Synovium-Derived MicroRNAs Regulate Bone Pathways in Rheumatoid Arthritis
ABSTRACT
Articular bone erosion in rheumatoid arthritis (RA) is mediated by the interaction between inflammation and pathways regulating bone metabolism. Inflammation promotes osteoclastogenesis and also inhibits osteoblast function, further contributing to the persistence of erosions. MicroRNAs (miRNAs) are important regulators of skeletal remodeling and play a role in RA pathogenesis. We therefore determined the expression of miRNAs in inflamed synovial tissue and the role they play in pathways regulating osteoblast and osteoclast function. Using the serum transfer mouse model of RA in C57BL/6 mice, we performed Fluidigm high-throughput qPCR-based screening of miRNAs from nonarthritic and arthritic mice. Global gene expression profiling was also performed on Affymetrix microarrays from these same synovial samples. miRNA and mRNA expression profiles were subjected to comparative bioinformatics. A total of 536 upregulated genes and 417 downregulated genes were identified that are predicted targets of miRNAs with reciprocal expression changes. Gene ontology analysis of these genes revealed significant enrichment in skeletal pathways. Of the 22 miRNAs whose expression was most significantly changed (p < 0.01) between nonarthritic and arthritic mice, we identified their targets that both inhibit and promote bone formation. These miRNAs are predicted to target Wnt and BMP signaling pathway components. We validated miRNA array findings and demonstrated that secretion of miR-221-3p in exosomes was upregulated by synovial fibroblasts treated with the proinflammatory cytokine TNF. Overexpression of miR-221-3p suppressed calvarial osteoblast differentiation and mineralization in vitro. These results suggest that miRNAs derived from inflamed synovial tissues may regulate signaling pathways at erosion sites that affect bone loss and potentially also compensatory bone formation. © 2016 American Society for Bone and Mineral Research.
Introduction
Rheumatoid arthritis (RA) is characterized by focal articular erosion that is mediated by osteoclasts. Cortical bone around the joints is the initial target of erosion. Progression of bone erosion causes loss of subchondral bone and contributes to destruction of articular cartilage. Repair of bone erosions in RA patients has been documented through control of inflammation; however, it is uncommon,1 suggesting an ongoing suppression of function of bone-forming osteoblasts. Bone formation is regulated at multiple levels by factors that control osteoblast function. The bone morphogenetic protein (BMP) and Wingless (Wnt) signaling pathways are critical in the regulation of osteoblast differentiation and function.2 Evidence suggests that the BMP pathway may also regulate inflammation in RA.3 BMP signaling is transduced by Smad1, Smad5, and Smad8, inducing expression of Runx2 and Osterix (Osx) to promote osteoblast differentiation. Activation of canonical Wnt signaling is transduced by β-catenin4 and promotes bone formation. Wnt signaling antagonists inhibit this pathway and have been shown to regulate the erosive process in RA.5, 6
Many studies have demonstrated a close relationship between synovial inflammation and pathways regulating bone formation.7 One of the mouse models of RA used to study bone erosion is the serum transfer arthritis (STA) model.8 Injection of serum from arthritic K/BxN mice to naive recipient mice induces an RA-like inflammation. Inflammation then peaks, and resolves after several weeks.5 In the STA model, inflammation was shown to impair bone formation and repair at sites of erosion.9 Once inflammation resolved, enhanced articular bone formation and repair of erosions was seen.5 In this model, mRNA expression of Sfrp-1 and Sfrp-2, Wnt signaling pathway antagonists, was upregulated in synovium at peak inflammation and resolution of inflammation was accompanied by a decrease in Sfrp-1 and Sfrp-2 expression and an increase in Wnt10b and Dkk2 expression.5, 9 Wnt10b promotes bone formation and Dkk2 promotes mineralization of bone.4, 10 Expression of Sfrp-1 and Dkk1 protein was also observed at bone erosion sites during peak inflammation.9 The Wnt signaling inhibitor Dkk1 is also induced in RA synovium by TNF, and a Dkk1 antagonist prevented bone erosion and increased periosteal bone formation in a TNF-driven RA model.6 Furthermore, targeting of another Wnt antagonist, Sclerostin, completely arrested bone erosion and systemic bone loss in this model.11
One potential mechanism regulating bone homeostasis is microRNA (miRNA)-mediated gene silencing.12 miRNAs are small, noncoding RNAs that are epigenetic regulators of cellular protein levels through destabilization of mRNA and/or inhibition of protein translation. miRNAs can target numerous mRNAs and thus affect several biological pathways simultaneously. miRNAs are known to be key posttranscriptional modulators of inflammation in RA.13, 14 Expression of the anti-inflammatory miR-23b (current ID: miR-23b-3p) was downregulated in RA fibroblast-like synoviocytes by IL-17,15 and administration of miR-23b suppressed inflammation in RA models by suppressing expression of NF-κB signaling components. In contrast, miR-155 (current ID: miR-155-5p) promoted inflammation and bone erosion by enhancing expression of TNF.16 Furthermore, miRNAs are essential for osteoclast differentiation17 and thus regulation of specific miRNAs could contribute to osteoclastogenesis and bone erosion in RA models. For example, inhibition of miR-223 (current ID: miR-223-3p) in the collagen-induced arthritis model suppressed osteoclast differentiation and bone erosion.18
It is not known whether synovium-derived miRNAs regulate osteoblast differentiation at bone erosion sites. It has been shown, however, that miRNAs can be exported out of cells and transported by various carriers including exosomes, miRNA-binding protein complexes, or high density lipoproteins to be taken up by other cells.19 Therefore, we hypothesized that miRNAs derived from synovial tissues not only regulate inflammation but also regulate skeletal pathways in RA. We used the STA model to generate synovial inflammation and performed global gene and miRNA analyses to better understand pathways regulating bone remodeling in this disease. We identified synovium-derived miRNAs that are predicted to regulate signaling pathways affecting both bone erosion and compensatory bone formation, and demonstrated that many miRNAs regulated by inflammation are linked to skeletal pathways. Examining the functional activity of a selected miRNA, which is predicted to regulate Wnt signaling, revealed that synovium-derived miRNAs likely contribute to regulation of osteoblast function.
