A rapid HPV typing assay to support global cervical cancer screening and risk-based management: A cross-sectional study
Abstract
The World Health Organization recommends human papillomavirus (HPV) testing for cervical screening. Extended genotyping can identify the highest-risk HPV-positive women. An inexpensive, rapid, mobile isothermal amplification assay (ScreenFire HPV RS test) was recently redesigned to yield four channels ordered by cancer risk (ie, hierarchical approach): HPV16, HPV18/45, HPV31/33/35/52/58 and HPV39/51/56/59/68. Stored specimens from 2076 women (mean age 30.9) enrolled in a colposcopy clinic, with high HPV prevalence, were tested with ScreenFire. We calculated hierarchical channel positivity and non-hierarchical channel and type positivity, according to histologic diagnosis (256 cancer, 350 cervical intraepithelial neoplasia [CIN]3, 409 CIN2, 1020 < CIN2) and known virologic reference results (Linear Array and TypeSeq). Additionally, we analyzed ScreenFire time-to-positive up to 60 min by channel and histology. Overall clinical sensitivity for CIN3+ was 94.7% (95% confidence interval 92.6-96.4), similar to Linear Array (92.3, 89.7-94.3) and TypeSeq (96.0, 93.9-97.6). Sensitivity was high for all types and channels. The hierarchical approach was well in line with HPV typing and histologic diagnosis, prioritizing higher risk women having HPV16 and precancer. For HPV16, time-to-positive was shorter in women with precancer. ScreenFire showed excellent agreement with research reference typing tests and detection of CIN2+. Risk-based type results could help guide clinical management of HPV-positive women. Time-to-positive combined with genotyping might be useful. ScreenFire is rapid, mobile, relatively inexpensive and designed for implementation of HPV-based screening and management, including in lower-resource settings. Further validation in screening by self-sampling and practical effectiveness merit evaluation.
Graphical Abstract
What's new?
With the 13 high-risk human papillomavirus (HPV) genotypes carrying different risks of progression to cervical cancer, extended genotyping is being included in triage strategies for management of HPV-positive women. ScreenFire is the first HPV genotyping assay specifically designed to quickly and inexpensively yield results grouped according to the risk of developing cervical cancer (HPV16, HPV18/45, HPV31/33/35/52/58 and HPV39/51/56/59/68). This study using a disease-enriched dataset shows that ScreenFire provides accurate genotype risk-level information that permits efficient primary HPV screening combined with risk-based clinical management of HPV-positive women in screen-and-treat or screen-triage-treat strategies. The formal validation, practicality and effectiveness of the assay for widespread dissemination are under study.
Abbreviations
-
- Adeno
-
- adenocarcinoma
-
- AI
-
- artificial intelligence
-
- AIS
-
- adenocarcinoma in situ
-
- ASCUS
-
- atypical squamous cells of undetermined significance
-
- AVE
-
- automated visual evaluation
-
- CIN
-
- cervical intraepithelial neoplasia
-
- ct
-
- cycle threshold
-
- HPV
-
- human papillomavirus
-
- hrHPV
-
- high-risk HPV
-
- HSIL
-
- high-grade squamous intraepithelial lesion
-
- IARC
-
- International Agency for Research on Cancer
-
- LLETZ
-
- large loop excision of the transformation zone
-
- LSIL
-
- low-grade squamous intraepithelial lesion
-
- NCI
-
- National Cancer Institute
-
- OUHSC
-
- University of Oklahoma Health Sciences Center
-
- RS
-
- risk stratification
-
- SCC
-
- squamous cell carcinoma
-
- SUCCEED
-
- Study to Understand Cervical Cancer Early Endpoints and Determinants
-
- UCSF
-
- University of California San Francisco
-
- VIA
-
- visual inspection with acetic acid
-
- WHO
-
- World Health Organization
1 INTRODUCTION
In 2021, the World Health Organization (WHO) first issued the recommendation that countries should ensure regular DNA-based testing for human papillomavirus (HPV) to identify women who have or are at risk of cervical precancer.1, 2 The last decade has therefore seen a move toward primary HPV screening, with countries either shifting from existing programs based on cytology or visual inspection with acetic acid (VIA) or looking for sustainable solutions to implement HPV screening ex novo.3 As each of the 13 high-risk HPV (hrHPV) genotypes (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68) carries a different risk of progression to cervical cancer,4, 5 the addition of HPV genotype to triage HPV-positive women has been well studied with substantial prospective epidemiologic support. Positivity for HPV16/18 is currently being included in clinical algorithms (eg, US, the Netherlands, Italy) as an indication of high-risk supporting direct referral for colposcopy2, 6 and the inclusion of extended genotyping is being piloted (eg, Sweden).7 Extended or full genotyping refer, respectively, to the ability of the HPV test to identify some or all the hrHPV types other than 16/18 in groups or individually. However, most commercial HPV assays (i) yield a pooled result for types other than 16/18 (ie, partial genotyping), neglecting differences in risk between HPV16-related alpha-9 types and other hrHPV types, (ii) include HPV66 or HPV53, whose classification has been rectified by IARC as extremely rarely carcinogenic to humans,5 (iii) do not permit testing a large or flexible number of samples per run or (iv) are too expensive to be used in large-scale screening settings, especially in lower resource settings.
