Cardiac troponin I sense-antisense RNA duplexes in the myocardium
Abstract
Natural antisense RNA is now thought to regulate, at least in part, a growing number of eukaryotic genes. It is becoming increasingly apparent that such endogenous antisense RNA molecules may modulate gene expression in a manner analogous to synthetic oligomers. Here, we report the detection of antisense-orientated RNA transcripts of cardiac specific troponin I in rat and human myocardium. Interestingly, the different sizes of the rat and human antisense cTNI transcripts suggest species-specific reverse transcription initiation sites. Moreover, for the first time in cardiomyocytes, we could demonstrate in vivo duplex formation between sense and antisense transcripts. The existence of antisense-sense duplexes represents compelling evidence and a potential mechanism for endogenous antisense transcript-mediated modulation of mRNA translation. The potential effect of attenuating translation was illustrated by in vitro and in vivo model systems. Testing several oligonucleotides based on the natural antisense sequences, the optimal region for inhibition of translation was identified as being close to the translational start codon. J. Cell. Biochem. 85: 198–207, 2002. © 2002 Wiley-Liss, Inc.
Gene expression in eukaryotes is regulated at each step from the gene to the native protein. Recently, attention has focused on regulatory mechanisms involving endogenous antisense RNA, since cis-encoded antisense RNA has been detected for an increasing number of genes [Kimelman and Kirschner, 1989; Kumar and Carmichael, 1998; Shi et al., 2000]. Antisense transcripts are thought to form duplexes with the complementary sense strands and hence modulate gene expression at transcriptional, post-transcriptional or translational levels [Knee and Murphy, 1997]. Work in our group previously revealed antisense transcription of both MHC-isogenes (α and β) in the rat's heart [Luther et al., 1998], and recently, we were able to demonstrate antisense transcripts for β- but not α-MHC in human myocardium [Luther et al., 2001].
In order to screen for further heart specific genes with antisense transcription as an additional regulatory mechanism, we investigated whether rat and human myocardium synthesize cardiac troponin I (cTNI) antisense RNA. TNI is part of the heterotrimeric troponin complex and plays a crucial role in actin/myosin interaction by regulating conformational changes in the tropomyosin complex [Tobacman, 1996; Solaro and Rarick, 1998]. Phosphorylation of cTNI by cAMP-dependent kinase (PKA) induces a significant increase in the relaxatory properties of cardiac muscle by reducing the calcium affinity of Troponin C [Holroyde et al., 1980], the protein that acts as the receptor for calcium ions released from the sarcoplasmatic reticulum.
Reduction of cTNI levels in the heart significantly alters the mechanical properties of cardiac muscle and is observed in cardiac-diseased-states in particular. A 40% decrease of cTNI protein was detected in rat myocardium subjected to complete ischemia for 1 h complex [Westfall and Solaro, 1992]. cTNI protein levels were also reduced by 40–80% after experimental myocardium infarction in rat and remained at this low level for 7 days [Li et al., 1997]. Since in an animal model, cTNI knockout mice were shown to suffer acute heart failure leading to death, it is clear that cTNI is essential for normal heart function. Ventricular myocytes of cTNI-deficient mice demonstrated elevated resting tension and deteriorated Ca2+ sensitivity of the regulatory system [Huang et al., 1999].
A cardiac-specific isoform of TNI (cTNI) accumulates in the heart during postnatal development and is exclusively expressed in the adult myocardium [Saggin et al., 1989; Ausoni et al., 1991; Murphy et al., 1991; Sasse et al., 1993], while co-expression of the fetal or slow skeletal isoform (ssTNI) decreases, becoming undetectable 1 month after birth. The fetal isoform lacks functionally important phosphorylation sites (two serines) for protein kinase A myocardium [Bartel et al., 1994; Westfall et al., 2001]. These are located at the N-terminus of cTNI and mediate the β-adrenergic-induced decrease in calcium sensitivity of the myofilament. In contrast to expression patterns of MHC isoforms, fetal TNI is not re-expressed in the senescent or failing heart [Wade and Kedes, 1989; Ball and Solaro, 1994]. Regulation of cTNI expression at transcriptional levels has already been characterized and several cis-acting DNA elements and transcription factors were identified [Murphy et al., 1997; Di Lisi et al., 1998].
