Interaction among long non-coding RNA, micro-RNA and mRNA in glioma
Abstract
With the rapid development and wide application of gene sequencing, biotechnology, and informatics about cancer, it has been found that the main causes of malignant gliomas occurrence not only consist of abnormal mutations of protein-coding genes but also abnormal expressions of non-coding RNA (ncRNA). In this review, we summarize the interaction and mechanism between lncRNA-miRNA-mRNA and gliomas in occurrence, development, aggression, and migration in depth.
Introduction
The proportion of gliomas in all invasive nervous system tumors is above 50% (Sosa E, et al., 2021) and increasing year by year (Rades D, et al., 2021; Zhang J, et al., 2021). With the continuous development and extensive application of neuroendoscopy, gliomas can be removed maximumly nowadays. However, due to the less obvious boundary between glioblastoma (GBM) and the surrounding normal tissue, it is critical to attain the entire excision, which has become a serious threat to human survival.
Over 98% of the nucleotide sequences in the human genome do not encode proteins, and their transcription products are non-coding RNAs (ncRNAs), which are divided into microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) according to fragments. LncRNAs can be used as decoy molecules to effectively inhibit the interaction between miRNAs and target mRNAs by directly absorbing miRNAs. Therefore, formulating the interaction among lncRNA, miRNA, and mRNA in glioma is crucial for enhancing the efficacy of glioma’s Diagnostics and Treatment, prolonging life, and early detection of recurrence.
LncRNAs and miRNAs
It has been found that more than 98% of the nucleotide sequences in the human genome do not encode proteins, and their transcription products are non-coding RNAs (ncRNAs), which can be divided into long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) according to fragments and regulate multiple protein-coding genes. LncRNAs are produced by RNA polymerase ˊ (Pisignano G, et al., 2021), and their length exceeds 200 nucleotides (nt). While MiRNAs are widely found in eukaryotes, with the length of 18~25nt only. Apart from that, iRNAs must go through a process from primary (pri-) to precursor (pre-) before finally mature. Firstly, in the nucleus, the genes encoding miRNAs are transcribed into primary miRNAs (pri-miRNAs) with cap and poly adenylate tail under the function of RNA polymerase IIⅡ. Then pri-miRNAs are sheared into precursor miRNAs (pre-miRNAs) by enzyme protein complex RNase III DROSHA-DGCR8, pre-miRNAs are a prominent hairpin structure with 2 nt at the 3` end. And pre-miRNAs are transferred to the cytoplasm by the transporter Exportin-5-RanGTP, and finally are sheared by RNase III DICER into mature miRNAs with 5` terminal phosphate group and 3` hydroxyl group (Balasubramanian S, et al., 2020). Most miRNA and lncRNA sequences are highly conserved, with temporal, spatial, tissue, and disease specificity. But the expression level of lncRNAs is considerably low, the number of nucleotides is relatively small, and the exons are longer but few.
Interaction among lncRNA, miRNA and mRNA
At the transcription level, lncRNA encoded by genes located in the promoter region can act as homeopathic components to interfere with the transcription of downstream genes, thus affecting the production of mRNA. At the post-transcription level, lncRNA binds to and pairs with pre-miRNA to form a double-stranded complex, which affects its splicing, nuclear transport, and degradation to regulate gene expression. At the same time, pre-miRNA can be attained through intracellular shear by lncRNA or when other genes can be transcribed into lncRNAs. However, further processing is needed to obtain mature miRNA to work. For example, although the precursor of miR-675 is H19, the bases of miR-675 are mainly C and G, while the bases of H19 are mainly G, which may be the structural basis for different functions (Alhelf M, et al., 2021). Since lncRNA also has 3'UTR, miRNA can bind to 3'UTR of lncRNA just as it acts with mRNA, thus directly inhibiting lncRNA. MiRNA can also indirectly regulate lncRNA by regulating DNA methylase (DNMT).
LncRNA and miRNA can competitively bind to the 3'UTR of target mRNA, thereby inhibiting the negative regulation of miRNA on target genes. One example is that beta-site APP cleaving enzyme 1 (BACE1) is a membrane-bound aspartic protease, which can transcribe Antisense lncRNA (BACE1-AS) about 2kb. The up-regulation of the BACE1-AS level increases the stability of BACE1 and competitively bound with BACE1 mRNA to inhibit miR-285-5p binding and silencing BACE1. In additionly, lncRNA as competing endogenous RNA (ceRNA) adsorb specific miRNA employing bait. After binding with lncRNA, miRNA can directly act on the lncRNAs through cis acting. It can also indirectly act on other RNAs through trans-action, and this way of indirectly inhibiting the expression of miRNA target genes is called “miRNA sponge” (Zhang G, et al., 2021) while the way of indirectly inhibiting the expression of lncRNA is called “molecular sponge” (Figure 1).