These limitations highlighted the need for more affordable and feasible extended genotyping tests. Extensive evidence supports categorizing HPV types globally into four groups reflecting the risk of progression to cervical precancer: HPV16, HPV18/45, HPV31/33/35/52/58 and HPV39/51/56/59/68.8, 9 Slightly different categorizations of genotypes have also been supported.10, 11 Given that new isothermal amplification techniques can lower the complexity of HPV testing without compromising sensitivity, HPV researchers from the US National Cancer Institute (NCI) collaborated, in the interest of public health, to help redesign a 15-type isothermal amplification assay (AmpFire) into a single-step 13-type assay (ScreenFire) that aims to target the four type groups in a hierarchical primer design: HPV16 is targeted as the highest design priority, else HPV18/45, else HPV31/33/35/52/58 and else HPV39/51/56/59/68.12 The new version of the assay is meant to be a screening test that also serves as a risk-stratification test, thus allowing primary HPV testing and management of HPV-positive women by genotyping. The genotype channels are ordered such that the result supports clinical management recommendations in cervical cancer screening. These efforts were undertaken to help identify who, among HPV-positive women, needs more attention and to create more precise screening and management recommendations depending on established cancer risk.13, 14
The aim of the present study is to evaluate the clinical accuracy of the novel HPV assay using a disease enriched dataset and to demonstrate the rationale for a risk-based HPV test.
2 METHODS
2.1 Study population and design
In this cross-sectional study we used aliquots from stored specimens of women who participated in the Study to Understand Cervical Cancer Early Endpoints and Determinants (SUCCEED). SUCCEED was conducted among women referred for colposcopy at the University of Oklahoma Health Sciences Center (OUHSC), Oklahoma City, due to an abnormal Pap diagnosis, HPV-positive equivocal cytology, or a biopsy diagnosis of cervical intraepithelial neoplasia (CIN). The design has been published previously.15-17 In brief, women were enrolled between November 2003 and September 2007. They were excluded if they were younger than 18 years of age, pregnant or living with HIV at the time of their visit, previously treated with chemotherapy or radiation for any cancer, or previously had a hysterectomy. Eligible consenting women underwent colposcopic examination by a gynecologist, during which a cervical sample was collected for cytology and HPV testing followed by biopsy, large loop excision of the transformation zone (LLETZ, aka LEEP), or surgery.
The data reflected a colposcopy clinic with a high proportion of confirmed precancer, as well as referrals for invasive cancer. We chose this population to test for sensitivity and cross-reactivity given multiple infections, which were common. Despite the high HPV prevalence, the type-specificity of ScreenFire (Atila Biosystems Inc., Sunnyvale, CA) was measurable (see below), because even women with multiple HPV infections are negative for most types.
We selected a random sample of 2087 stored cervical samples originally derived from pelleted cells collected in Thin-Prep PreservCyt solution (Hologic Inc., Marlborough, MA), to be resuspended, lysed, aliquoted and re-tested for HPV using ScreenFire at two laboratories: 1043 at the NCI laboratory for Cancer Genomics Research (CGR) (Frederick National Laboratory for Cancer Research, Leidos Biomedical Research Inc., Frederick, MD),* 989 at University of California San Francisco (UCSF, San Francisco, CA), and 55 at both locations. Of the samples tested at both laboratories, HPV results by ScreenFire were very similar, with all but three yielding completely concordant results on the individual channel level. ScreenFire results by UCSF were used for samples tested at the two laboratories. For the present analysis, of the total set of 2087 samples, we excluded five due to missing (HPV, cytology and/or histology) data in the SUCCEED dataset and six due to missing ScreenFire HPV results, thus leaving a total analytical sample of 2076.
2.2 SUCCEED procedures yielding reference standard diagnoses and HPV results
At the time of SUCCEED, cervical samples were tested for Thin-Prep cytology (Hologic, Boxborough, MD) and HPV genotyping using the Linear Array HPV Genotyping Test (Roche Molecular Diagnostics, Branchburg, NJ), which was used as reference standard typing test for many US-funded epidemiologic projects.18-22 SUCCEED cervical samples were subsequently tested for HPV by TypeSeq, which is a next-generation sequencing-based assay that detects 51 HPV genotypes,17, 23 created by NCI to perform epidemiologic HPV typing studies. For the present study, the residual stored SUCCEED cervical samples were re-tested by ScreenFire HPV RS test.
Specimen collection, colposcopy and biopsy protocols for SUCCEED have been previously described.15, 16, 24 Analyses presented here were based on the worst overall diagnosis, including biopsy results, LLETZ, or surgical outcomes. Adenocarcinoma in situ was added to CIN3, and cancers were combined, unless presented separately.
