Two Rare Mutations in the COL1A2 Gene Associate With Low Bone Mineral Density and Fractures in Iceland
ABSTRACT
We conducted a genome-wide association study of low bone mineral density (BMD) at the hip and spine utilizing sequence variants found through whole-genome sequencing of 2636 Icelanders. We found two rare missense mutations, p.Gly496Ala and p.Gly703Ser, in the COL1A2 gene that associate with measures of osteoporosis in Icelanders. Mutations in COL1A2 are known to cause the autosomal dominant disorder osteogenesis imperfecta. Both variants associate with low BMD and with osteoporotic fractures. p.Gly496Ala (frequency of 0.105%) shows the strongest association with low BMD at the spine (p = 1.8 × 10−7, odds ratio [OR] = 4.61 [95% confidence interval (CI) 2.59, 8.18]), whereas p.Gly703Ser (frequency of 0.050%) is most strongly associated with low BMD at the hip (p = 1.9 × 10−8, OR = 9.34 [95% CI 4.28, 20.3]). Association with fractures was p = 2.2 × 10−5, OR = 3.75 (95% CI 2.03, 6.93) and p = 0.0023, OR = 4.32 (95% CI 1.69, 11.1), respectively. The carriers of these variants do not have signs of osteogenesis imperfecta other than low BMD, demonstrating that similar mutations in COL1A2 can affect skeletal phenotypes in more than one way. © 2015 American Society for Bone and Mineral Research.
Introduction
Osteoporosis is a common disease and a major public health problem worldwide with more than 9 million osteoporosis-related fractures suffered per year.1 Bone mineral density (BMD) is the single best predictor of osteoporotic fractures and is a good tool in evaluation of the risk of fractures.2, 3 There is abundant evidence for a genetic contribution to variation in BMD with heritability estimates between 0.6 and 0.8.4 Clearly, environmental and medical factors also influence BMD.
Genome-wide association (GWA) studies in large sample sets have in recent years yielded numerous common sequence variants of small effects that predispose to osteoporosis.5-13 The more than 60 loci that have been found to robustly associate with BMD account for only 6% of the total variation in BMD. Hence, more sequence variants remain to be discovered. With larger sample sizes, more common variants will be found, although these are expected to have even smaller effect than those already found. Whole-genome sequencing (WGS) offers an opportunity to study all variations in the genome of an individual and to find rare variants of large effect that associate with a disease. We are conducting a WGS project in Iceland aimed at identifying rare variants in the genome that associate with various human diseases and other traits. Through this project, we previously described a rare stop codon variant in the LGR4 gene that associates strongly with low BMD and fractures with considerable effect.14
Here, we report results from genome-wide association scan of low BMD at the hip and spine using sequence variants detected through a large-scale whole-genome sequencing effort in the Icelandic population. Through this effort, we identified rare variants in a priori strong candidate gene, collagen type I alpha 2 (COL1A2) previously linked to the osteogenesis imperfecta disorder.
Materials and Methods
Study populations
The Icelandic samples have previously been described in detail.14 The BMD (dual-energy X-ray absorptiometry [DXA]; Hologic QDR4500A, Hologic, Inc., Waltham, MA, USA) values were age and weight corrected and standardized in each sex separately. The low BMD groups consist of individuals who have standardized BMD 1 SD or more below the mean at either hip (total hip), lumbar spine (L2 to L4), or whole body. The control group consisted of individuals who had BMD above –1 SD and individuals who were recruited into other studies at deCODE genetics and did not have BMD information available (approximately 20% of the control group had BMD information). Fracture assessments were as previously described,5, 7, 14 excluding high-trauma fractures, corticosteroid users, early menopause, and fractures of the hands and feet. All participants gave informed consent, and the study was approved by the Data Protection Commission of Iceland (DPC) and the National Bioethics Committee of Iceland.
The Danish samples are postmenopausal women aged 55 to 86 years, taking part in the Prospective Epidemiological Risk Factor (PERF study).15 The study was approved by the Ethics Committee of Copenhagen County and was in accordance with the principles of the Helsinki Declaration. The Australian samples were derived from the Dubbo Osteoporosis Epidemiology Study (DOES),16 including subjects aged 60 to 99 years. All are of white ethnicity. The study was approved by the St. Vincent's Ethics Review Committee (Sydney, Australia), and all subjects gave written informed consent.