Materials and Methods
K/BxN model of RA
Preparation of serum, generation of arthritis in C57BL/6 mice, and scoring methods were as described.5 Because male mice are more susceptible to arthritis, males were used at 12 weeks of age to avoid the juvenile growth spurt and any hormonal influence on bone. Mice were injected with 200 μL of serum from K/BxN mice intraperitoneally on days 0, 2, and 7. Mice were housed in specific pathogen-free conditions and arthritic mice had full access to food and water. All animal experiments were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC).
Tissue RNA preparation and qPCR
Synovium and navicular bones were isolated from mice in separate experiments, and processed for RNA as described.9 Synovium from six (day 10 [D10]) and twenty (day 0 [D0]) mice represented two pools (biological replicates) at each time point (three [D10] and 10 [D0] mice per pool). Total RNA was used for iScript cDNA synthesis (Bio-Rad Laboratories, Hercules, CA, USA). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed on diluted cDNA samples using iScript SYBR Green RT-PCR mix (Bio-Rad Laboratories, Hercules, CA, USA). Primers were purchased from Qiagen (Valencia, CA, USA). qPCR for miRNAs was performed using QuantiMir kits (System Biosciences [SBI], Palo Alto, CA, USA) and SYBR Green PCR mix (Bio-Rad Laboratories, Hercules, CA, USA). Forward primer sequences were obtained from miRBase.20 Gene and miRNA expression was normalized to hydroxymethylbilane synthase (HMBS) and U6 (U6 spliceosomal RNA) expression, respectively. Gene expression in nonarthritic controls was stable over time, and similar variability in gene expression among individual mice was observed at each time point. Data were analyzed using the delta-delta comparative threshold cycle algorithm (2ΔΔCt) method.21
Cell culture
Fibroblast-like synoviocytes (FLS) were isolated by incubation of three C57BL/6 mouse hindlimbs with 1 mg/mL type IV collagenase and pooled (Worthington, Reading, UK) for 2 hours. Cells were maintained in DMEM supplemented with 10% FBS (Atlanta, GA), 2 mM L-glutamine (Corning, VA), 0.1 mM nonessential-amino acids (Mediatech Ink., VA), 1× essential amino acids (Mediatech Ink., VA), 50 mM 2-ME (MP Biomedicals, OH), 10 μg/mL gentamicin 100 μg/mL (Corning, VA), penicillin/streptomycin (Corning, VA). Cells were maintained at 37°C in a humidified 10% CO2 environment and passaged every 3 days. After six passages, cells were plated in six-well plates, and once confluent, treated with 10 ng/mL TNF (R&D Systems, Minneapolis, MN, USA), 50 ng/mL IL-17 (R&D Systems, Minneapolis, MN, USA) or 10 ng/mL IL-1 (R&D Systems, Minneapolis, MN, USA) for 72 hours. RNA was collected for miRNA expression analysis using Trizol (Thermo Fisher Scientific, MA).
Calvarial osteoblasts were isolated from neonatal mice (postnatal day 0 [P0] to postnatal day 2 [P2]) by collagenase digestion (Roche, Basel, Switzerland) and maintained as described.22 Fifty nanomoles (50 nM) miRNA and anti-miRNAs (Thermo Fisher Scientific, Waltham, MA, USA) were introduced using oligofectamine (Invitrogen, Carlsbad, CA, USA), and cells were harvested for histochemical detection of alkaline phosphatase (Sigma-Aldrich, St. Louis, MO, USA) and Von Kossa staining (Sigma-Aldrich, St. Louis, MO, USA).