Post-translational modifications of cTNI—including phosphorylation, proteolysis, and formation of crosslinks—which were observed in hypertrophy and ischemia/reperfusion injury, compromise the control of Ca2+-dependent contraction myofilaments [Solaro, 1999]. The aim of this study was to identify and analyze endogenous antisense transcripts of cTNI and their possible regulatory role in gene expression in rat or human myocardium.
MATERIALS AND METHODS
Animals
Female Wistar rats with an approximate weight of 200 g were used. After excision, the hearts were immediately frozen in liquid nitrogen and stored at −80°C.
Total heart RNA from normal human myocardium (three men, age 34.7 ± 10.9 years; range, 26–50 years, from persons without known heart disease), was obtained commercially from Invitrogen®, Groningen.
Neonatal Heart Cell Culture
To isolate neonatal rat myocytes, minced ventricles of 1- or 2-day-old Wistar rats were digested with 50 μg/ml trypsin (Cell systems) in 10 ml calcium and magnesium-free Hanks' Balanced Salt Solution (Gibco) for 18 h at 4°C. After stopping the digestion and oxygenation for 1 min, 5 ml of crude collagenase (Worthington) was added, and the cells were incubated for 45 min at 37°C. The cells were then washed twice with Leibovitz L-15 medium, centrifuged and resuspended in Medium 199 with Earle's Salt, 10% newborn calf serum (NBCS), 2 mM l-glutamine, 100 U/100 μg penicillin/streptomycin, 0.01 mM cytosine-d-arabinofuranoside. The cells were cultured as monolayers at a density of 200,000 cells/cm2. The medium was changed daily.
RNA Preparation
Total and pure mRNA was prepared as described elsewhere [Luther et al., 1998, 2001]. The ratio of optical density at 260 and 280 nm was ≥ 1.8 in all cases. After isolation, RNA was treated DNase I (Ambion). DNase I efficacy was checked by using DNA-specific primers in RT-PCR reactions.
Detection of Duplex RNA
For the detection of duplex cTNI transcripts, RNA was isolated under nondenaturing conditions from neonatal rat cardiomyocytes cells cultured for 4 days. Briefly, the cells were washed three times with PBS, transferred to a reaction tube, and centrifuged for 5 min at 300g at 4°C. The pellet was lysed with 125 μl ice-cold buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 0.5% Igepal) and incubated for 5 min on ice. To prepare exclusively cytoplasmic RNA, nuclei were removed by centrifugation. The supernatant was carefully vortexed in the presence of 0.2% SDS and proteins were degraded by adding proteinase K for 15 min at 37°C. Subsequently, cytoplasmic RNA was extracted by phenol/chloroform followed by ethanol precipitation. The RNA pellet was dissolved in water.
In addition to DNase I degradation, cytoplasmic or total RNA was treated with 1.5 μl RNase A (Roche Molecular Biochemicals 1 μg/μl) for 20 min at 37°C, and the reaction stopped by adding EDTA, before performing RT-PCR.
Reverse Transcription and Polymerase Chain Reaction (RT-PCR)
RT-PCR was carried out under standard conditions [Luther et al., 2001] using primer pairs listed in Table I and Figure 1. All PCR products were sequenced in both directions.
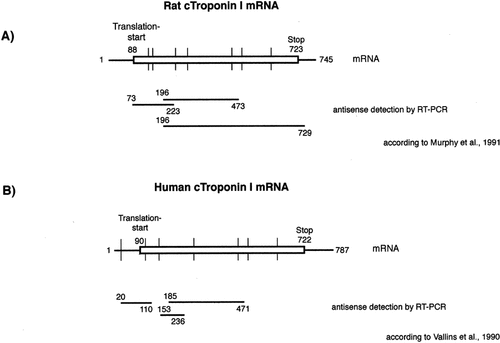
Cardiac TNI spliced transcript sequence and antisense fragments amplified by RT-PCR in rat (A) and human myocardium (B). The vertical lines spanning the transcript represent the positions of spliced-out introns.