Genotyping and association analysis
Genotyping and imputation methods and the association analyses method in the Icelandic samples were as described.17 In short, we sequenced the whole genomes of 2636 Icelanders using Illumina technology to a mean depth of at least 10× (median 20×). Single-nucleotide polymorphisms (SNPs) and indels were identified, and their genotypes called for all samples simultaneously using the Genome Analysis Toolkit (GATK version 2.2-13).18 Genotype calls were improved by using information about haplotype sharing, taking advantage of the fact that all the sequenced individuals had been genotyped with SNP chips and their genotypes phased using long-range phasing.19, 20 A total of 25 million SNPs and indels that met stringent quality criteria were identified in the 2636 sequenced Icelanders.17 These variants were then imputed into 104,220 Icelanders who had been genotyped with various Illumina SNP chips and their genotypes phased using long-range phasing.19, 20 In brief, phasing is achieved using an iterative algorithm, which phases a single proband at a time given the available phasing information about everyone else that shares a long haplotype identically by state with the proband. Given the large fraction of the Icelandic population that has been genotyped on SNP chips, accurate long-range phasing is available genome-wide for all chip-typed Icelanders. Genealogical deduction of genotype probabilities of 294,212 untyped relatives of chip-typed individuals further increased the sample size for association analyses and increased the power to detect associations. Cases and controls were derived from the chip-typed individuals and untyped relatives. Association testing for case-control analyses was performed using logistic regression, and a generalized form of linear regression was used to test for association of quantitative traits.
To account for relatedness and stratification within the case and control sample sets, we applied the method of genomic control.21 The inflation λg in the χ2 statistic in each genome-wide analyses was estimated on the basis of a subset of about 300,000 common variants, and p values were adjusted by dividing the corresponding χ2 values by this factor. For the traits reported here, the estimated inflation factors were 1.32 for low BMD at the hip, 1.32 for low BMD at the spine, 1.46 for low BMD for the whole body, 1.16 for the WHO criteria for osteoporosis, 1.37 for any osteoporotic fracture, 1.27 for forearm fractures, 1.20 for hip fractures, 1.11 for vertebral fractures, 1.23 for hip BMD, 1.23 for spine BMD, 1.67 for height, and 1.49 for weight.
We used the weighted Holm-Bonferroni method22 to allocate familywise error rate of 0.05 equally between three annotation-based classes of sequence variants. For this data set, this yields significance thresholds of 3.1 × 10−6 for loss-of-function (LoF) variants (including stop-gained, frameshift, splice acceptor or donor) (n = 5432), 1.5 × 10−7 or missense, splice-region variants and in-frame indels (n = 109,370), and 6.7 × 10−10 for other (non-coding) variants (n = 24,873,426).23
Single-SNP genotyping of the variants described here was carried out on the Centaurus (Nanogen, Bothell, WA, USA) platform24 and by Sanger sequencing.
We validated and improved the imputation by directly genotyping likely and possible carriers and predicted non-carriers (n = 5353 for p.Gly703Ser and n = 439 for p.Gly496Ala). Directly assessed genotypes were added to the genotypes for the 2636 sequenced individuals, and the combined set was used as a training set for re-imputation of the variants. Both the imputation information and association of p.Gly496Ala with osteoporosis remained very similar before and after the re-imputation, whereas the imputation of the p.Gly703Ser variant improved, resulting in slightly stronger association.
Genealogical analysis
We used the extensive nationwide genealogical database at deCODE genetics for the genealogical analysis.