Gene array, miRNA array, and bioinformatic analysis
Gene array, miRNA array, and bioinformatic analysis are provided in the Supplemental Materials and Methods. The raw data are uploaded to GEO (http://www.ncbi.nlm.nih.gov/geo/; accession number: GSE71601).
Western blotting
Protein was extracted from calvarial osteoblasts in radioimmunoprecipitation assay (RIPA) buffer. Fifty micrograms (50 μg) were loaded onto a 10% SDS-PAGE gel, transferred to polyvinylidene fluoride (PVDF) membrane and probed with antibodies against Dkk2 (Abcam, Cambridge, UK) and beta-actin (Cell Signaling Technology, Beverly, MA, USA) and detected by chemiluminescence (Thermo Fisher Scientific, Waltham, MA, USA). Protein bands were measured by Image J software (NIH, Bethesda, MD, USA; https://imagej.nih.gov/ij/).23
Exosome collection from FLS culture media
Exosomes were isolated from FLS culture media. Cells were cultured in exosome-depleted FBS (System Biosciences [SBI], Palo Alto, CA, USA) medium with or without 10 ng/mL TNF for 3 days. For the isolation of exosomes, 50 mL of culture medium was centrifuged at 3000g for 15 min to remove cells and cell debris. The resulting supernatant was concentrated using the Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-100 membrane (Millipore, Billerica, MA, USA). Exosomes were precipitated using ExoQuick-TC reagent (System Biosciences [SBI], Palo Alto, CA, USA) as described by the manufacturer. The exosome pellet was resuspended in PBS. Protein volume of exosomes was measured by Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA). RNA was collected from 5 μg of exosomes using the RNeasy Micro Kit (Qiagen, Hilden, Germany).
3′ Untranslated region reporter luciferase assay
The 3′ untranslated region (3′UTR) reporter plasmid for Dkk2 was purchased from GeneCopoeia (Rockville, MD, USA). The 1074-bp fragment of the Dkk2 3′UTR region that contains the miR-221-3p binding site (TGTAGCA) was cloned into the pEZX-MT06 vector. The binding site is deleted in the mutant Dkk2 3′UTR reporter plasmid. Transfection of MC3T3 cells was performed in 96-well plates. The 3′UTR reporters are co-transfected with 25 nM miR-221-3p, or negative control (Thermo Fisher Scientific, Waltham, MA, USA) using Lipofectamine (Invitrogen, Carlsbad, CA, USA). The transfected cells were incubated for 24 hours to determine firefly luciferase activity and renilla luciferase activity using Dual-Glo luciferase assay system (Promega, San Luis Obispo, CA, USA). The experiment was repeated three times.
Statistical analysis
For bioinformatic analysis, principal component analysis (PCA) and ANOVA methods were used (see Supplemental Materials and Methods). Biological replicate average, fold change, and ANOVA p value were calculated between groups in Spotfire DecisionSite. Statistical significance was calculated using t tests for qPCR data. Data are presented as means ± SD or SE (for qPCR data from bone samples from six individual mice).
Results
Gene and miRNA expression profiles in synovium
K/BxN STA replicates many features of chronic RA in humans in a synchronized manner.24 Therefore, to examine the possibility that synovium-derived miRNAs regulate skeletal pathways in RA, we analyzed expression of both miRNAs and mRNAs in whole synovial tissues from the STA model.5 We injected serum from K/BxN mice into 33 wild-type C57BL/6 mice. Thirty mice (91%) developed arthritis in paws and ankle joints. Peak inflammation occurred at approximately 10 days (D10) after initial serum injection (Fig. 1A). Expression levels at D10 were compared with those in nonarthritic mice (D0, uninjected). Gene array analysis confirmed upregulation at D10 of mRNA for the expected proinflammatory cytokines TNF and IL-1β and genes that inhibit Wnt signaling (eg, Sfrp-1, Sfrp-2), as well as downregulation of the Wnt inhibitor Dkk2, which augments osteoblast mineralization, confirming previous studies (Supplemental Fig. 1A).5
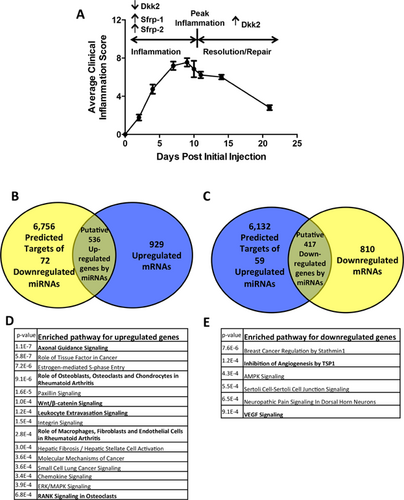
High-throughput TaqMan-based qPCR25 identified 168 miRNAs that passed quality control (QC) criteria (Supplemental Fig. 1B). We found few significant changes in miRNA expression between D10 and D21, when inflammation was resolving (data not shown). This may be because inflammation was not completely resolved at the D21 time point. We therefore pursued expression analysis between nonarthritic synovium and synovium at peak inflammation. Consistent with other RA models, miRNAs that regulate disease pathogenesis were identified in synovium at peak inflammation, including miR-23b-3p (downregulated 10-fold) and miR-221-3p (upregulated eightfold).15, 26 Having confirmed that miRNAs related to inflammation in our model were similar to previous reports, we focused on those miRNAs with differential expression during inflammation (D0 versus D10) that could affect bone.