| Primer set | cTNI | Forward | Position | Reverse | Position |
|---|---|---|---|---|---|
| 1 | Rat M57679 | 5′ CCT CTG GAG ATC AGC ATG GCG | 73 | 5′ TGG AGG CGG AGA TCT TAG ACT | 223 |
| 2 | M57679 | 5′ GCC ACA TGC CAA GAA AAA GTC | 186 | 5′ GAT CTG CAA TCT CAG TGA TGT TC | 473 |
| 3 | M57679 | 5′ GCC ACA TGC CAA GAA AAA GTC | 186 | 5′ CAT GGC TCA GCC CTC AAA C | 729 |
| 4 | Human X54163 | 5′ CCC CCT CAC TGA CCC TCC AAA C | 20 | 5′ ATC GCT GCT CCC ATC CGC CAT | 110 |
| 5 | X54163 | 5′ CGC TCC TCC AAC TAC CGC GC | 153 | 5′ CAG CTG CAA TTT TCT CGA GGC G | 236 |
| 6 | X54163 | 5′ GCC GCA CGC CAA GAA AAA ATC | 185 | 5′ GAT CTG CAA TCT CCG TGA TGT TC | 471 |
Northern Blot Hybridization
Northern blotting using total RNA (10 μg/lane) from adult rat or human hearts were performed as described earlier [Luther et al., 2001]. For sense and antisense cTNI detection in the rat DIG-labeled oligonucleotide hybridization probes (for antisense forward 174: 5′ CTA TGC CAC CGA GCC ACA TGC CAA GAA AAA GTC TAA GAT CTC CGC; for sense reverse 485: 5′ TCT TCT GGG TCA GAT CTG CAA TCT CAG TGA TGT TCT TGG TGA CTT) were used. Hybridization was carried out overnight with 5 pmol/μl of oligonucleotide at 50°C for sense, and 10 pmol/μl oligonucleotide at 40°C for antisense detection. After several washing steps, the hybrids were detected by incubation with an anti-digoxygenin/alkaline phosphatase conjugate and subsequent color reaction of the substrate 5-bromo-4-chloro-3-indolyl phosphate/nitroblue-tetrazolium (BCIP/NBT).
The detection of human cTNI sense and antisense RNA was performed using radioactive α 32p-CTP-labeled RNA probes transcribed from the pGEM3Z vector described below.
In Vitro Transcription/Translation
In vitro transcription/translation experiments were performed using TNT® T7 Quick Coupled Transcription/Translation System (T7 TNT, Promega) with [35S]-labeled methionine/ cysteine (10 μCi), as described elsewhere [Luther et al., 2001]. The template vector pGEM3Z (Promega) contains the cDNA of human cTNI from position 87 to 730 according to Vallins et al. [1990], which we confirmed by DNA sequencing in both directions. Antisense oligonucleotides (Table II) were added to the reaction mix at a concentration between 1 and 8 μM. Controls contained either no added oligonucleotide or a scrambled sequence, and protein products were analyzed by SDS–PAGE followed by autoradiography.
| Antisense | Positions | Scrambled | |
|---|---|---|---|
| 1 | 5′ ATC GCT GCT CCC ATC CGC CAT | 90–110 | 5′ CCA TAT CGT CCG CTG CCA TCC |
| 2 | 5′ CAG CTG CAA TTT TCT CGA GGC G | 215–236 | |
| 3 | 5′ TCT TCT GGG TCA GAT CTG CAA TCT CAG TGA TGT TCT TGG TGA CTT | 438–483 |
- * According to Vallins et al. [1990].
Cardiac Troponin I Expression in Cardiomyocytes
Rat cardiomyocytes cells were cultured for 2 days in the presence of 10 μM phosphorothioate oligonucleotides (antisense 108: 5′ ATC GCT GCT CTC ATC CGC CAT, according to Murphy et al. [1991]; scrambled: 5′ CCA TAT CGT CCG CTG CCA TTC) in serum-free medium for 2 days. The medium and oligonucleotides were changed daily. Timed controls were treated with the same volume of medium without oligonucleotides. Cardiomyocytes were harvested and protein was isolated by adding 0.1 μl RIPA-buffer (9.1 mmol/L dibasic sodium phosphate, 1.7 mmol/L monodibasic sodium phosphate, 150 mmol/L NaCl, pH 7.4, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS) with freshly added inhibitor mix (Boehringer Manneheim). The cells were homogenized, incubated on ice for 1 h, and centrifuged for 30 min at 4°C. Protein concentration of the supernatant was measured by Biorad reagents before analysis by Western blotting.