Results
To search for rare- and low-frequency sequence variants of large effect that associate with low BMD, we performed a GWA study of variants that were found in the whole genomes of 2636 Icelanders. We focus on low BMD as a dichotomous trait, rather than BMD as a quantitative trait, with the aim of finding variants that may have a direct effect on the risk of pathologically low BMD rather than on the regulation of BMD in the healthy population. We imputed the identified SNPs and indels, by long-range phasing,19, 20 into 104,220 Icelanders genotyped with Illumina SNP chips and further into 294,212 untyped relatives by familial imputation. We then selected cases and controls from this data set and examined association between the 25 million sequence variants found through WGS that passed stringent quality control17 and low BMD at the spine and low BMD at the hip. Individuals with standardized BMD below −1 standard deviation were included in the low BMD groups; controls included those who had a measured BMD above –1 SD and, for increased power, population samples that lacked BMD information.
For criterion of genome-wide significance, we use a weighted variant class-specific Bonferroni correction where the variants tested are weighted according to their prior probability of affecting gene function: 3.1 × 10−6 for loss-of-function variants, 1.5 × 10−7 for missense variants, and 6.7 × 10−10 for all other non-coding variants (Materials and Methods).
For low hip BMD (Supplemental Fig. S1A), a significant association was observed for a missense SNP in the COL1A2 gene with p = 1.9 × 10−8 and odds ratio (OR) = 9.31 (95% confidence interval [CI] 4.28, 20.3), whereas no loss-of-function or non-coding variants reached the criterion for their significance. This missense SNP is a G to A transition at c.2107 (hg18 position chr7:93887508), resulting in a replacement of glycine at amino acid (aa) position 703 of the COL1A2 protein with serine (p.Gly703Ser).
For low spine BMD (Supplemental Fig. S1B), common variants at the previously reported 13q14 locus5, 13 and the stop codon variant in LGR414 were significantly associated, whereas no missense SNP reached the criterion for significance. However, another missense SNP in the COL1A2 gene, a G>C transversion at c.1487 (hg18 position chr7:93879914), replacing glycine with alanine at position 496 (p.Gly496Ala), was associated with p = 1.8 × 10−7 and OR = 4.61 (95% CI 2.59, 8.18), just above the significant threshold of 1.5 × 10−7. Seven other rare non-coding variants at this locus were more strongly associated; however, none were significant after accounting for the effect of c.1487G>C (Supplemental Table S1). The dbNSFP database (v.2.7), which compiles prediction scores from 11 prediction algorithms (SIFT, Polyphen2, LRT, MutationTaster, MutationAssessor, FATHMM, VEST3, CADD, MetaLR, MetaSVM, PROVEAN),25, 26 predicts both COL1A2 mutations to be very deleterious, with RadialSVM_normalized rank score of 0.977 and 0.975.
Because these variants are rare in our data set, we validated them by directly genotyping likely and possible carriers and predicted non-carriers. The genotyping results were then used to re-impute the variants into the low hip and spine BMD cases and controls. The association results presented (Table 1) are derived from the re-imputed data set.
| p.Gly496Alaa (0.105%) chr7:93879914 [C] | p.Gly703Serb (0.050%) chr7:93887508 [A] | |||||
|---|---|---|---|---|---|---|
| Phenotype | p Value | OR (95% CI) | p Value | OR (95% CI) | Cases | Controls |
| Low BMD (< −1 SD)c | ||||||
| Hip | 0.0021 | 2.84 (1.46, 5.52) | 1.9 × 10−8 | 9.31 (4.28, 20.3) | 2678 | 206,701 |