2.3 ScreenFire HPV RS test
The ScreenFire HPV risk-stratification (RS) test is designed to detect and group all 13 hrHPV genotypes into four groups with different cervical cancer risk: (i) HPV16, (ii) HPV18/45, (iii) HPV31/33/35/52/58 and (iv) HPV39/51/56/59/68, and simultaneously the internal control as sample quality guidance in a single tube reaction. ScreenFire HPV assay is an isothermal, multiplex nucleic acid amplification method. The assay takes advantage of 3NT technology (US patent 11 091 799, Atila Biosystems Inc., Sunnyvale, CA) in primer design to reduce the primer-primer interaction which is a major barrier for multiplex nucleic acid isothermal amplification. Primers containing only three out of four conventional nucleotides are used. Such primers generally cannot form stable hybridization with each other, therefore preventing nonspecific primer interaction. The 3NT technology greatly reduces non-specific primer interaction and consequently false positivity.
Multiple sequence-specific primers targeting each of the 13 hrHPV genotypes are used in an isothermal amplification system to amplify targeted sequences in the different HPV genotype regions. The amplified products, tagged with a specific molecular fluorescent probe, are measured in real-time to detect the increase in specifically targeted sequences (in the same way as in a traditional real-time PCR assay). Rounds of primer set optimization focused on sensitive detection of the known most carcinogenic types, particularly HPV16. This was intended to help avoid missed detection of those types in case of amplification competition (eg, in earlier assay versions, HPV16 was occasionally negative when multiple infections were present, and redesign aimed to assure that this did not occur).
The ScreenFire HPV testing was carried out slightly modified as described in the kit user manual due to specimens being previously pelleted and testing conducted in two locations. Briefly, previously pelleted cells, from a variable volume of specimen in Thin-Prep PreservCyt solution, were resuspended with 500 μl of 1X lysis buffer and heated in a 95°C incubator for 30 min. 5 μl of the crude lysate was added to 96-well microplates and frozen at −20°C prior to testing. 20 μl of the prepared reaction master mix was added to each well to carry out the assay. Real-time fluorescence data were collected using Powergene9600 software (v1.0.14RC) connected to the Atila Powergene 9600 Plus Real-Time PCR system (Atila Biosystems Inc.).
2.4 Statistical analyses
Our main analysis focused on assessing the performance of the ScreenFire HPV test. For that purpose, we utilized existing data from SUCCEED, including histologic diagnosis and HPV results by Linear Array and TypeSeq which we used to define histologic and virologic reference standards, respectively. Because of its design, HPV results by ScreenFire were analyzed hierarchically. First, the HPV16 channel result was tabulated. If positive, the result was HPV16-positive regardless of other channels. Then among the HPV16-negative specimens, the results for HPV18/45 were tabulated, and so on. The HPV16-related type channel (HPV31/33/35/52/58) was assessed among specimens negative for HPV16 and HPV18/45. The last channel (HPV39/51/56/59/68) was tabulated when no higher-risk channel was positive.
For comparability, HPV results by Linear Array and TypeSeq were also analyzed hierarchically and showed similar results (Table 1). Therefore, a combination of the two was used as virologic reference standard when either of the tests was positive with the exception of HPV52, whose positivity was based on TypeSeq only (Linear Array does not detect HPV52 directly25). We tabulated the hierarchical HPV results by ScreenFire against the histologic and virologic reference standard and calculated the new assay's overall clinical sensitivity. Clinical specificity was not emphasized due to the colposcopy setting of the study sample, which was chosen to maximize HPV positivity, particularly with multiple HPV types.
| Characteristics | N | % |
|---|---|---|
| Total | 2076 | 100 |
| Age (mean, SE) | (30.9, 0.3) | |
| <30 years | 1219 | 61.2 |
| 30 to 49 years | 591 | 29.7 |
| >49 years | 181 | 9.1 |
| Unknown | 85 | |
| Race/ethnicity | ||
| Black | 188 | 9.8 |
| White non-Hispanic | 1278 | 66.4 |
| Hispanic | 225 | 11.7 |
| Other | 233 | 12.1 |
| Unknown | 152 | |
| Cytologic result | ||
| HSIL | 827 | 40.4 |
| LSIL | 308 | 15.0 |
| ASCUS | 471 | 23.0 |
| Negative | 442 | 21.6 |
| Unknown | 28 | |
| Histologic diagnosis | ||
| Adeno | 43 | 2.1 |
| SCC | 213 | 10.5 |
| AIS | 13 | 0.6 |
| CIN3 | 337 | 16.6 |
| CIN2 | 409 | 20.1 |
| CIN1 | 473 | 23.2 |
| Negativea | 547 | 26.9 |
| Unknown | 41 | |
| HPV assay (analyzed hierarchically) | ||
| Linear arrayb | ||
| HPV16 | 780 | 37.6 |
| else HPV18/45 | 254 | 12.2 |
| else HPV31/33/35/58 | 316 | 15.2 |
| else HPV39/51/56/59/68 | 281 | 13.5 |
| else hrHPV negative | 445 | 21.4 |
| TypeSeqb | ||
| HPV16 | 718 | 37.8 |
| else HPV18/45 | 243 | 12.8 |
| else HPV31/33/35/52/58 | 413 | 21.7 |
| else HPV39/51/56/59/68 | 222 | 11.7 |
| else hrHPV negative | 305 | 16.0 |
| Unknown | 175 | |
| Virologic reference standard (hierarchical)bc | ||
| HPV16 | 803 | 38.7 |
| else HPV18/45 | 267 | 12.9 |
| else HPV31/33/35/52/58 | 420 | 20.2 |
| else HPV39/51/56/59/68 | 256 | 12.3 |
| else hrHPV negative | 330 | 15.9 |
- Abbreviations: Adeno, adenocarcinoma; AIS, adenocarcinoma in situ; ASCUS, atypical squamous cells of undetermined significance; CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; hrHPV, high-risk HPV; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; SCC, squamous cell carcinoma.