Electrophoresis and Western Blotting
Samples containing 40 μg of protein homogenate were electrophoretically separated by SDS–PAGE and blotted to a PVDF-membrane. To detect cTNI protein levels polyclonal cTNI-antibodies (Santa Cruz Biotechnologies, 1:1,000 dilution) was used as primary antibodies and horseradish peroxidase conjugated anti-goat-antibodies (Santa Cruz Biotechnologies, 1: 8,000 dilution) as secondary antibodies. Protein visualization by chemiluminescence was performed using the ECL-Kit (Amersham). The optical density of the detected signals was measured by densitometric analysis.
Statistics
Values are expressed as means ± SD. Significance was tested either by Student's t-test for unpaired groups or by one-way ANOVA including Bonferroni Posthoc Test for selected pairs.
RESULTS
Detection and Characterization of Antisense cTNI Transcripts
Sense and antisense cTNI transcripts in rat and human heart tissue were detected by performing RT-PCR as well as Northern blotting. For amplification of antisense RNA by RT-PCR, sense-sequence primers were added to the reverse transcription instead of reverse primers. RT-PCR using different sequence overlapping primer pairs resulted in three antisense signals from position 73 to 223, 186 to 473, and 186 to 729 in the rat heart (Fig. 1A). Since primer pairs were located in different exons, amplification of contaminating genomic DNA, or unspliced mRNA, would have generated longer PCR-products. Therefore, both DNA and pre-spliced transcripts can be definitively excluded as the sources of antisense signals. Analyzing each PCR product by sequencing revealed the antisense RNA to be completely complementary to the sequence of the corresponding sense mRNA. Furthermore, both rat and human antisense transcripts were apparently polyadenylated, since the amounts of antisense products generated were comparable using poly(A) RNA preparations or total RNA as templates in RT-PCR amplification (data not shown).
Northern blotting also revealed the existence of antisense RNA transcripts in the rat's heart of a size identical to the sense transcripts (around 800 bp) (Fig. 2A). When human myocardium was investigated, Northern blotting experiments showed sense cTNI RNA with a size of approximately 1,000 bp (Fig. 2B). The detected antisense cTNI transcripts of 450 bp were clearly shorter than the sense transcripts, which correlates well with our failure to amplify an antisense transcript complementary to the 3′ end of the human cTNI mRNA (Fig. 1B). RT-PCR produced antisense signals only with primer pairs spanning regions 20–110, 153–236, and 186–471 of the human cTNI cDNA. As with the rat products, the human antisense RNAs were identical in sequence to the spliced mRNA and were polyadenylated.
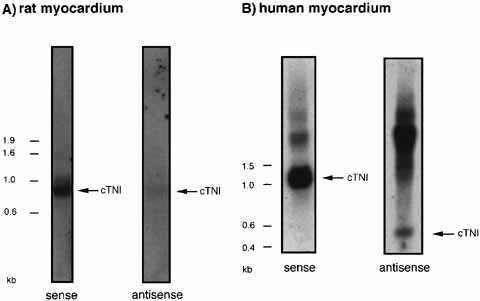
Northern blot analysis of rat (A) and human (B) myocardium. A 10 μg of total RNA were hybridized with the appropriate DIG-labeled (A) or α 32p-CTP-labeled (B) oligonucleotides (forward orientation for antisense detection; reverse orientation for sense detection). Specific signals indicated the length of the sense transcript, which is different in rat (ca. 800 bp) and human (1,100 bp). Antisense signals show a different amount of antisense transcript compared to the sense counterpart in both species. The high background of unspecific signals in human sense and antisense samples (B) are due to the long exposure time of the autoradiograph (1 week).
cTNI Antisense/Sense Ratio
The proportion of antisense to sense cTNI was determined by RT-PCR with using primer pairs 73-223 and 185-471 for rat and human, respectively. Semi-quantification revealed a ratio of antisense to sense cTNI RNA of 52 ± 10% in the adult rat heart and 38 ± 8% in the human myocardium of normal controls. Since cTNI expression is developmentally regulated during the first days after birth, we measured the antisense/sense ratio of cTNI in heart tissue of neonatal rats. In the neonatal rat heart relative levels of antisense cTNI corresponded approximately to sense cTNI levels (94 ± 17% of sense cTNI). Thus, comparing antisense/sense ratios in neonatal and adult myocardium revealed that relative antisense cTNI levels (normalized to sense cTNI mRNA) were significantly higher (P < 0.01) in neonatal hearts of the rat (Fig. 3).