Synovium-derived miRNAs regulate skeletal pathways
To analyze potential pathways regulated by synovial miRNAs, we compared gene and miRNA array results. Of the 168 miRNAs passing QC analysis, 72 miRNAs were significantly downregulated and 59 upregulated (≥1.5 fold) between nonarthritic and arthritic synovium. We compared the predicted 6756 target genes of the 72 downregulated miRNAs with the 929 upregulated genes identified in the gene array analysis (≥1.5 fold) (Fig. 1B). We similarly compared the predicted 6132 target genes of the 59 upregulated miRNAs with the 810 downregulated genes (≥1.5 fold) (Fig. 1C). We found 536 upregulated and 417 downregulated genes to be in the overlap of each respective comparative analysis (Venn diagram). To identify the biological pathways to which these genes belong, we performed ontology analysis using DAVID v6.7 (Fig. 1D, E).27 Interestingly, pathways identified in the upregulated gene group showed significant enrichment of skeletal pathways, including “Role of Osteoblasts, Osteoclasts, and Chondrocytes in Rheumatoid Arthritis” (p = 9.1E–06), “Wnt/β-catenin signaling” (p = 1.0E–04), and “RANK Signaling in Osteoclasts” (p = 6.8E–04) (Fig. 1D). These pathways included inhibitors of Wnt signaling, such as Dkk2, Dkk3, Sfrp-1, and Sost (Supplemental Table 1).
As expected, we also found inflammation-related pathways, including “Role of Macrophages, Fibroblasts, and Endothelial Cells in Rheumatoid Arthritis” (p = 2.8E–04), “VEGF Signaling” (p = 9.1E–04), and “Leukocyte Extravasation Signaling” (p = 1.2E–04), among others (Fig. 1D, E). The most enriched pathway was “Axonal Guidance Signaling” (p = 1.1E–07) (Fig. 1D). Notably, some genes in this pathway, such as ADAMTS7,28 HHIP,29 Sema3A,30 and MMP9,31 are also important regulators of skeletal development and bone formation (Supplemental Table 1). These results support the hypothesis that synovium-derived miRNAs not only regulate inflammation but also regulate skeletal pathways in RA.
Using statistical analyses, we constricted our definition of significant changes to determine the most critical candidate miRNAs regulating bone. Of the 168 miRNAs that passed QC analysis, 22 exhibited a greater than fourfold change (D10 versus nonarthritic) with an ANOVA p value <0.01 (Fig. 2A). Some of these miRNAs are known to play a role in inflammation. For example, miR-224 (current ID: miR-224-5p) is upregulated and miR-145 (current ID: miR-145a-5p) downregulated in T cells from systemic lupus erythematosus patients32 and miR-16 (current ID: miR-16-5p) is decreased in serum from early stage RA patients.33 We also found that miR-224-5p is upregulated and miR-145a-5p and miR-16-5p are downregulated at peak inflammation (Fig. 2A).
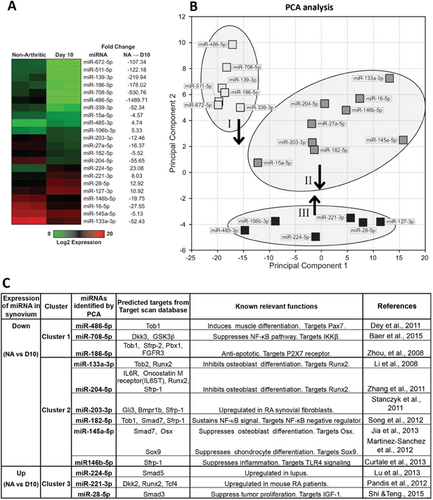
We performed PCA on the 22 candidate miRNAs to identify similarities and determine functional miRNA clusters affecting biological processes. Candidate miRNAs segregate into three clusters based on expression changes (Fig. 2B). Target genes of miRNAs in each cluster are enriched in distinct skeletogenesis pathways (Table 1). miRNAs in cluster I are downregulated and their target genes are enriched in cartilage matrix pathways and osteogenic signaling, including JAK/STAT,34 IGF-1,35 and HIF1α signaling.36 miRNAs in cluster II are downregulated and their target genes are enriched in Wnt/β-catenin signaling and cell cycle–related pathways. miRNAs in cluster III are upregulated and their target genes are enriched in pathways regulating bone metabolism, such as glucocorticoid receptor37 and eNOS signaling.38 Within these 22 miRNAs, several are identified that have been reported to regulate skeletal pathways. For example, miR-133a-3p and miR-204-5p (cluster II) inhibit osteoblast differentiation by suppressing expression of Runx2.39, 40 miR-145a-5p (cluster II) inhibits differentiation of osteoblasts and chondrocytes by suppressing Osx and Sox9.41, 42 Downregulation of these miRNAs may contribute to maintaining chondrocyte and osteoblast progenitors during bone erosion as a compensatory mechanism. Thus, the most highly regulated miRNAs in inflamed synovium are commonly involved in regulation of skeletal pathways.