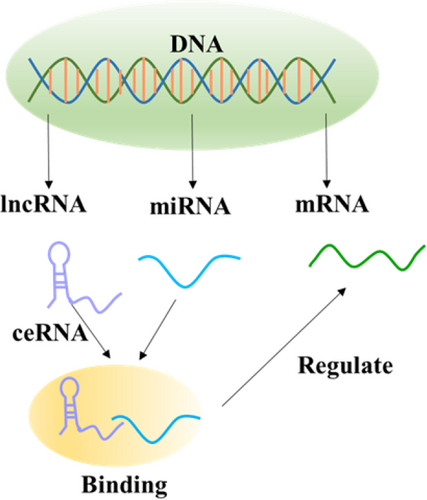
lncRNA as competing endogenous RNA (ceRNA) adsorbs specific miRNA by means of bait, after binding with lncRNA. MiRNA can also indirectly act on other RNAs through the transaction. This way of indirectly inhibiting the expression of miRNA target genes and lncRNA is called “miRNA sponge” and “molecular sponge”.
Interaction among lncRNA, miRNA and mRNA in glioma
Although both lncRNA and miRNA can act on mRNA alone, their regulatory networks are not independent of each other. During the occurrence and development of gliomas, the regulatory mechanisms of lncRNA and miRNA interweave, and they form a huge regulatory loop together.
LncRNA positively correlates with the malignant degree of gliomas. It not only promotes the occurrence and development of gliomas but also significantly related to the poor prognosis of patients. Metatasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a nuclear lncRNA, which was first discovered in the study of its relationship with metastasis of human lung cancer. MALAT1 increases the expression of its target genes STMN1, RAB5A and ATG4D by inhibiting miR-101. Namely, autophagy of glioma cells is activated by MALAT1-miR-101-STMN1/RAB5A/ATG4D regulatory network and promotes the proliferation of glioma cells by up-regulating the expression of STMN1, RAB5A, and ATG4D in glioma cells (Wu B, et al., 2021).
Furthermore, knockout of MALAT1 could up-regulate miR-140 and weaken the immune barrier function of glioma cells, thus effectively improving the immune response of glioma cells to drug chemotherapy (Li M, et al., 2021). MALAT1 can also act on glioma cells by inhibiting the expression of miR-155 (Syllaios A, et al., 2021). The high expression of X inactive-specific transcript (Xist) transcribed from the X chromosome inactivation center in malignant gliomas promotes the proliferation and invasion of glioma cells, and increases the resistance of glioma cells to temozolomide. The possible mechanism is that Xist, as a ceRNA of miR-137, binds and inhibits miR-137, and accelerates the expression of the target gene Rac1 (Wang J, et al., 2021). H19 is a maternally imprinted gene. It was found that the expression levels of H19 and miRNA-675 derived from H19 were significantly higher in some high-grade malignant gliomas than in low-grade malignant gliomas. H19 promotes the invasion of glioma cells through its miRNA-675. The over-expression of H19 and the down-regulation of miR-140 were also detected by luciferase assay, indicating that miR-140 as a target of H19 takes part in promoting the growth of glioma cells (Ghafouri-Fard S, et al., 2021). Vasohibin 2 (VASH2) is up-regulated through the H19-miR-29a pathway (Wang F, et al., 2021) to promote angiogenesis in gliomas. Moreover, lncRNA-ATB promotes the expression of TGR-β2 by inhibiting miR-200a (Yin Y, et al., 2021). Hox transcript antisense intergenic RNA (HOTAIR) as the ceRNA of miR-15b acts through HOTAIR-miR-15b-p53 axis, and miR-15b can not only independently inhibit HOTAIR, but also promote the expression of the p53 gene (Feng Q, et al., 2021).
Plasmacytomas variant translocation gene 1 (PVT1) is highly expressed in vascular endothelial cells of gliomas. By binding to miR-186, PVT1 inhibits the binding of miR-186 to the 3` UTR of autophagy proteins Atg7 and Beclin1 and up-regulates Atg7 and Beclin1 (Luo YY, et al., 2021). Small nucleolar RNA host gene 7 (SNHG7) exerts an influence on proto-oncogene, binds to miR-5095, directly inhibits miR-5095 binding to the 3` UTR of target CTNNB1 mRNA, and works by down-regulating the Wnt/β-catenin signaling pathway (Chen W, et al., 2021). However, other lncRNAs inhibit the proliferation of malignant glioma cells, and their expression in GBMs is significantly lower than that in normal brain nerve tissue (Tasaki Y, et al., 2021; Ueharai T, et al., 2021).