| Spine | 1.8 × 10−7 | 4.61 (2.59, 8.18) | 1.2 × 10−4 | 5.42 (2.29, 12.8) | 2894 | 206,485 |
| Whole body | 5.5 × 10−4 | 4.41 (1.90, 10.2) | 1.2 × 10−4 | 8.50 (2.85, 25.4) | 1198 | 202,147 |
| Osteoporosis BMDd | 2.5 × 10−7 | 5.03 (2.72, 9.29) | 0.048 | 2.88 (1.01, 8.22) | 2504 | 206,875 |
| Fractures | ||||||
| Any OP fracture | 2.4 × 10−5 | 3.75 (2.03, 6.93) | 0.0023 | 4.32 (1.69, 11.1) | 2641 | 205,351 |
| Forearm fractures (including young) | 0.014 | 3.16 (1.27, 7.87) | 6.7 × 10−4 | 7.14 (2.30, 22.1) | 1102 | 196,718 |
| Hip fractures | 0.079 | 1.83 (0.93, 3.57) | 0.50 | 0.67 (0.20, 2.17) | 6239 | 115,587 |
| Vertebral fractures | 0.0054 | 6.51 (1.74, 24.4) | 0.45 | 0.02 (0.00, 625) | 263 | 176,455 |
| Other fracturese | 3.0 × 10−5 | 4.42 (2.20, 8.88) | 1.2 × 10−4 | 7.02 (2.60, 18.9) | 1490 | 200,292 |
| Effect | Effect | Individuals | ||||
| BMD | ||||||
| Hip | 0.0025 | −0.47 (−0.77, −0.16) | 4.6 × 10−5 | −0.93 (−1.37, −0.48) | 20,162 | |
| Spine | 1.4 × 10−4 | −0.60 (−0.91, −0.29) | 0.050 | −0.47 (−0.93, 0.00) | 20,132 | |
| Height | 0.027 | −0.24 (−0.45, −0.03) | 8.1 × 10−4 | −0.53 (−0.84, −0.22) | 77,055 | |
| Weight | 0.46 | −0.08 (−0.29, 0.13) | 0.33 | 0.15 (−0.15, 0.46) | 76,631 | |
- OR = odds ratio; CI = confidence interval; BMD = bone mineral density; SD = standard deviation; OP = osteoporosis.
- The association results presented for both markers are derived from the re-imputed data set (Materials and Methods).
- All p values are corrected for relatedness using the method of genomic controls (Materials and Methods).
- a The results are shown for the C allele at hg18_chr7:94041978 (0.105%) in exon 25, resulting in p.496Ala.
- b The results are shown for the A allele at hg18_chr7:93887508 (0.050%) in exon 35, resulting in p.703Ser.
- c The low BMD phenotypes are defined as those BMD values that are below –1 SD from the mean. These values are corrected for age, weight, and sex.
- d World Health Organization definition of osteoporosis BMD: < –2.5 SD in young women at the spine or hip, uncorrected for age or weight.
- e Other fractures refer to low-trauma fracture at any other site than hip, spine, forearm, skull, and hands and feet.
The COL1A2 p.Gly703Ser variant is in 0.05% allelic frequency in the Icelandic population. It shows the strongest association with low BMD at the hip (less than –1 SD, age and weight corrected), with p = 1.9 × 10−8 and OR = 9.31 (95% CI 4.28, 20.3) (Table 1). The p.Gly703Ser variant also associates with low BMD at other sites, spine and the whole body, although less significantly. The mean BMD values are 0.46 to 0.93 SD lower in carriers than non-carriers, depending on site. An association is also observed with osteoporotic fractures, p = 0.0023, OR = 4.32 (95% CI 1.69, 11.1), although no association was observed with vertebral fractures or hip fractures individually, p = 0.45 and p = 0.50, respectively. The variant is most strongly associated with forearm fractures, p = 1.2 × 10−4, OR = 7.02 (95% CI 2.3, 22.1), and with fractures at sites other than hip, spine or forearm, p = 6.7 × 10−4, OR = 7.14 (95% CI 2.60, 18.9).
The COL1A2 p.Gly496Ala variant is at 0.105% allelic frequency in the Icelandic population. It shows the strongest association with low BMD at the spine (less than –1 SD, age and weight corrected), with p = 1.8 × 10−7 and OR = 4.61 (95% CI 2.59, 8.18) (Table 1). It also associates with low BMD at other sites, hip and the whole body. The mean BMD values are 0.46 to 0.60 SD lower in carriers than non-carriers, depending on site. An association is also observed with osteoporotic fractures, p = 2.2 × 10−5, OR = 3.75 (95% CI 2.03, 6.93), and with the WHO definition of osteoporosis BMD,27 p = 2.3 × 10−7, OR = 5.01 (95% CI 2.72, 9.29).