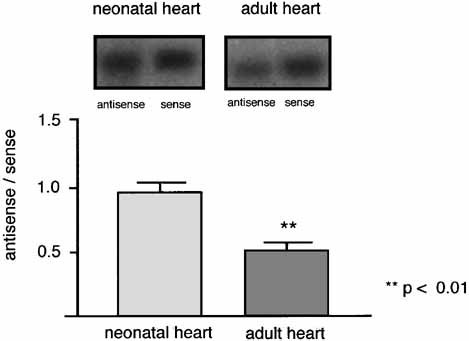
Relation of antisense to sense cTNI in rat heart. Significant higher relative amount of antisense TNI RNA in neonatal rat heart compared to adult heart. Data were obtained by semi-quantitative RT-PCR using primer pair 73-223.Values are means ± SD. Student's t-test for unpaired groups, levels of significance are indicated.
Detection of Antisense-Sense Duplexes of cTNI
In order to demonstrate RNA antisense-sense duplexes, we performed a modified RNase A protection assay. Since RNase A exclusively digests single-stranded RNA, double-stranded RNA is resistant to treatment. Following RNase A treatment of cytoplasmic, subsequent analysis of rat cTNI by RT-PCR generated specific sense and antisense products (Fig. 4A), indicating that a certain fraction of both RNA transcripts must be in a mutually duplex state. The efficacy of the RNase A treatment was confirmed by analyzing genes showing no evidence of natural antisense RNA and thus existing solely in the single-stranded form. For example, RT-PCR of beta-actin or GAPDH mRNA demonstrated complete absence of product generation following RNase A digestion (Fig. 4B).
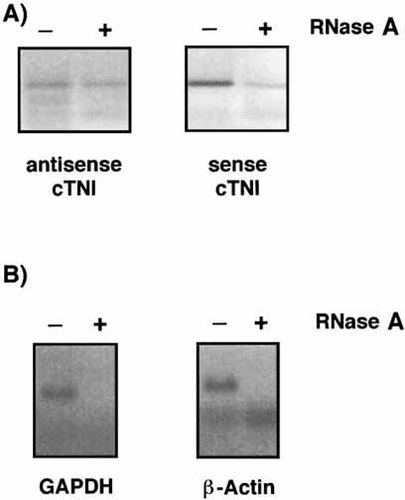
Duplex cTNI sense and antisense transcripts present in vivo detected by RNase A protection assay. Following RNase A digestion of single stranded RNA from cytoplasmic or total RNA isolated from neonatal rat cardiomyocytes cell cultures, standard RT-PCR of cTNI produced specific signals of equal intensity for both sense and antisense transcripts (A). The efficacy of RNA digestion was confirmed by the effect of RNase A on the RT-PCR signals of control genes GAPDH and beta-actin (B).
Modulation of cTNI Gene Expression by Antisense Oligonucleotides
To investigate the ability of antisense transcripts to inhibit specific protein synthesis, we performed in vitro transcription/translation experiments, adding three different antisense oligonucleotides (ASO, Table II) to the assay. ASO 90-110 spanning the AUG initiation codon of the human cDNA clone (pGEM3Z; see Materials and Methods) exerted the most significant inhibitory effect on protein synthesis (Fig. 5). ASO 215–236 and 438–483 slightly reduced the protein levels (100 ± 11.0% for control; 17.7 ± 15.7% with ASO 90–110, P < 0.001; 89.8 ± 29.5% with ASO 215–236; 76.5 ± 3.8% with ASO 438–483 as quantified by densitometry. The specificity of the ASO 90–110-mediated reduction of protein levels was tested by comparing the effect of adding a scrambled oligonucleotide to the reaction. The scrambled oligonucleotide did not significantly decrease the cTNI protein amount compared to the control, in stark contrast to ASO 90–110 (100 ± 14.9% for control; 17.7 ± 15.7% with ASO 90–110; 73.7 ± 27.7% with scrambled oligo (Fig. 6A). The attenuation of protein synthesis by ASO 90–110 showed a concentration dependency within a range of 1–8 μM, protein synthesis being almost completely suppressed by 8 μM ASO 90–110 (9 ± 5% of the control: data not shown).
