic substrate for certain protein-tyrosine kinases. Both moesin (Fig. 1, D21) and ezrin belong to the so-called ERM proteins. They act as linkers between the plasma membrane and the actin cytoskeleton. MAPK3 phosphorylation and activity were reduced when ezrin was suppressed. Ezrin-mediated early metastatic survival was partially dependent on the activation of MAPK. In this study, both ezrin and moesin were downregulated.
Mitogen-activated protein kinase kinase 6 (Fig. 1, U9), which is related with the MAPK family, was upregulated in this research.
Hence, it is reasonable to postulate that the MAPK family may be involved in apoptotic effects induced by sodium selenite. Continued mechanistic studies are needed to provide vital information regarding the MAPK pathway in the apoptotic process induced by sodium selenite.
Some proteins listed in Table I and Figure 1, such as fumarate hydrataseprecursor (D2), Ribosomal protein P0 (D4, 8), long-chain acyl-CoA synthetase (D15), uracil DNA glycosylase (D16), Ribosomal protein P2 (D18), are related with metabolism. Their downregulation shows that sodium selenite (≥5 µmol/L) suppressed the NB4 cell growth, and then induced apoptosis. Both Ribosomal protein P0 (Fig. 1, D4/8) and Ribosomal protein P2 (Fig. 1, D18) belong to acidic phosphoprotein component of the large 60S ribosomal subunit. And Ribosomal protein P2 plays an important role in the elongation step of protein synthesis. Fumarate hydratase precursor (Fig. 1, D2) is an enzymatic component of the tricarboxylic acid (TCA) cycle, and catalyzes the formation of L-malate from fumarate. The long-chain acyl-coenzyme A synthetase (LACS) (Fig. 1, D15), which is also known as palmitoyl-CoA ligase, plays a key role in both the synthesis of cellular lipids and the degradation of fatty acids.
Some previous study showed that DNA damage was also involved in selenite-induced apoptosis [Zhou et al., 2003]. The apoptosis of NB4 cells induced by sodium selenite was determined by DNA ladder electrophoresis [Li et al., 2002b]. The dUTP pyrophosphatase (Fig. 1, D17) is involved in nucleotide metabolism. It produces dUMP, the immediate precursor of thymidine nucleotides, and it decreases the intracellular concentration of dUTP so that uracil cannot be incorporated into DNA, and unusually high levels of dUMP incorporation into DNA during replication and repair, in turn, cause DNA fragmentation and cell death [Lindahl, 1982].
The downregulation of ubiquitin-conjugating enzyme E2N (UBE2N) (Fig. 1, D13) also contributes to DNA damage. In yeast [Hoffman and Liebermann, 1998], UBE2N, which referred to as UBC13, formed a specific heteromeric complex with MMS2, ubiquitin-conjugating enzyme variant (UBE2V2). A model in which an MMS2/UBC13 complex assembles novel polyubiquitin chains for signaling in DNA repair was supported. Hoege et al. [2002] demonstrated that some modifications in which UBC13 is involved differentially affect resistance to DNA damage, and that damage-induced PCNA (proliferating cell nuclear antigen) ubiquitination is elementary for DNA repair and occurs at the same conserved residue in yeast and human.
Lamin B1 (Fig. 2, U10) belongs to Lamins which are the major components of the nuclear lamina that underlies the nuclear envelope of eukaryotic cells. The upregulation of Lamin B1 showed that in this research, it might play certain roles.
ACTB (Fig. 1, U6) was identified as upregulated protein. As a cytoskeleton protein, the disruption and reorganization of actin filaments to form apoptotic blebs [Hoffman and Liebermann, 1998] were previously observed during apoptosis. It is feasible that actin acts as a positive regulator of apoptosis [Koya et al., 2000; Wan et al., 2001].
The upregulation of tubulin alpha 6 (Fig. 1, U1) was also found. Chen et al. [2002] reported that Tat (a small trans-acting regulatory protein encoded HIV-1) binds tubulin/microtubules leading to the alteration of microtubule dynamics and the activation of mitochondria-dependent apoptosis. Bim, a proapoptotic Bcl-2 relative and a transducer of death signals initiated by perturbation of microtubule dynamics, facilitates the Tat-induced apoptosis. The disruption of microtubules can induce cell-cycle arrest in M-phase, formation of abnormal mitotic spindles, and final triggering of the signals for programmed cell death [Mitchison, 1988]. In addition, Li and Broome [1999] reported that As2O3 treatment resulted in cell-cycle progression arrest at metaphase in myeloid leukemia cells through non-competitive disruption of GTP binding to tubulin and inhibition of GTP-induced tubulin polymerization [Ling et al., 2002]. Agents such as the vinca alkaloids and the taxanes, which affect the dynamics of microtubules, have emerged as useful therapeutic agents for the treatment of human cancer [Rowinsky and Donehower, 1991; Hadfield et al., 2003]. So the altered tubulin may show that the perturbation of microtubule dynamics facilitates the Se-induced apoptosis in NB4 cells.
Annexin I (ANX AI) (Fig. 1, D1) is a member of the calcium-dependent phospholipid-binding protein family. Mechanistic study demonstrates a complex regulatory role of caspase-dependent apoptosis where ANX AI is processed at the N-terminal region which could give susceptibility to apoptosis upon ceramide treatment [Debret et al., 2003].
Splicing factor, arginine/serine-rich 3 (SRp20) (Fig. 1, D7) was also identified. Jumaa et al. [1997] demonstrated that in vivo SRp20 mRNA levels are cell cycle regulated and that the SRp20 mRNA is itself alternatively spliced, apparently in a cell cycle-specific manner. Sequence analysis revealed that the SRp20 promoter contains two consensus-binding sites for E2F, a transcription factor thought to be involved in regulating the cell cycle. This suggests that cellular pre-mRNA splicing may be regulated during the cell cycle, perhaps in part by regulated expression of SR proteins.
Although more effort is required to provide vital information regarding the role of c-Myc, c-Jun, c-Fos, and MAPK pathway in the apoptotic process induced by sodium selenite, this work lays the foundation for the future studies aimed to further validate some potential regulators.
Acknowledgements
The authors thank Professor Liuyu Huang, Professor Guofu Su, Professor Kaihua Wei, and Mr. Jie Wang for technical assistance and helpful discussions. We thank Mr. Douglas Allan for editorial assistance.