Many mutations in the COL1A2 gene are known to cause autosomal dominant osteogenesis imperfecta (OI), a generalized connective tissue disorder characterized mainly by bone fragility from early childhood and low BMD.28 Other characteristics are short stature, blue sclerae, dentinogenesis imperfecta, hearing deficit, fragile skin, and sometimes joint hyperlaxity.28-30 Osteogenesis imperfecta presents with variable degree of severity. To evaluate whether carriers of p.Gly496Ala or p.Gly703S could be diagnosed as having OI, we physically reexamined two of the homozygous carriers of p.Gly496Ala, two heterozygous carriers of p.Gly496Ala, and four heterozygous carriers of p.Gly703Ser, for additional phenotypic signs of osteogenesis imperfecta. No signs of osteogenesis imperfecta, such as blue sclera, dentinogenesis imperfecta, impaired hearing, joint hyperlaxity, fragile skin, or early childhood fractures, were observed in these individuals. Furthermore, the carriers of these mutations are not related to known osteogenesis imperfecta patients in Iceland, and neither of the mutations are present in the osteogenesis imperfecta (OI) variant database31, 32 (http://www.le.ac.ul/ge/collagen). For both the BMD cases and controls, a large fraction had information on height. Interestingly, both carriers of p.Gly703Ser and p.Gly496Ala were, on average, shorter than non-carriers; p.Gly703Ser carriers are 4.7 cm shorter than the non-carriers (p = 8.1 × 10−4) and p.Gly496Ala carriers 2.09 cm shorter than non-carriers (p = 0.027).
Neither of these variants are present in the dbSNP database,33 the Exome Variant Server of 13,000 sequenced individuals34 nor the Exome Variant Browser of 60,706 sequenced individuals.35 They are likewise not present in two BMD sample sets of European descent that we genotyped directly; the PERF study of 3032 Danish postmenopausal women15 and 1393 individuals in the Australian DOES study.16 These data demonstrate that if these variants are present in other European populations, they are in very low frequency (<0.001%).
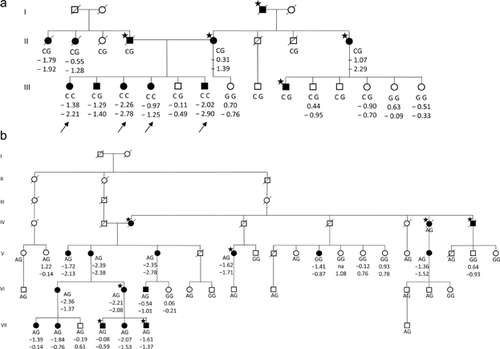
Genealogical analyses revealed that the ancestry of all the carriers of the p.Gly496Ala variant can be traced to common ancestors, a couple born in 1590 to 1592, and the p.Gly703Ser variant to another couple also born around 1590. Seven individuals are homozygous for the p.Gly496Ala variant, four of whom are siblings. As expected given the frequency of p.Gly703Ser in our data set, no homozygous individual was observed in our data. Two pedigree examples are shown in Fig. 1, demonstrating incomplete penetrance and possible phenocopies in these families. The four homozygous individuals for whom we have BMD information (the siblings) seem to have a more extreme phenotype than the eight heterozygous carriers in their families (median spine BMD = –2.50 SD [range –1.25 to –2.78] versus –1.34 SD [range –0.49 to –2.29]). One of the four siblings has attained five vertebral compression fractures, in her thirties and early forties. Two of the other siblings had suffered high-trauma fractures as young adults. The siblings are all of normal height.
COL1A2 is a priori strong candidate gene because of its role in bone biology and the many known mutations that cause osteogenesis imperfecta. A relatively common variant, rs1800012 (19%), an SP1 binding site polymorphism in the intron 1 of the COL1A1 gene, is one of the most widely studied variant in a candidate gene for osteoporosis and associates modestly with reduction in BMD and an increased risk of fracture.36-38 We, therefore, subsequently investigated if mutations in other known osteogenesis imperfecta genes, ie, COL1A1, WNT1, LEPRE, CRTAP, PPIB, FKBP10, SERPINF1, IFITM5, BMP1, TMEM38B, SP7, SERPINH1, and PLOD2, also associate with low BMD at the spine or low BMD at the hip in our data set. We selected all markers within a region 5 kb upstream and 5 kb downstream of each of these genes, 850 markers in total, and tested for association with low BMD. No variant in these genes met our criterion of a weighted Bonferroni variant class-specific genome-wide significance for association with low BMD at the hip or spine. Many variants were, however, nominally associated with low hip BMD or low spine BMD in our data and warrant further investigation in additional samples (Supplemental Table S2).