| Cluster 1 | p | Cluster 2 | p | Cluster 3 | p |
|---|---|---|---|---|---|
| IL-9 signaling | 0.0051 | Estrogen-mediated S-phase Entry | 0.0001 | Estrogen-dependent breast cancer signaling | 0.0100 |
| Insulin receptor signaling | 0.0085 | HIPPO signaling | 0.0002 | Reelin signaling in neurons | 0.0158 |
| Heparan sulfate biosynthesis (late stages) | 0.0107 | Dermatan sulfate biosynthesis (late stages) | 0.0010 | TR/RXR activation | 0.0182 |
| 3-Phosphoinositide biosynthesis | 0.0129 | Chondroitin sulfate biosynthesis (late stages) | 0.0012 | Glioma signaling | 0.0224 |
| Glioma invasiveness signaling | 0.0138 | Heparan sulfate biosynthesis (late stages) | 0.0017 | Neuropathic pain signaling in dorsal horn neurons | 0.0245 |
| Heparan sulfate biosynthesis | 0.0138 | Wnt/1β-catenin signaling | 0.0017 | Glucocorticoid receptor signaling | 0.0257 |
| ErbB4 signaling | 0.0151 | Chondroitin sulfate biosynthesis | 0.0022 | Hereditary breast cancer signaling | 0.0324 |
| Role of JAK1 and JAK3 in γc cytokine signaling | 0.0166 | Heparan sulfate biosynthesis | 0.0027 | Insulin receptor signaling | 0.0427 |
| Erythropoietin signaling | 0.0186 | Dermatan sulfate biosynthesis | 0.0027 | eNOS signaling | 0.0468 |
| Growth hormone signaling | 0.0200 | Cyclins and cell cycle regulation | 0.0081 | Regulation of eIF4 and 70S6K Signaling | 0.0490 |
| ERK/MAPK signaling | 0.0209 | Isoleucine degradation 1 | 0.0087 | ||
| JAK/Stat signaling | 0.0214 | D-myo-inositol (1,4,5,6)-tetrakisphosphate biosynthesis | 0.0095 | ||
| Prolactin signaling | 0.0219 | D-myo-inositol (3,4,5,6)-tetrakisphosphate biosynthesis | 0.0095 | ||
| Superpathway of inositol phosphate compounds | 0.0224 | Cell cycle: G2/M DNA damage checkpoint regulation | 0.0135 | ||
| HIPPO signaling | 0.0302 | Breast cancer regulation by Stathmin1 | 0.0135 | ||
| IGF-l signaling | 0.0372 | Superpathway of inositol phosphate compounds | 0.0138 | ||
| Amyotrophic lateral sclerosis signaling | 0.0380 | GADD45 signaling | 0.0155 | ||
| p53 signaling | 0.0380 | DNA damage-induced 14-3-30 signaling | 0.0155 | ||
| Telomerase signaling | 0.0389 | D-myo-inositol-5-phosphate metabolism | 0.0155 | ||
| Neuropathic pain signaling in dorsal horn neurons | 0.0389 | 3-Phosphoinositide degradation | 0.0158 | ||
| HIF1α signaling | 0.0407 | Glioblastoma multiforme signaling | 0.0166 | ||
| HGF signaling | 0.0427 | Unfolded protein response | 0.0174 | ||
| Pancreatic adenocarcinoma signaling | 0.0437 | Role of CHK proteins in cell cycle checkpoint control | 0.0182 | ||
| Regulation of cellular mechanics by calpain protease | 0.0204 | ||||
| 3-Phosphoinositide biosynthesis | 0.0209 | ||||
| ATM signaling | 0.0219 | ||||
| cAMP-mediated signaling | 0.0245 | ||||
| Cell cycle: Gl/S checkpoint regulation | 0.0275 | ||||
| Antiproliferative role of TOB in T cell signaling | 0.0282 | ||||
| Synaptic long-term potentiation | 0.0331 | ||||
| Caveolar-mediated endocytosis signaling | 0.0372 | ||||
| STAT3 pathway | 0.0380 | ||||
| ERK/MAPK signaling | 0.0427 | ||||
| G-protein coupled receptor signaling | 0.0468 | ||||
| Insulin receptor signaling | 0.0479 | ||||
| Cell cycle regulation by BTG family proteins | 0.0490 | ||||
| Role of JAK2 in hormone-like cytokine signaling | 0.0490 |
- Target genes of miRNAs in each cluster (Fig. 2B) are identified by Target Scan. To find enriched pathways, each gene group is analyzed by Ingenuity Pathway Analysis. A few examples of the spectrum of pathways that can effect cell proliferation, cartilage and bone are bolded.