- a Including atypical metaplasia.
- b HPV results analyzed hierarchically (HPV16, else 18/45, else 31/33/35/52/58, else HPV39/51/56/59/68, else hrHPV negative).
- c The virologic reference standard is based on HPV results of Linear Array and TypeSeq, analyzed hierarchically and defined as positive when either of the tests result positive (with the exception of HPV52, whose positivity was based on TypeSeq only).
We repeated the analysis after excluding the equivocal positive results (eg, positive by Linear Array but not TypeSeq or vice versa), restricting to single-type infections, and stratifying by age group (<30, 30 to 49, >49 years). We also analyzed the HPV results in a non-hierarchical fashion, for each type group as well as for each of the 13 hrHPV types individually, for all observations and for single-type infections.
We performed ancillary analyses on the time-to-positive by ScreenFire (non-hierarchical), which is analogous to a cycle threshold (Ct) value in a real-time PCR-based test and a proxy for viral load. First, we plotted the distribution of time-to-positive by ScreenFire when both Linear Array and TypeSeq gave a positive type-specific result. Second, we performed an infection-level analysis of ScreenFire HPV results, in which we categorized the time-to-positive using tertiles of the distribution (hereafter: first tertile “fast”, second tertile “medium” and third tertile “slow”) and tabulated those against the HPV type channel, separately for CIN2+ and <CIN2. We calculated the odds of having a CIN2+ vs <CIN2, arbitrarily using medium time-to-positive for channel HPV31/33/35/52/58 as reference category.
3 RESULTS
The characteristics of the study population consisting of 2076 women, including the histologic and virologic reference standards used for the evaluation of ScreenFire HPV test, are displayed in Table 1. The mean age was 30.9 years and most women were below age 50. The sample included 256 cervical cancer cases (43 adenocarcinomas and 213 squamous cell carcinomas), 350 CIN3 (13 AIS and 337 CIN3), 409 CIN2 and 1020 < CIN2.
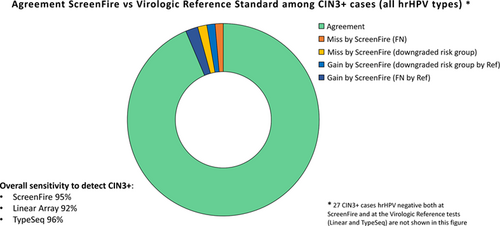
The hierarchical HPV results by ScreenFire are shown in Table 2 stratified by histology and in Table 3 stratified by the histologic and virologic reference standards. The concordance of ScreenFire by the virologic reference standard was high for all type groups and higher for cancer, CIN3, and CIN2 than for <CIN2. The highest priority cases were those with HPV16. Among HPV16-positives by the virologic reference standard, ScreenFire detected HPV16 in 140 out of 143 cancers, in 236 out of 238 CIN3, in 177 out of 185 CIN2, and in 180 out of 219 < CIN2. ScreenFire missed no HPV16 cancer case (ie, 3 cancers HPV16-positive by the virologic reference standard and negative for ScreenFire HPV16 were positive for other ScreenFire channels) and picked up HPV16 in five additional cancers missed by the reference HPV tests (ie, hrHPV negative by the virologic reference standard and HPV16-positive by ScreenFire). Although ScreenFire was designed to have highest sensitivity for HPV16, the sensitivity of the HPV18/45 channel, tabulated in HPV16-negative specimens, was virtually as high for CIN2+ (Table 3). The other channels were similarly very sensitive among women with CIN2+. There was, as expected, more variation in channel positivity among women with <CIN2, who are established to have lower viral loads than those with CIN2+ for HPV16 and the HPV16-related types.26 Overall, Table 3 shows a strong association of the hierarchical approach with the histologic diagnosis. HPV16 with precancer and cancer were the highest priority and, moving from the highest priority HPV16 channel to the lower risk channel, a lower number of precancer and cancer cases were observed.