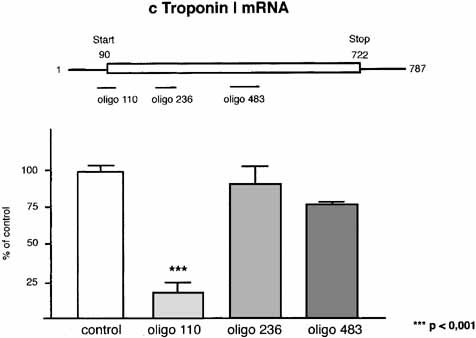
Effect of three different antisense oligonucleotides on cTNI translation in vitro. Circular plasmid DNA (pGEM3Z, Promega) with cloned full-length cDNA sequences from human cardiac TNI provided the template for the in vitro transcription/translation assay (see Materials and Methods). Antisense oligonucleotides corresponding to positions 90–110 (2 μM), 215–236 (2 μM) or 438–483 (4 μM) were added to the reaction. Protein products were separated by SDS–PAGE, and the radioactive bands were visualized by autoradiography. Semi-quantitative data represent densitometry measurements of the protein bands shown as a bar chart. Each value is based on three experiments. The percentage effect of inhibition refers to the control, which was set at 100%. Values are means ± SD one-way ANOVA with Bonferroni Posthoc test, with levels of significance indicated.
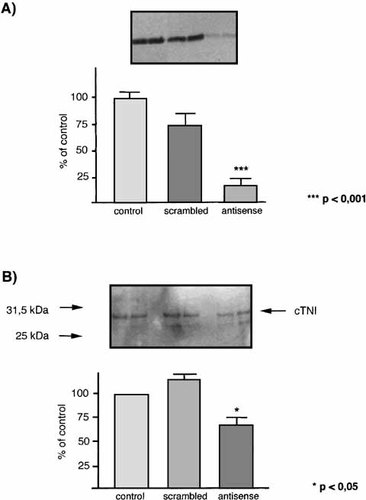
A: Inhibition of human TNI translation in vitro by antisense oligonucleotide 90-110. Circular plasmid DNA (pGEM3Z, Promega) with cloned full-length cDNA from human cardiac TNI sequences provided the template for the transcription/translation assay as in Figure 4. Top: The effect of an antisense oligonucleotide (2 μM) directed against position 90–110 compared to the control with no added oligonucleotide and a scrambled oligonucleotide (2 μM) on cTNI translation. Bottom: Data from densitometry measurements of the protein bands are shown as a bar chart. Each value is based on three experiments. The percentage effect of inhibition refers to the control, which was set at 100%. Values are means ± SD one-way ANOVA with Bonferroni Posthoc test, levels of significance are indicated. B: Inhibition of rat TNI translation in vivo by antisense oligonucleotides. The effect of an antisense oligonucleotide corresponding to position 88–108 of cTNI mRNA from rat on cTNI protein expression was tested in rat neonatal cardiomyocytes cell culture. Following treatment, total cell protein was extracted and separated by SDS–PAGE followed by Western blotting as described in Materials and Methods. The semi-quantified data represent densitometry measurements of the protein bands shown as a bar chart. A 2-day treatment of the cells with 10 μM antisense oligonucleotide led to a significant reduction in cTNI protein levels, down to 66.7 ± 12.7% of control amounts (P < 0.05). A scrambled oligonucleotide had no effect on the cTNI protein steady-state after 2-day exposure of the cells (113.5 ± 9.9%).