Downregulated miRNAs are predicted to target inhibitors of skeletal pathways
We and others have previously observed expression of mRNA and protein for the Wnt signaling inhibitors, Sfrp-1 and Dkk1 in bones at erosion sites in models of RA.6, 9 Although Runx2-expressing immature osteoblasts were present at bone erosion sites, maturation of these osteoblasts is not frequently observed.9 Here we examined expression of Wnt and BMP signaling components using RNA prepared from navicular bone, a reproducible site of erosion in STA, and found mRNA expression of Gsk3β, Sfrp-2, and Tob1, as well as an early osteoblast marker, Osx, to be upregulated in bones at erosion sites during peak inflammation (Supplemental Fig. 2). Upregulation of Gsk3β, Sfrp-2, and Tob1 would be predicted to inhibit osteoblast differentiation, whereas upregulation of Osx would promote early osteoblast differentiation. These results suggest that BMP signaling, Wnt signaling, and maturation of osteoblasts are suppressed in bone at erosion sites. Upregulation of Osx mRNA expression may provide a counter-regulatory mechanism, or may be due to an inhibition of osteoblast maturation at this site. To identify candidate miRNAs that may regulate skeletal pathways at erosion sites, we searched for miRNAs that are predicted to target components of major skeletal pathways using TargetScan v6.2. Among the 22 candidate miRNAs showing a greater than fourfold change in expression, we identified 12 miRNAs predicted to target components of major skeletal pathways (Fig. 2C). These miRNAs have known roles in inflammation,26, 32, 43-46 proliferation,47 apoptosis,48 or musculoskeletal systems39-42, 49 (Fig. 2C). Notably, nine downregulated miRNAs in clusters I and II target numerous inhibitors of Wnt and BMP signaling, including Dkk3, GSK3β, Sfrp-1, Sfrp-2, Smad7, Tob1, and Tob2,50 suggesting that decreased expression of these miRNAs in synovium may contribute to the induction of BMP and Wnt signaling inhibitors and limit bone formation at erosion sites.
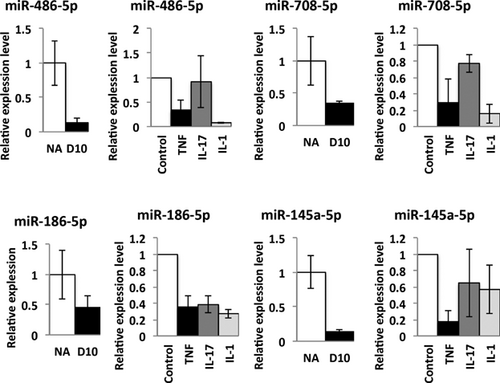
To validate expression of these 12 miRNAs in synovial fibroblasts, we isolated FLSs from wild-type mice, stimulated with proinflammatory cytokines including TNF, IL-1, or IL-17 and examined expression of candidate miRNAs. We confirmed that miR-486-5p, miR-708-5p, miR-186-5p, and miR-145a-5p are downregulated following stimulation with TNF treatment (Fig. 3). We also found that IL-1 suppresses expression of miR-486-5p, miR-708-5p, and miR-186-5p, and IL-17 suppresses expression of miR-186-5p. Expression of miR-133a-3p, miR-204-5p, and miR-182-5p is suppressed only by TNF (data not shown). We validated expression of these miRNAs (miR-708-5p, miR-186-5p, miR-486-5p, miR-145a-5p) by qPCR in whole synovial tissues at D10 compared to nonarthritic tissues using the same synovial samples used for array experiments. Taken together, we found numerous FLS-derived miRNAs that are predicted to target inhibitors of BMP and Wnt signaling and that are downregulated by inflammatory cytokines. These miRNAs may negatively regulate bone formation at erosion sites.