| ScreenFire | N totala | Adeno | SCC | AIS | CIN3 | CIN2 | <CIN2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | col % | N | col % | N | col % | N | col % | N | col % | N | col % | ||
| HPV16 | 836 | 21 | 48.8 | 128 | 60.1 | 9 | 69.2 | 231 | 68.5 | 194 | 47.4 | 253 | 24.8 |
| else HPV18/45 | 232 | 8 | 18.6 | 36 | 16.9 | 4 | 30.8 | 26 | 7.7 | 43 | 10.5 | 115 | 11.3 |
| else HPV31/33/35/52/58 | 374 | 0 | 0.0 | 17 | 8.0 | 0 | 0.0 | 66 | 19.6 | 112 | 27.4 | 179 | 17.5 |
| else HPV39/51/56/59/68 | 266 | 0 | 0.0 | 16 | 7.5 | 0 | 0.0 | 9 | 2.7 | 40 | 9.8 | 201 | 19.7 |
| else hrHPV negative | 327 | 14 | 32.6 | 16 | 7.5 | 0 | 0.0 | 5 | 1.5 | 20 | 4.9 | 272 | 26.7 |
| Total | 2035 | 43 | 100.0 | 213 | 100.0 | 13 | 100.0 | 337 | 100.0 | 409 | 100.0 | 1020 | 100.0 |
- Abbreviations: Adeno, adenocarcinoma; AIS, adenocarcinoma in situ; CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; hrHPV, high-risk HPV; SCC, squamous cell carcinoma.
- a N total includes observations with histologic diagnosis.
| ScreenFire (hierarchical) | Virologic reference standard (hierarchical)a | |||||
|---|---|---|---|---|---|---|
| HPV16 | else HPV18/45 | else HPV31/33/35/52/58 | else HPV39/51/56/59/68 | else hrHPV negative | total | |
| Cancerb | ||||||
| HPV16 | 140 | 1 | 1 | 2 | 5 | 149 |
| else HPV18/45 | 2 | 40 | 1 | 0 | 1 | 44 |
| else HPV31/33/35/52/58 | 1 | 0 | 15 | 1 | 0 | 17 |
| else HPV39/51/56/59/68 | 0 | 3 | 0 | 9 | 4 | 16 |
| else hrHPV negative | 0 | 1 | 4 | 1 | 24 | 30 |
| Total | 143 | 45 | 21 | 13 | 34 | 256 |
| CIN3c | ||||||
| HPV16 | 236 | 0 | 1 | 1 | 2 | 240 |
| else HPV18/45 | 0 | 30 | 0 | 0 | 0 | 30 |
| else HPV31/33/35/52/58 | 1 | 1 | 64 | 0 | 0 | 66 |
| else HPV39/51/56/59/68 | 0 | 0 | 1 | 8 | 0 | 9 |
| else hrHPV negative | 1 | 0 | 1 | 0 | 3 | 5 |
| Total | 238 | 31 | 67 | 9 | 5 | 350 |
| CIN2 | ||||||
| HPV16 | 177 | 2 | 9 | 4 | 2 | 194 |
| else HPV18/45 | 1 | 42 | 0 | 0 | 0 | 43 |
| else HPV31/33/35/52/58 | 5 | 6 | 97 | 1 | 3 | 112 |
| else HPV39/51/56/59/68 | 1 | 0 | 4 | 30 | 5 | 40 |
| else hrHPV negative | 1 | 0 | 1 | 3 | 15 | 20 |
| Total | 185 | 50 | 111 | 38 | 25 | 409 |
| <CIN2 | ||||||
| HPV16 | 180 | 13 | 25 | 14 | 21 | 253 |
| else HPV18/45 | 3 | 105 | 0 | 3 | 4 | 115 |
| else HPV31/33/35/52/58 | 8 | 5 | 152 | 5 | 9 | 179 |
| else HPV39/51/56/59/68 | 11 | 3 | 24 | 147 | 16 | 201 |
| else hrHPV negative | 17 | 8 | 16 | 21 | 210 | 272 |
| Total | 219 | 134 | 217 | 190 | 260 | 1020 |
- Note: Channel agreement is shaded.
- Abbreviations: AIS, adenocarcinoma in situ; CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; hrHPV, high-risk HPV.
- a The virologic reference standard is based on HPV results of Linear Array and TypeSeq, analyzed hierarchically, and defined as positive when either of the tests result positive (with the exception of HPV52, whose positivity was based on TypeSeq only).
- b Cancer includes adenocarcinoma.
- c CIN3 includes AIS.
Similar results were obtained when repeating the analysis after excluding equivocal positive results and after restricting to single-type infections by the virologic reference standard. Moreover, the level of agreement between ScreenFire and the virologic reference standard was excellent when the analysis was restricted to women aged 30 to 49 years, the main targeted screening age. The concordance of ScreenFire with the virologic reference standard was high also when the HPV results were analyzed non-hierarchically, both for each type group (Table S1) and for each individual type (Table S2).