Of previously reported mutations listed in the OI variant database,31, 32 we only observe p.Asn1394Ser (rs147266928, freq. 0.48%) in COL1A1 to be weakly associated with low BMD at the spine (p = 0.017, OR = 0.48). This variant is not associated with fractures and is not believed to be causal for osteogenesis imperfecta.39
Discussion
In this study, we discovered two rare mutations in the COL1A2 gene that associate with measures of BMD and osteoporosis that is a common disorder. Mutations in the COL1A2 gene are known to cause osteogenesis imperfecta, which is a distinctly rare disease. The prevalence of osteogenesis imperfecta in Iceland is estimated to be around 0.014%, with 82% of the cases belonging to the same extended family (OI-type I).40 Another family (OI-type I) with fewer cases is known, as well as cases representing recessive mode of inheritance or believed to carry de novo mutations. The carriers of the COL1A2 gene variants p.Gly496Ala and p.Gly703Ser that we present here are not related to any of these patients. Neither do these carriers show additional signs of osteogenesis imperfecta, such as blue sclera, dentinogenesis imperfecta, impaired hearing, joint hyperlaxity, fragile skin, nor early childhood fractures upon physical reexamination. However, both variants associate with reduced height, p.Gly703Ser more strongly that p.Gly496Ala, but osteogenesis imperfecta is also characterized by short stature.
Both COL1A2 mutations are within a triple helical region of the type I collagen protein structure at positions that are likely to result in the disruption of the helical structure and of similar nature as the most frequent type of mutations that cause osteogenesis imperfecta. Helical glycine mutations (Gly-X-Y) in COL1A1 and COL1A2 are the most frequent, in particular, substitutions of glycine with serine (Gly-X-Y), as in the p.Gly703Ser variant that we report here. Alanine substitutions, as in our p.Gly496Ala variant, are also reported. These glycine (Gly-X-Y) substitutions in the collagen type I chains exert a dominant negative effect on the protein, leading to destabilization of the collagen helix, impaired formation, or effects on its interactions with the extracellular matrix.41
There are more than 2000 different collagen type I mutations described in osteogenesis imperfecta patients (http://www.le.ac.ul/genetics/collagen); however, there is no clear-cut genotype-phenotype correlation between the type of collagen mutations and the osteogenesis imperfecta phenotype. Interestingly, mutations closer to the carboxy-terminal end of the triple helical domain of α-2 chain seem to have the largest effect on height.41 The p.Gly703Ser variant is closer to the carboxy end than p.Gly496Ala and has a larger effect on height, 4.7 cm versus 2.09 cm, on average. There is a subtle difference in skeletal manifestations between p.Gly703Ser and p.Gly496Ala; p.Gly703Ser is more strongly associated with BMD at the hip, whereas p.Gly496Ala is more strongly associated with BMD at the spine. In both instances, the effect on BMD is substantial, –0.9 SD and –0.6 SD, respectively. The fracture pattern is also somewhat dissimilar; p.Gly703Ser has an effect on the risk of forearm fractures and not on the typical osteoporotic fractures at the spine and the hip, whereas p.Gly496Ala increases risk of all types of osteoporotic fractures. This pattern of association may reflect differences in effect on cortical versus trabecular type of bone, the p.Gly703Ser influencing cortical bone rather than trabecular, and p.Gly496Ala trabecular rather than cortical bone.
Although the associated ORs of both p.Gly703Ser and p.Gly496Ala are high, up to 9.3 for p.Gly703Ser (low hip BMD) and 6.51 for p.Gly496Ala (vertebral fractures), they do not confer Mendelian type of inheritance with full penetrance. There were a few instances of incomplete penetrance and of phenocopies in the families. This is to be expected given the frequency of low BMD in the population. There are also many individuals in the families for whom we do not have information on osteoporosis related traits.