Cancer susceptibility candidate 2 (CASC2) is effective by inhibiting the expression of miR-21. Taurine Up-Regulated 1 (TUG1) is the molecular sponge of miR-R26a. MiR-R26a can activate the TUG1-miR-R26a-PTEN axis by binding with 3` UTR of PTEN to inhibit gliomas (Tasaki Y, et al., 2021). However, glioma cells need nutrients. Therefore, TUG1-miR-299 promotes angiogenesis of glioblastoma by regulating VEGF (Zhang J, et al., 2021). LncRNA Cas5 plays a role by inhibiting miR-222. LncRNA RP5-833A20.1 increases the methylation of the NFIA promoter to increases the expression of miR-382-5p by inhibiting the mRNA and protein levels of NFIA (Uehara T, et al., 2021). MiR-19a mainly acts as an oncogene and miR-93 is highly expressed in gliomas. Menternally expressed gene 3 (MEG3) can inhibit the proliferation of glioma cells by inhibiting the expression of miR-19a, resulting in cell cycle arrests in G0 / G1 phase (Ebrahimpour A, et al., 2021). In addition, MEG3 can inhibit the phosphatidylinositol 3 kinase/protein kinase B (P13K/AKT) signaling pathway by inhibiting miR-93 and finally contribute in inhibiting the occurrence of malignant gliomas (Xia Z, et al., 2021). If colorectal neoplasia differentially expressed (CRNDE) found in colorectal cancer is knocked out or miR-384 is over-expressed, PIWIL4 will be reduced and eventually lead to GBMs (Zhang R, et al., 2021). However, CRNDE has tissue specificity and presents different modes of action in different tissues.
Glioma stem cells (GSCs) often have a poor prognosis and relapse in the short term due to their strong self-renewal and differentiation ability. MALAT1 can bind to miR-129 and improve the expression of its target gene SOX2 by reducing miR-129, so as to regulate the characteristics of GSCs and the formation, proliferation, and migration of glioma cells (Gao X, et al., 2021).
Nuclear-enriched autosomal transcript 1 (NEAT1), which is highly expressed in GSCs, activates miR-107 through the miR-107-CDK6 pathway to inactivate cyclin-dependent kinases (CDK). After NEAT1 knockout, the cell cycle could be arrested in the G1 phase (Zhang J, et al., 2021). Besides, Knockout of Xist can up-regulate miR-152 to inhibit GBMs.
Conclusion
With high recurrence and mortality, glioma is the most common primary malignant tumor of the central nervous system. The 5-year average survival rate of glioma patients is 20% only, and the most bio-invasive GBM patients is less than 3%. Even with aggressive surgery, radiotherapy, and chemotherapy, the prognosis of GBM patients are still critically poor, with a median OS of 12 to 15 months after diagnosis. At present, the gene and molecular mechanism of GBM are not completely clear. Consequently, it is crucial to grasp the occurrence and development mechanism of glioma at the genetic and molecular level. From the discovery of H19 and Xist in mice, the role of lncRNAs in the development of cancer was not proposed until the last decade (Stone JK, et al., 2021); moreover, lncRNAs can be easily detected in body fluids, so lncRNAs have considerable application prospect as a key biomarker for early diagnosis, prognosis, and treatment of cancer (Xiao QQ, et al., 2021; Fattahi, et al., 2021). After binding to lncRNAs, miRNAs regulate mRNAs (Pisignano G, et al., 2021) and finally regulate various biological processes. Hence, the abnormal expression of lncRNAs will eventually interfere with the mRNAs, leading to the occurrence of tumors (Monroe JD, et al., 2021).
By analyzing the published literature, studies on the interaction among lncRNA-miRNA-mRNA are still rather limited. So it is very important to study the regulation of related lncRNAs and miRNAs on gliomas. To provide a reference for the studies on the mechanism of action between lncRNA-miRNA-mRNA and gliomas, this paper reviews researches about lncRNA-miRNA-mRNA in recent years. For example, MALAT1 binds to different types of miRNAs, either by up-regulating miRNAs or by inhibiting miRNAs to act as an oncogene. In the process of tumor genesis and development, autophagy plays a two-side role, which can not only inhibit cell cancerization but also provide more abundant nutrients for cancer cells to grow.
Ethical statement
Not applicable.
Acknowledgements
Not applicable.
Conflict of interest
No conflict of interest was declared.
Funding
This study was supported by grant from Science and Technology Support Project of Zunyi: 1). Zunyi science and technology foundation HZ (2019) No. 79; 2). Zunyi science and technology foundation HZ (2019) No. 100; 3). Zunyi science and technology foundation HZ (2020) No. 240; 4). Zunyi science and technology foundation HZ (2020) No. 241; 5). Zunyi science and technology foundation HZ (2019) No. 102. Science and Technology Fund project of Guizhou Provincial Health Commission in 2020: gzwjkj2020-1-107.
Transparency statement
All the authors affirm that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Authors' contribution
Yang Chai and Ming-Xiang Xie conceived and designed the work that led to the submission; Shun Liu revised the manuscript; Ming-Xiang Xie approved the final version.