The overall clinical sensitivity of the three HPV assays, also stratified by age group, is displayed in Table 4. For all assays, positivity was higher for CIN2+ than for <CIN2, and among women with CIN2+, for endpoints CIN2 and CIN3 (the best-established screening target) than for cancers. A subset of cases, particular adenocarcinomas, were negative for all assays. For endpoint CIN3+, the sensitivity was 94.7% (92.6-96.4) for ScreenFire, 92.3% (89.7-94.3) for Linear Array, and 96.0% (93.9-97.6) for TypeSeq. The CIN3+ sensitivity was higher for younger women (<30 years: 98.7% [96.2-99.7] for ScreenFire, 97.8% [95.0-99.3] for Linear Array, and 98.2% [95.4-99.5] for TypeSeq; 30 to 49 years: 96.0% [92.5-98.2] for ScreenFire, 93.3% [89.2-96.2] for Linear Array, and 96.9% [93.4-98.9] for TypeSeq) than for women >49 years of age (84.1% [76.0-90.3] for ScreenFire, 78.8% [70.1-85.9] for Linear Array and 89.1% [80.9-94.7] for TypeSeq).
| HPV assay | Sensitivity % (95% CI)a | |||
|---|---|---|---|---|
| Adeno | SCC | CIN3+ | CIN2+ | |
| All ages | ||||
| ScreenFire | 65.8 (48.7-80.4) | 93.9 (89.6-96.8) | 94.7 (92.6-96.4) | 95.1 (93.5-96.4) |
| Linear array | 65.8 (48.7-80.4) | 89.8 (84.8-93.7) | 92.3 (89.7-94.3) | 91.3 (89.4-93.0) |
| TypeSeq | 80.6 (62.5-92.6) | 95.0 (90.3-97.8) | 96.0 (93.9-97.6) | 95.5 (93.9-96.8) |
| Age group ≤ 30 | ||||
| ScreenFire | 100.0 (47.8-100.0) | 100.0 (73.5-100.0) | 98.7 (96.2-99.7) | 97.2 (95.4-98.5) |
| Linear array | 100.0 (47.8-100.0) | 100.0 (73.5-100.0) | 97.8 (95.0-99.3) | 93.9 (91.5-95.8) |
| TypeSeq | 100.0 (47.8-100.0) | 90.0 (55.5-99.8) | 98.2 (95.4-99.5) | 96.6 (94.5-98.1) |
| Age group 30 to 49 | ||||
| ScreenFire | 80.0 (56.3-94.3) | 97.1 (91.8-99.4) | 96.0 (92.5-98.2) | 95.8 (93.0-97.7) |
| Linear array | 80.0 (56.3-94.3) | 95.2 (89.1-98.4) | 93.3 (89.2-96.2) | 92.4 (89.0-95.0) |
| TypeSeq | 88.9 (65.3-98.6) | 98.8 (93.5-100.0) | 96.9 (93.4-98.9) | 96.3 (93.5-98.1) |
| Age group ≥ 49 | ||||
| ScreenFire | 30.8 (9.1-61.4) | 88.9 (80.0-94.8) | 84.1 (76.0-90.3) | 84.2 (76.4-90.2) |
| Linear Array | 30.8 (9.1–61.4) | 81.5 (71.3-89.3) | 78.8 (70.1-85.9) | 77.5 (69.0-84.6) |
| TypeSeq | 50.0 (15.7-84.3) | 90.9 (81.3-96.6) | 89.1 (80.9-94.7) | 87.9 (79.8-93.6) |
- Abbreviations: Adeno, adenocarcinoma; CI, confidence interval; CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus; SCC, squamous cell carcinoma.
- a Exact confidence intervals.
We performed ancillary analyses on time-to-positive by ScreenFire. Figure S1 depicts the distribution of the time-to-positive by ScreenFire (non-hierarchical) separately for each of the 13 hrHPV types as detected by the virologic reference standard. Most HPV infections detected by ScreenFire gave a positive result within 30 min. The infections detected after 30 min were mostly with types HPV51 or HPV59. Higher odds of having a CIN2+ vs <CIN2 were seen for fast time-to-positive than for medium or slow time-to-positive in the three higher risk channels (Table S3). A markedly increased odds of CIN2+ was seen for the HPV16 channel, especially when fast-positive (high load). Lower odds of CIN2+ were seen for the lowest risk channel found alone as compared with the others, regardless of time-to-positive.
4 DISCUSSION
In the cross-sectional study presented here, we showed excellent agreement of ScreenFire HPV test designed for risk-based screening with two research reference typing tests that NCI has depended on for epidemiologic work on type-specific HPV natural history. The test population was a colposcopy clinic, highly enriched for cases of cervical cancer/precancer, and the comparison was based on stored provider-collected specimens. Most importantly, the read-out of the assay is redesigned for risk-based screening. The channels are read as hierarchical under multiple channel positivity, stressing the importance of types that cause most cervical cancers worldwide.