Overexpression of miR-221-3p suppresses differentiation of primary osteoblasts
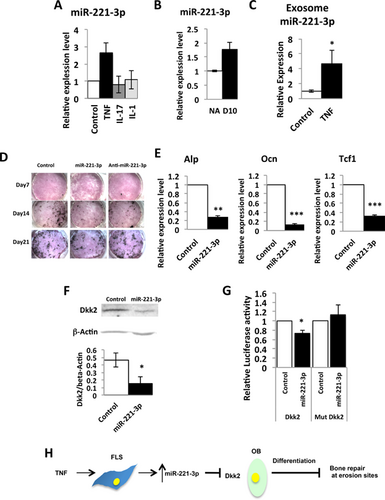
miR-224-5p, miR-221-3p, and miR-28-5p are among the most highly upregulated miRNAs in cluster III (Fig. 2B) that target components of the BMP and Wnt pathways. (Fig. 2C). We confirmed that miR-221-3p is induced in TNF-treated FLS (Fig. 4A) and in inflamed synovium (Fig. 4B)26. miR-224-5p and miR-28-5p did not show relevant expression patterns in FLS and may be regulated in other synovial cell types such as hematopoietic or endothelial cells, or by other cytokines. miR-221-3p was also reported to be induced in a TNF-driven model of arthritis and in FLS from RA patients.26 We therefore focused on miR-221-3p and questioned whether miR-221-3p might affect skeletal pathways at the erosion site. Since miRNAs can be exported from one cell type and taken up to regulate gene expression in a distant cell type, we examined expression of miR-221-3p in FLS-derived exosomes. We found that with treatment of FLS cells with TNF (10 ng/mL), miR-221-3p is significantly upregulated in FLS-derived exosomes (Fig. 4C). This result suggests that secretion of miR-221-3p into exosomes from synovium is also upregulated by inflammation and that synovium-derived miR-221-3p may regulate bone formation at erosion sites. Ontology analysis implicated miR-221-3p as targeting the Wnt pathway26; however, Wnt reporter activity was not affected by miR-221-3p in HEK 293T cells.26 Using TargetScan v6.2, miR-221-3p is predicted to target bone anabolic genes including Runx2, TCF4, Esr1,51 and Dkk2.10 We therefore transfected miR-221-3p into mouse primary calvarial osteoblasts. This inhibited differentiation and mineralization, as indicated by decreased staining for alkaline phosphatase (Alp) and mineralization (von Kossa) (Fig. 4D). We did not find significant change in osteoblast differentiation by anti-miR-221-3p (qPCR data for osteoblast markers not shown). This may indicate that only excess miR-221-3p affects osteoblast differentiation but not loss of miR-221-3p expression. We further observed a significant decrease in mRNA expression of Alp and a mature osteoblast marker, Osteocalcin (Ocn) by miR-221-3p (Fig. 4E). mRNA expression of Tcf-1, a readout of Wnt signaling,52 was also suppressed by miR-221-3p (Fig. 4E), confirming that miR-221-3p inhibits osteoblast differentiation. Among the predicted pro-osteoblastic target genes of miR-221-3p, we found the protein level of Dkk2 was suppressed in the transfected osteoblasts (Fig. 4F). miR-221-3p is known to bind the 3′UTR region of Dkk2 mRNA in 293T cells.53 We confirmed the binding of miR-221-3p to the 3′UTR region of Dkk2 mRNA in the osteoblastic cell line MC3T3 (Fig. 4G), supporting Dkk2 as a target gene of miR-221-3p. These results suggest that upregulation of synovium-derived miR-221-3p may suppress expression of Dkk2 in osteoblasts, and subsequent osteoblast maturation, and may, at least in part, contribute to the inhibition of bone formation in RA erosions (Fig. 4H).
Discussion
For the treatment of RA, disease-modifying agents such as methotrexate and TNF inhibitors are currently available. These agents are proven to be effective in suppressing inflammation and progression of focal articular erosions. However, repair of focal bone erosions through the formation of new bone in patients is rare, suggesting an ongoing disturbance of bone formation. In addition, systemic bone loss is still a significant clinical issue that is not adequately treated with current disease-modifying agents. Therefore, additional anabolic therapy to maintain osteoblast function and bone formation is needed.
A role for miRNAs expressed in synovium that regulate inflammation in RA,13 and that may have therapeutic potential, is emerging. miRNAs are also known to regulate bone homeostasis12 and are essential for osteoclast, osteoblast, and chondrocyte differentiation.17, 54, 55 miRNAs also regulate the BMP and Wnt signaling pathways12 that promote osteoblast differentiation from immature precursors, or counterbalance differentiation by inhibiting this process. It is not known, however, whether synovium-derived miRNAs regulate these pathways in RA, and if so, what are the cellular sources of these miRNAs. In this study we identified synovium-derived miRNAs that may regulate bone formation at erosion sites.