Another mutation in COL1A2 (p.Gly751Ser [legacy change p.Gly661Ser]) was previously reported to cause postmenopausal osteoporosis in one subject,42 who was later found to have blue sclerae, slight hearing loss, and fracture history consistent with osteogenesis imperfecta. A p.Gly526Arg mutation (legacy change Gly436Arg) was found in two siblings who were diagnosed with juvenile osteoporosis of the spine and in their asymptomatic father.43 One of the siblings was of moderately short stature and had a mild neurosensory hearing loss. In our study, the subjects with the p.Gly703Ser and p.Gly496Ala mutations do not show signs of osteogenesis imperfecta, apart from the overlapping phenotypes of low BMD, risk of fracture, and decreased height. This indicates a much wider spectrum of skeletal phenotypes associated with mutations in the COL1A2 gene than previously considered, ranging from very severe osteogenesis imperfecta phenotypes to a reduced BMD without the associated osteogenesis imperfecta characteristics. It is both surprising and interesting that such phenotypic spectrum is observed with mutations of seemingly very similar character, ie, substitutions of the important Gly in Gly-X-Y in the collagen triple helix that all should lead to instability of the protein. Such difference may be explained by epistasis between the coding variants and presently unknown cis-regulatory variants, ie, the impact of the rare coding variant is modified by regulatory variation as suggested in Lappalainen and colleagues.44
Mutations in both the WNT1 and the LRP5 genes have shown diverse bone phenotype characteristics depending on their genotype state; homozygotes, or compound heterozygotes, result in osteogenesis imperfecta or osteoporosis pseudoglioma syndrome, respectively, whereas heterozygote carriers have been shown to have low BMD or osteoporosis.45-48 The homozygous carriers of p.Gly496Ala in COL1A2 in our samples do have lower BMD than the heterozygotes, representing an effect on a phenotype that is additive in nature rather than a fundamental difference in phenotype characteristics as for WNT1 and LRP5.
Here, we show that mutations of very similar character, helical glycine mutations, in the COL1A2 gene can either lead to the common osteoporosis in the population or to the rare osteogenesis imperfecta disease of various severity. The helical glycine mutations in COL1A2 now range from a lethal form of osteogenesis imperfecta to the much milder, and different overall phenotype, of osteoporosis/osteopenia. Further studies of type 1 collagen production in the subjects carrying these mutations as well as histomorphometry analyses before and after medical intervention could give valuable information on the function of these mutations. Our study may contribute to the understanding of the common disease of osteoporosis and its causes in the general population.
Disclosures
US, GT, SAG, UT, and KS are employed by deCODE genetics/Amgen. CC is chairman of Nordic Bioscience A/S, board member of CCBR/Synarc/BioClinical, and has consulted for Roche, Eli Lilly, Novartis, Novo Nordisk, Procter and Gamble, Groupe Fournier, Besins EscoVesco, MSD, Chiesi, Boehringer Mannheim, Pfizer, and GSK. JAE has consulted for Amgen, Eli Lilly, Merck Sharp & Dohme, Novartis, Sanofi-Aventis, and Aspen. All other authors state that they have no conflicts of interest.
Acknowledgments
We thank the subjects of the Icelandic deCODE study, the Danish PERF study, and the Australian DOES study for their participation. We also thank the staff at deCODE genetics core facilities, the staff at the Research Service Center, and all our colleagues for their important contribution to this work.
Authors' roles: Study design: US, GT, and KS. Study conduct: US and UT. Data collection: GS, BE, CC, JAE, JRC, TVN, US, UT, and KS. Data analysis: US, SAG, and GT. Data interpretation: US, GT, and KS. Drafting manuscript: US, GT, and KS. Revising manuscript content: US, GT, JAE, GS, UT, and KS. Approving final version of manuscript: US, GT, UT, BE, SAG, TI, JAE, JRC, TVN, GS, and KS. US, GT, and KS take responsibility for the integrity of the data analysis.