The agreement between ScreenFire, Linear Array and TypeSeq was excellent, for each of the four type groups defined by ScreenFire as well as for each of the 13 hrHPV types individually. ScreenFire had an overall clinical sensitivity for CIN3+ of 94.7%, similar to that of the other two tests. When the analyses were repeated in the subset of women aged 30 to 49 years, ScreenFire performed even better (96.0% overall clinical sensitivity for CIN3+), indicating that the newly redesigned HPV assay would work very well in screening settings. Our ancillary analysis on the time-to-positive by ScreenFire revealed markedly increased odds of CIN2+ for the HPV16 channel, especially when fast-positive (high load), and lower odds of CIN2+ for the lowest risk channel found alone regardless of time-to-positive. In other words, in particular for HPV16, time-to-positive was shorter in women with precancer. This suggests that time-to-positive could be used in combination with HPV genotyping to evaluate the risk of developing cervical precancer, but further evaluations are needed to support this indication.
We helped redesign ScreenFire to fit a risk-based screening strategy in low-resource settings, thus for use as primary HPV test on self-collected samples also allowing clinical management by genotyping. The reasoning behind this can be explained by giving more details on the hierarchical approach. The hierarchical approach presented in this article was designed to manage the reality that all HPV typing tests have at least a slight amount of error in type assignment. Slight differences in sensitivity by type are typically seen, and at least slight type-type competition occurs when multiple types are present. ScreenFire was designed by primer optimization to favor detection of the HPV types that cause most of the cervical cancer cases worldwide and particularly to detect HPV 16, 18 and 45. This means that the reading of ScreenFire HPV results should follow the hierarchical design (HPV16, else HPV18/45, else HPV31/33/35/52/58, else HPV39/51/56/59/68). This approach has clear consequences; hierarchical reading aims to direct the clinical management based on the risk channel. Subject to the type of triage under use and the follow-up capacity, positivity for HPV16 confers a much higher risk of progression to cancer and therefore closer monitoring is warranted as compared to a positive result for the HPV39/51/56/59/68 channel. Similarly, efforts to detect HPV18 have omitted the high risk of progression of HPV45, particularly for adenocarcinomas. Adding both types at the same level of risk is consistent with prospective data. In a sub-analysis presented in this article, the performance of ScreenFire was evaluated at the type-level and shown to be very good. Nevertheless, ScreenFire is not meant to be used as a genotyping test (apart from HPV16) but rather as a risk-stratification screening test. The risk of HPV infections is known to be measured best by the most carcinogenic type present. Multiple infections do not convey additive risk; in fact, HPV16 found alone is the highest risk result.27 When used hierarchically, the four HPV type groups reflect different cervical cancer risks, thus they can be used to manage HPV-positive women accordingly giving always maximum risk level to HPV16 and the lowest (not null) to the channel detecting HPV39/51/56/59/68. Risk-based screening strategies, including the use of HPV genotyping, are gaining importance worldwide because they offer the possibility to maximize screening efficiency by better allocating available resources and directing them to women at highest-risk. A risk-based strategy combining extended genotyping and age is being piloted in Sweden7 and it is likely that more countries will adopt similar strategies as, in the near future, more HPV tests used for primary screening will also provide extended/full genotyping information without additional costs.
This independent evaluation of a commercial product supports the following conclusions. As compared to the currently available HPV typing tests, ScreenFire (i) provides extended genotyping sufficient for screening, (ii) focuses on the most carcinogenic hrHPV types without including HPV66 or HPV53, (iii) can process a 96-well plate without requiring batch testing (1 to 96 samples/controls without waste of reagents) and (iv) is relatively low-cost (6 USD per test not including the collection device, laboratory supplies, or labor).28, 29 Compared to many typing tests, the assay is simpler to run requiring basic pipetting skills for sample preparation, is rapid (60 min run time plus approximately 60 min of hands-on preparation time, depending on experience, for 96 samples/controls), and does not require DNA purification or collection medium,30 which makes it compatible with self-sampled swabs transported dry to the laboratory. Additionally, reagents and equipment are low-volume making them easy to ship. Finally, the testing platform is suitable for other pathogens (eg, other sexually transmitted infections, COVID-19).28, 29 As for the assay's limitations, 6 USD per test is still expensive for some settings, pipetting skills are required, along with careful laboratory technique. ScreenFire is a table-top molecular test, thus an adequately clean environment is still needed and contamination is still possible especially with post-amplification products if technique is flawed and the seals or disposal are compromised. Efforts are ongoing to reduce the number of pipetting steps and the risk of contamination. For example, the use of reaction tube strips with pre-aliquoted reagents instead of an empty 96-well plate could be a technical solution to both those limitations.