Among the 22 significantly changed miRNAs between nonarthritic and peak inflammation, 12 miRNAs target osteogenic genes. We confirmed that eight of these 12 miRNAs are regulated by TNF in isolated FLS. It is possible that the other four miRNAs identified by PCA analysis are regulated by other inflammatory cytokines or in other synovial cell types. Of the upregulated miRNAs, we focused on miR-221-3p because it is known to be upregulated in the synovium of TNF-transgenic mice and in FLS from RA patients.26 Furthermore, miR-221-3p was reported to promote migration and invasion of FLS by promoting the expression of inflammatory cytokines and vascular endothelial growth factor, matrix metalloproteinase (MMP)-3 and MMP-9.56 Although ontology analysis predicted that miR-221-3p regulates Wnt signaling, miR-221-3p does not regulate Wnt reporter activity in HEK 293T cells.26 We found that miR-221-3p inhibits expression of Dkk2, as well as inhibiting osteoblast differentiation. These findings of the effectiveness of miR-221-3p overexpression in osteoblasts may be the result of both direct and indirect effects, whereas depletion of miR-221-3p showed no effect. It is known that Dkk2 is required for osteoblast maturation.10 miR-221-3p may inhibit osteoblast differentiation by suppressing expression of Dkk2. miR-221-3p is also known to suppresses both p27 and p57 in various cancer cell lines. This would block differentiation by preventing cells from becoming quiescent, thereby forcing apoptosis.57-59 Interestingly, p27 and p57 also suppress proliferation of osteoblasts60, 61 and chondrocytes.62, 63 These mechanisms may also underlie the regulation of osteoblast differentiation by miR-221-3p in inflammatory arthritis.
miRNAs can be packaged in vesicles or bound to miRNA-binding protein complexes, or high density lipoproteins to regulate genes within target cells.19, 64 Exosomes are one example of vesicles that allow for transfer of miRNAs from secreting cells to target recipient cells and it has been shown that miRNAs transferred via exosomes are functional.65 Examples include transfer of miRNA-loaded exosomes from T cells to antigen-presenting cells66 and bone mesenchymal stromal cell–derived exosomes promoting tumor growth in myeloma cells.67 Furthermore, a recent report showed that osteoclast derived exosomes are taken up by osteoblasts and suppress osteoblast differentiation.68 In bone, synovium-derived miRNAs including miR-221-3p may control skeletal pathways that inhibit osteoblast differentiation to augment bone erosion in RA. Our results with miR-221-3p provide one possible mechanism by which synovium-derived miRNAs could regulate bone. miR-221-3p, produced by synovial fibroblasts, may be secreted and subsequently taken up by osteoblasts in bone, where miR-221-3p regulates Dkk2 protein expression (Fig. 4H). Transfer of miR-221-3p to osteoblasts has not been proven in vivo, but this will be the topic of future studies. In contrast, we found that miR-708-5p, miR-186-5p, miR-486-5p, and miR-145a-5p are downregulated in synovial fibroblasts by pro-inflammatory cytokines and in whole inflamed synovium. It is possible these miRNAs promote osteoblast differentiation in nonarthritic states through similar mechanisms of exosomal transfer, and that downregulation of expression of these miRNAs in inflammatory arthritis thereby inhibits osteoblast differentiation.
We have studied synovial samples from male mice only, because male mice develop more severe arthritis in the STA model. Limitations of this study include the fact that female mice are not studied, and that we have focused on regulation of miRNAs at only one time point, peak inflammation, and in only one arthritis model. We did find, however, that expression patterns of several miRNAs previously reported in other mouse RA models or in human RA were confirmed, including miR-221-3p, miR-23b, miR-224-5p, miR-145a-5p, and miR-16-5p.15, 26, 32, 33 It will be important to repeat these studies in other models, in female mice, and at other time points during the inflammatory process.
Our studies focused on pathways documented in bone and inflammation. We have also found enrichment of pathways that relate to proliferation, migration, and cellular metastasis. Although some pathways appear unrelated, the individual genes/pathway revealed are potentially involved in bone erosion and can be pursued in future studies (see Table 1). For example, IL-9 signaling (cluster 1, p = 0.051),69 estrogen-mediated S-phase entry (cluster 2, p = 0.0001), and estrogen-dependent breast cancer signaling (cluster 3, p = 0.01) are the most highly enriched pathways among the significantly regulated miRNAs. These pathways regulate processes that are also active during inflammation, as inflammatory cells migrate to synovium and proliferate.
Taken together, these results support the hypothesis that synovium-derived miRNAs regulate bone pathways in RA. Future studies to determine whether these miRNAs are secreted from synovial cells to regulate bone remodeling in vivo will be of significant interest.
Disclosures
EMG receives grant support from AbbVie, consults for GSK, Novo Nordisk, Lilly, Sanofi, and Flexion, and receives Royalties from UptoDate. The remaining authors state that they have no conflicts of interest.
Acknowledgments
This project was funded by a Musculoskeletal Center of Excellence Grant from University of Massachusetts Medical School (EMG) and by a Disease Targeted Innovative Research Award from the Rheumatology Research Foundation of the American College of Rheumatology (EMG). JBL is supported by NIH R37DE012528. We thank Catherine Manning for her technical assistance and discussions.
Authors’ roles: Study design: YM and EMG. Study conduct: YM. Data collection: YM and MMM. Data analysis: YM, NHF and EMG. Data interpretation: YM, NHF, PJF, JL and EMG. Drafting manuscript: YM, NHF, MMM, PJF, JBL and EMG. EMG takes responsibility for the integrity of the data analysis.