The effect of antisense oligonucleotides on cTNI protein steady-state synthesis in vivo was investigated in rat neonatal cardiomyocytes cell culture. To increase uptake from the medium, phosphorothioate oligonucleotides were used since they are more resistant to endogenous nucleases [Shaw et al., 1991]. Two-day treatment of the cardiomyocytes with 10 μM of an ASO corresponding to position 88–108 of the cTNI mRNA led to a significant reduction of cTNI protein levels, down to 66.7 ± 12.7% of the control (P < 0.05; Fig. 6B). A scrambled oligonucleotide had no effect on the cTNI steady-state after 2-day exposure of the cells (113.5 ± 9.9%). Since cTNI protein is upregulated during postnatal development, time-matched controls were used in these experiments.
DISCUSSION
Two classes of antisense RNA, which differ in length and homology to the sense transcript, have been described in eukaryotes [Nellen and Sczakiel, 1996]. One class comprises short (100 nucleotides and less) antisense transcripts with incomplete homology to their targets, while the other category is characterized by a size of several hundred nucleotides with stringent homology to their corresponding sense sequence. Our sequenced PCR products indicated full sequence complementarity to the spliced sense transcript, and were supported by the results of Northern blotting, which detected RNA species of approximately 800 bp in rat myocardium. The size of the antisense cTNI RNA in the rat is consistent with the known mRNA [Murphy et al., 1991]. In contrast, human heart RNA analysed by Northern blotting revealed an antisense transcript shorter than the mRNA, which is in agreement with our RT-PCR results where no amplification of antisense could be detected in the 3′ region of the human cTNI mRNA.
Different sized antisense transcripts in rat and man support the idea of various transcription initiation sites, as shown by Krystal et al. [1990] for c-myc. Antisense cTNI RNA also seems to exist as a polyadenylated population in the rat and human heart, since antisense PCR products were efficiently detected in RNA pools purified with oligo(dT)-cellulose. This was also recently shown for human β-MHC antisense [Luther et al., 2001] and the antisense M40 subunit of the insect V-ATPase [Merzendorfer et al., 1997]. Merzendorfer et al. suggested that the poly (A) tail is localized at the 3′-end of the antisense transcripts, analogous to sense transcripts. Semi-quantification by RT-PCR revealed a relative amount of antisense to sense cTNI RNA of around one half (52 ± 10%) or one-third (38 ± 8%) in the rat or human heart. Antisense/sense ratios vary considerably between different genes and species. Antisense RNA of c-myc, for example, comprises only 5% of the total sense RNA [Krystal et al., 1990], in contrast to antisense of β-MHC in rat and human myocardium, whose levels were 50 and 15% of the sense RNA, respectively [Luther et al., 1998, 2001].
Regarding the origin of antisense transcripts, three different mechanisms can be considered: antisense synthesis could occur by transcription of the opposite strand of a gene [Tommasi and Pfeifer, 1999] or a pseudogene [Korneev et al., 1999], or by transcription of the sense mRNA by an RNA-dependent RNA polymerase in the cytoplasm. The latter was previously postulated for those antisense RNAs assumed to be complementary to the spliced sense transcripts (M40 subunit of insect V-ATPase and urocortin) [Merzendorfer et al., 1997; Shi et al., 2000].
Despite increasing reports of the detection of endogenous antisense RNA, evidence for a regulatory function of antisense RNA in terms of gene expression is rare. The predicted duplex formation of antisense-sense RNA could be detected in only a very few cases. First demonstrated by electron microscopy when investigating heterogeneous nuclear RNAs in HeLa cells [Fedoroff et al., 1977], Krystal and co-workers (c-myc in neuroblastoma cells) [Krystal et al., 1990] and Korneev and co-workers (nNOS in snail neuronal cells) [Korneev et al., 1999] could demonstrate RNA duplex formation using an RNase protection technique. The problem of duplex detection was explained by the observation that cells contain an apparatus capable of recognizing and cleaving dsRNA [Wu et al., 1998]. Duplex RNA in the nucleus was shown to be processed by deaminases that convert many of the adenosine residues to inosine [Bass and Weintraub, 1988]. Depending on the degree of modification, RNAs are either degraded, retained in the nucleus or transported to the cytoplasm [Kumar and Carmichael, 1998]. Other enzymes with double-stranded RNA as their potential substrate are protein kinase R (PKR) and the 2-5A synthetase [Kerr and Brown, 1978; Proud, 1995]. Since modification may also interfere with their detection by molecular probes [Kumar and Carmichael, 1998], and since duplex-formation was shown to be a complicated and fragile process [Wagner and Simons, 1994], demonstration of in vivo RNA-duplex formation is incidental rather than obligatory.