Our study has two important limitations with regards to the study population and type of samples used. First, the study sample was a colposcopy referral population and not a general screening one. We chose this population with a high proportion of confirmed precancer and multiple infections because we were interested to test for sensitivity, also among women living with HIV who are known to have multiple infections. The study sample also had an age range wider than the targeted screening age. We repeated the analyses by age groups and showed that ScreenFire had an excellent performance in the age group 30 to 49 years, which is the main targeted screening age. Second, this study was done using provider-collected cervical samples and not self-collected vaginal samples. By using a specific protocol for dry swab and ensuring best practice of vaginal self-collection, we expect ScreenFire to perform well on self-collected material, but sensitivity could be an issue given the small volume of specimens assayed. Additionally, ScreenFire was run on a residual cell pellet while Linear Array and TypeSeq were performed on the same extracted DNA, thus, technically, the relative amounts of HPV could have been different. If that was the case, a bias in sensitivity could have been possible, but the three assays showed very high similar sensitivities.
Taking these limitations into account, this study does not provide validation of the assay for cervical cancer screening by self-sampling. This cross-sectional comparison shows that the assay has sufficient accuracy to move forward with it. ScreenFire is currently being used in nine low- and middle-income countries (LMICs), as part of a large study that we are conducting, called PAVE, aiming at validating a “screen-triage-treat” strategy comprising HPV genotyping of self-samples and artificial intelligence (AI)-assisted visual evaluation (AVE) on 100 000 women.31 The PAVE study will provide absolute risk (positive and negative predictive values) estimates for every test result (channel and time-to-positive as a measure of viral load), and show what the yield of precancer/cancer is for every result, alone and combined with visual assessment. We will validate the assay on self-collected samples and conduct extensive evaluations, including but not limited to: optimization of the workflow, optimization of the cutpoints to define how to guarantee sensitivity and specificity yielding good risk stratification, repeatability experiments, evaluation of the assay usability in LMIC settings. By the end of 2024, we will be able to confirm the ability of ScreenFire to be used as screening and management HPV typing test in real-world screening settings. Additionally, in the interest of making a screening test available for public health use, the data from PAVE, if convincing, will be submitted to scientific and regulatory bodies for formal consideration along with other trial data as required for approval and implementation. For the present, we can conclude that ScreenFire has the potential to bring an accurate HPV screening test to an increased number of women, irrespective of geography.
In conclusion, ScreenFire showed excellent agreement with other HPV assays and high sensitivity for each of the types individually and grouped. The risk-based type groups can be managed hierarchically for cancer risk allocation, thus facilitating clinical management of HPV-positive women. The potential role of time-to-positive as a risk stratifier in combination with genotyping should be further evaluated. ScreenFire could support implementation of HPV-based screening and management, particularly in low-resource settings, if it proves to be equally accurate and practically effective in real-life screening by self-sampling.
AUTHOR CONTRIBUTIONS
The study reported in the article has been performed by the authors, unless clearly specified in the text. Study conceptualization and supervision was carried out by Silvia de Sanjosé, Nicolas Wentzensen, Joel M. Palefsky, Mark Schiffman. Sample collection and clinical characterization was done by Rosemary E. Zuna, Joan L. Walker, Nicolas Wentzensen, Mark Schiffman. HPV testing was performed by Casey Dagnall, Amanda Hoffman, Sepideh Farhat Nozzari. Statistical, epidemiologic and/or clinical expertise was provided by all coauthors. Data analyses and writing of the original draft were done by Federica Inturrisi. The data were accessed and verified by Brian Befano and Mark Schiffman. The manuscript was reviewed and edited by all coauthors.
ACKNOWLEDGEMENTS
We acknowledge the participants and providers involved in the SUCCEED study. We thank the team from Information Management Services (IMS) for the data management.
FUNDING INFORMATION
The study was funded by the NCI Intramural Research Program. NCI chose the specimen collection and the two laboratories for the ScreenFire HPV testing (NCI-CGR and UCSF). NCI invited UCSF to test half to rule out laboratory variability due to the NCI laboratory having previously tested these same specimens. The reported data were masked to prior results at NCI, and the half tested at UCSF were totally masked and previously unknown to that laboratory. UCSF was provided with equipment and reagents by Atila Biosystems for the sole purpose of this study. The authors declare no intellectual property or conflict of interest with regard to the manufacturer, whose scientists had no input into the masked testing, analysis, or reporting of the results.
CONFLICT OF INTEREST STATEMENT
Joel M. Palefsky reports institutional grant support from Merck & Co., Roche Diagnostics, Antiva Biosciences, Vir Biotechnologies and Virion Therapeutics, as well as honoraria for attending meetings, giving lectures, presentations, consultations from Merck & Co., Roche Diagnostics, Antiva Biosciences, Vir Biotechnologies, Gilead Pharmaceuticals and Janssen Pharmaceuticals. He has stock or stock options in Virion Therapeutics and he has received resources/services from Atila Biosystems. All other authors declare no competing interests.
ETHICS STATEMENT
Written informed consent was obtained from all women enrolled into the SUCCEED study and Institutional Review Board approval was provided by OUHSC and the NCI.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
REFERENCES
- * The subset of samples assigned to CGR was tested in July 2022 after which changes in the composition of reagents occurred in order to increase the sensitivity, especially for the HPV18/45 channel. Both subsets of samples were tested in January 2023 using the new reagents.