By using an approach similar to that described by Korneev et al. [1999], we were able to demonstrate duplexes of cTNI antisense and sense RNA for the first time in myocardium. In view of protein synthesis, double-stranded RNA can either act as a target for double-strand-activated RNases or lead to an arrest of translation by different processes, including steric alterations of the sense RNA or by preventing the initiation of protein synthesis [Nellen and Lichtenstein, 1993]. Therefore, using antisense oligonucleotides, we investigated the ability of antisense RNA to affect cTNI protein synthesis in vitro and in vivo. In vitro translation experiments showed that the potency of antisense oligonucleotide inhibition of protein synthesis depends on the location of the selected oligonucleotides in the mRNA sequence. Since it is known that interaction of RNA is—due to intricate secondary and tertiary structure—a complex process [Delihas et al., 1997], we were interested in the region of cDNA, which is a crucial for regulating translation functions of natural sense RNA. We synthesized oligonucleotides corresponding to three different sectors of antisense RNA and monitored their effect on translation in vitro. The oligonucleotide directed against the translation initiation site was effective in a dose-dependent manner, whereas the oligonucleotides closer to the 3′ end had significantly less or no effect.
Since our oligonucleotides comprise desoxyribonucleotides, one should note that the interaction may be different from RNA–RNA interactions. However, addition of RNase H, considered necessary for an oligonucleotide effect in most, but not all cases, was not required in our system, indicating a true translation arrest mechanism. The high efficacy of oliogonucleotides directed against a translation initiation site is in agreement with a report by Chen et al. [1997] who analyzed various antisense oligonucleotides in vitro.
In addition to the inhibitory properties of antisense oligucleotides in vitro, we studied steady-state levels of cTNI in cell cultures of neonatal rat cardiomyocytes. To imitate the effect of natural antisense, cardiomyocytes were treated with the most effective oligonucleotide directed against the translation start. We observed a significant reduction in cTNI protein levels after a 2-day incubation with the antisense oligonucleotide, but no effect with a scrambled version.
In mammalian heart, troponin I isoforms are differentially regulated during development by switching from ssTNI to cTNI. In adult rats, cTNI is expressed as the unique isoform of troponin I in heart tissue [Murphy et al., 1991]. A number of transcription factors and cis-acting elements including GATA-4 and GATA regulatory elements have already been identified in the rat cTNI promotor, and are thought to be responsible for the upregulation of cTNI during postnatal development [Murphy et al., 1997; Di Lisi et al., 1998]. Post-transcriptional regulation of cTNI expression is reported to happen at the protein level [Solaro, 1999]. However, we observed a significantly higher ratio of antisense/sense cTNI RNA in neonatal rat compared to adults, implying a possible role of endogenous antisense cTNI in post-transcriptional regulation during development. An inverse correlation between antisense/sense ratios and protein expression was recently detected for the human atrial essential myosin light chain (ALC-1) [Ritter et al., 1999]. As shown in the hypertrophied heart, a higher ALC-1 RNA antisense/sense ratio was associated with lower ALC-1 protein expression, suggesting that ALC-1 translation is disturbed by high levels of antisense RNA expression. Thus, naturally occurring antisense transcripts of cTNI may provide a potential means for negative modulation of gene expression.
In summary, we describe here how two independent methods, RT-PCR and Northern blotting, successfully detected antisense cTNI transcripts in rat and normal human myocardium. Moreover, we show for the first time that a certain fraction of cTNI RNA extracted from neonatal rat cardiomyocytes is double stranded, suggesting that the formation of antisense-sense RNA duplexes can apparently take place in vivo. In addition, the significant inhibitory effects of antisense oligonucleotides on cTNI protein expression levels both in vitro and in vivo point to potential mechanisms of antisense post-transcriptional control, illuminating an important aspect of how antisense RNA may interfere with gene regulation in the myocyte.
Acknowledgements
We thank Marc Eigen and Anke Stach for excellent technical assistance.